Proliferative Diabetic Retinopathy Diagnosis Using Varying-Scales Filter Banks and Double-Layered Thresholding
Abstract
:1. Introduction
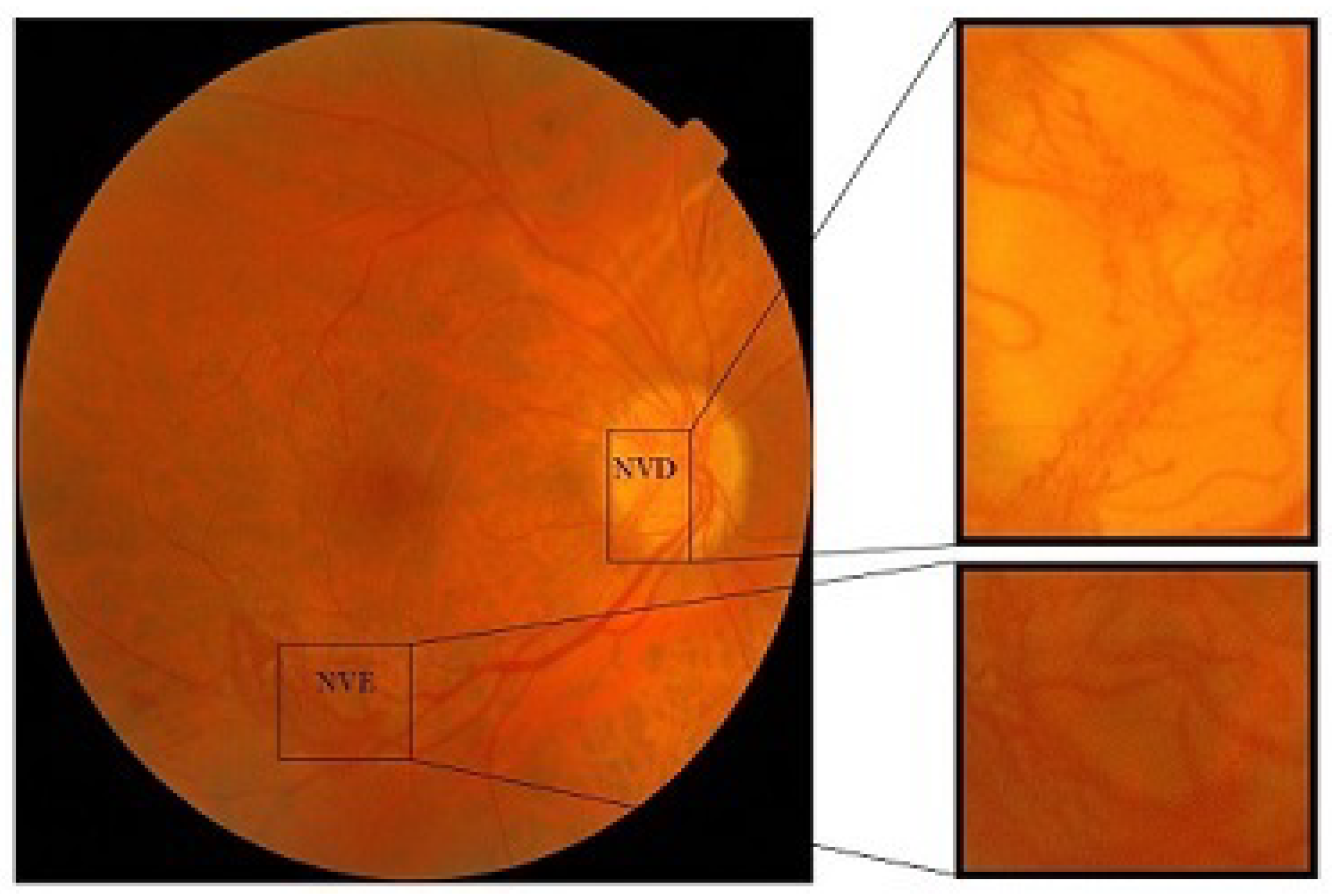
- Neovascularization on disc (NVD): If the new vessel formation occurs within one disc diameter of the optical disc then this is categorized as NVD or neovascularization on disc
- Neovascularization elsewhere (NVE): If new vessel formation occurs elsewhere on the surface of the retina, then this is called neovascularization elsewhere (NVE).
2. Related Work
2.1. Machine-Learning-Based Algorithms
2.2. Deep-Learning-Based Algorithms
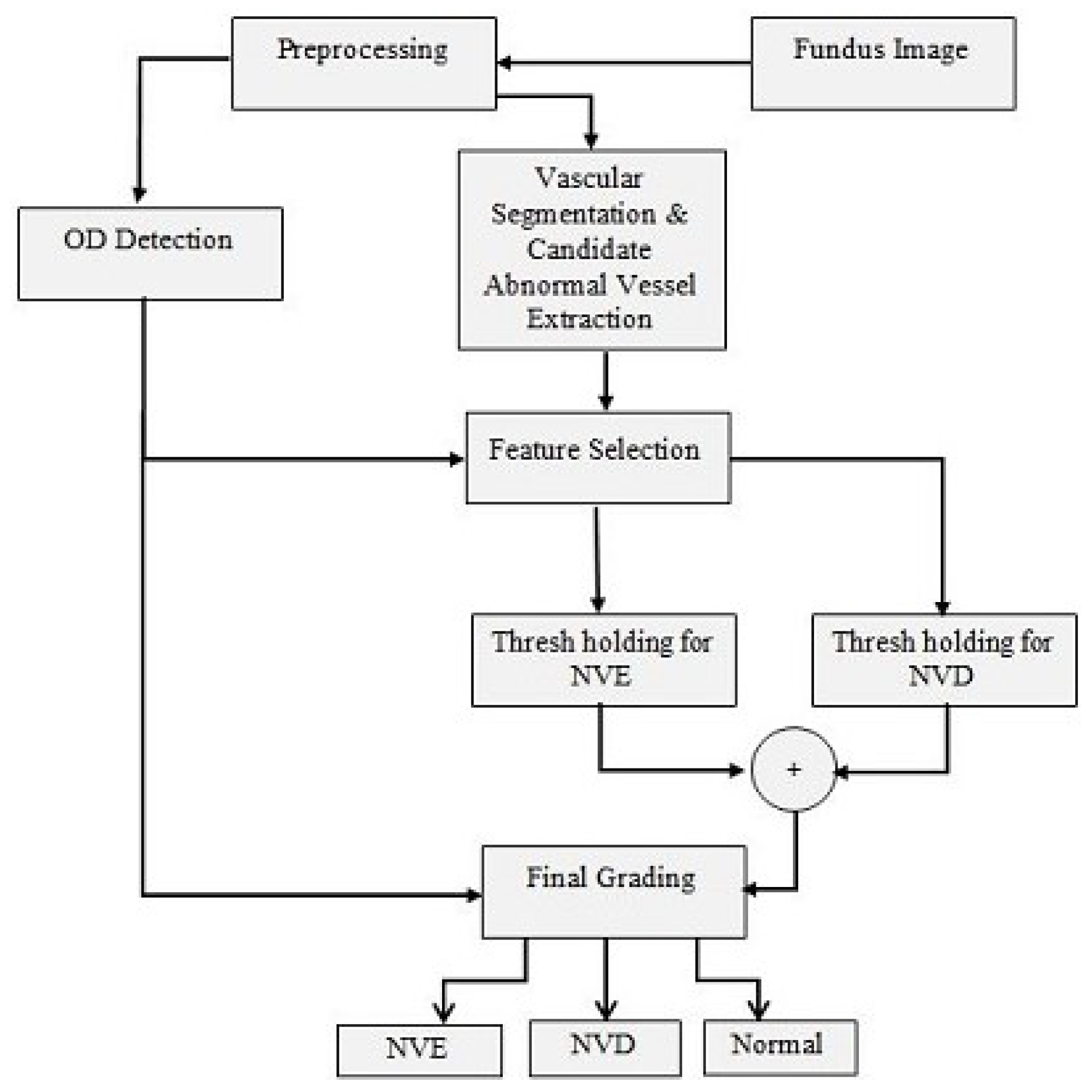
3. Materials and Methods
3.1. Preprocessing
3.2. Vessel Segmentation
3.3. Abnormal Vessel Detection
3.4. Optic Disc Detection
3.5. Feature Selection and Thresholding
- Entropy: Entropy is the measure of uncertainty in a system. Abnormal blood vessels are fragile and follow no proper pattern. Thus, the regions that contain abnormal blood vessels have a high entropy value. If is the probability of occurrence of a grey level k and M is the number of grey levels in the image, then entropy is calculated as:
- Energy: Energy is the sum of squares of all pixel intensities within a candidate region of interest. The energy of the region containing the abnormal blood vessels lies in between those of the regions that contain normal blood vessels and the bright lesions or exudates. If is the pixel value in an image then the energy is calculated as:
- Homogeneity: Homogeneity returns a value that tells the closeness of the distribution of elements. The homogeneity of abnormal blood vessels lies very close to that of normal blood vessels but it is away from that of the lesions and exudates. The abnormal blood vessels originate near normal vessels, while the exudates and bright lesions can be found anywhere on the retina. The homogeneity is calculated as:
- Energy: As a smaller window size was chosen for NVE, it shows a relatively high energy value in that small area.
- Gradient: The mean gradient magnitude in the candidate region of interest is calculated by using the Sobel gradient operator. Separate measurements of the gradient component in each orientation, called and , are calculated. Then the magnitude of the gradient is given by:The mean of the gradient magnitude is used as a feature, which is:where m and n are the dimensions of the region of interest.
- Gradient Direction: The directional gradient is the standard deviation of the Sobel gradient in the candidate region of interest. As the abnormal vessels are much less defined, are less homogeneous, and have more contrast variation than normal vessels, this feature is taken in to account. The direction can be calculated as:and the mean of its standard deviation is:
3.6. Post-Processing
- Mean Intensity : It is the mean value of pixels within the green plane of the candidate region.
- Maximum Intensity : It is the maximum value of pixels within the green channel of the candidate region.
- Mean Skewness : It is the measure of the lack of symmetry in a candidate region. It is computed as:where is the candidate pixel value, is the mean value pixels, s is the standard deviation, and N is the number of pixels in the candidate region.
- Entropy : It is the value of all pixels in a candidate region and its neighboring pixels. It is the measure of unpredictability in an ROI.
- Energy : It is the sum of the squares of all the pixel values of the green plane inside a candidate region.
- Mean Gradient : It is the mean of the pixels of the edges detected using the Sobel gradient within the candidate region.
- Gradient Direction : It is the standard deviation of the direction of the Sobel gradient in a candidate region.
- Mean Intensity of red plane : It is the mean value of pixels within the red plane of the candidate region.
- Mean Intensity of blue plane : It is the mean value of pixels within the blue plane of the candidate region.
- Mean Intensity lightness in LAB color space : It is the mean value of pixels within the lightness plane in the LAB color space of the candidate region.
3.7. Grading of PDR as NVD or NVE
4. Results
- are true positives, meaning abnormal blood vessel regions correctly classified as abnormal.
- are true negatives, meaning normal blood vessel regions correctly classified an normal.
- are false positives, meaning normal blood vessel regions wrongly classified as abnormal.
- are false negatives, meaning abnormal blood vessel regions wrongly classified as normal blood vessel regions.
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PDR | Proliferative Diabetic Retinopathy |
| DR | Diabetic Retinopathy |
| NVD | Neovascularization on Disc |
| NVE | Neovascularization Elsewhere |
| SVM | Support Vector Machine |
| OD | Optic Disc |
References
- Leontidis, G.; Al-Diri, B.; Hunter, A. Diabetic retinopathy: Current and future methods for early screening from a retinal hemodynamic and geometric approach. Expert Rev. Ophthalmol. 2014, 9, 431–442. [Google Scholar] [CrossRef] [Green Version]
- Arden, G.B.; Ramsey, D.J. Diabetic retinopathy and a novel treatment based on the biophysics of rod photoreceptors and dark adaptation. In Webvision: The Organization of the Retina and Visual System; Kolb, H., Fernandez, E., Nelson, R., Eds.; University of Utah Health Sciences Center: Salt Lake City, UT, USA, 2015. Available online: https://www.ncbi.nlm.nih.gov/books/NBK310272/ (accessed on 29 June 2023).
- Wong, T.; Cheung, C.; Larsen, M.; Sharma, S.; Simó, R. Diabetic retinopathy. Nat. Rev. Dis. Prim. 2016, 2, 16012. [Google Scholar] [CrossRef] [PubMed]
- Shukla, U.V.; Tripathy, K. Diabetic Retinopathy. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK560805/ (accessed on 22 February 2023).
- Nentwich, M.M.; Ulbig, M.W. Diabetic retinopathy—Ocular complications of diabetes mellitus. World J. Diabetes 2015, 6, 489–499. [Google Scholar] [CrossRef]
- Salamat, N.; Missen, M.M.; Rashid, A. Diabetic retinopathy techniques in retinal images: A review. Artif. Intell. Med. 2019, 97, 168–188. [Google Scholar] [CrossRef] [PubMed]
- Akram, M.U.; Khalid, S.; Tariq, A.; Javed, M.Y. Detection of Neovascularization in Retinal Images using Multivariate m-Mediods based Classifier. Comput. Med Imaging Graph. 2013, 37, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Leontidis, G.; Al-Diri, B.; Wigdahl, J.; Hunter, A. Evaluation of geometric features as biomarkers of diabetic retinopathy for characterizing the retinal vascular changes during the progression of diabetes. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 5255–5259. [Google Scholar] [CrossRef] [Green Version]
- Mendes, L.; Marques, I.P.; Cunha-Vaz, J. Comparison of Different Metrics for the Identification of Vascular Changes in Diabetic Retinopathy Using OCTA. Front Neurosci. 2021, 15, 755730. [Google Scholar] [CrossRef]
- Das, S.; Kharb, a.K.; Suchetha, M.; Raman, R.; Dhas, E. Deep learning architecture based on segmented fundus image features for classification of diabetic retinopathy. Biomed. Signal Process. Control 2021, 68, 102600. [Google Scholar] [CrossRef]
- Goatman, K.A.; Fleming, A.D.; Philip, S.; Williams, G.J.; Olson, J.A.; Sharp, P.F. Detection of new vessels on the optic disc using retinal photographs. IEEE Trans. Med. Imaging 2011, 30, 972–979. [Google Scholar] [CrossRef]
- Agurto, C.; Murray, V.; Barriga, E.; Murillo, S.; Pattichis, M.; Davis, H.; Russell, S.; Abramoff, M.; Soliz, P. Multi-scale AM-FM methods for diabetic retinopathy lesion detection. IEEE Trans Med Imaging 2010, 29, 502–512. [Google Scholar] [CrossRef] [Green Version]
- Jelinek, H.F.; Cree, M.J.; Leandro, J.J.G.; Soares, J.V.B.; Cesar, R.M., Jr.; Luckie, A. Automated segmentation of retinal blood vessels and identification of proliferative diabetic retinopathy. J. Opt. Soc. Am. A 2007, 24, 1448–1456. [Google Scholar] [CrossRef]
- Mudigonda, S.; Oloumi, F.; Katta, K.M.; Rangayyan, R.M. Fractal analysis of neovascularization due to diabetic retinopathy in retinal fundus images. In Proceedings of the E-Health and Bioengineering Conference (EHB), IEEE, Iasi, Romania, 19–21 November 2015. [Google Scholar]
- Saranya, K.B.; Mohideen, S.K. A Novel Approach for the Detection of New Vessels in the Retinal Images for screening Diabetic Retinopathy. In Proceedings of the 2012 International Conference on Communication and Signal Processing: IEEE Advancing Technology for Humanity, Chennai, India, 4–5 April 2012. [Google Scholar] [CrossRef]
- Roychowdhury, S.; Koozekanani, D.D.; Parhi, K.K. Automated detection of neovascularization for proliferative diabetic retinopathy screening. In Proceedings of the 2016 IEEE 38th Annual International Conference: Engineering in Medicine and Biology Society (EMBC), IEEE, Orlando, FL, USA, 16–20 August 2016. [Google Scholar]
- Lee, J.; Zee, B.C.Y.; Li, Q. Detection of Neovascularization Based on Fractal and Texture Analysis with Interaction Effects in Diabetic Retinopathy. PLoS ONE 2013, 8, e75699. [Google Scholar] [CrossRef]
- Welikalaa, R.A.; Dehmeshki, J.; Hoppe, A.; Tah, V.; Mann, S.; Williamson, T.H.; Barman, S.A. Automated detection of proliferative diabetic retinopathy using a modified line operator and dual classification. Comput. Methods Programs Biomed. 2014, 114, 247–261. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Xiao, D.; Kanagasingam, Y. Automatic detection of neovascularization on optic disk region with feature extraction and support vector machine. In Proceedings of the 2016 IEEE 38th Annual International Conference of the Engineering in Medicine and Biology Society (EMBC), IEEE, Orlando, FL, USA, 16–20 August 2016. [Google Scholar]
- Coelho, D.F.G.; Rangaraj, M.R.; Vassil, S.D. Detection of neovascularization near the optic disk due to diabetic retinopathy. In Proceedings of the 2016 24th European Signal Processing Conference (EUSIPCO), IEEE, Budapest, Hungary, 29 August–2 September 2016. [Google Scholar]
- Yu, S.; Xiao, D.; Kanagasingam, Y. Machine learning based automatic neovascularization detection on optic disc region. IEEE J. Biomed. Health Inform. 2018, 22, 886–894. [Google Scholar] [CrossRef]
- Argyrios, C.; Hurtut, T.; Cheriet, F. Proliferative diabetic retinopathy characterization based on the spatial organization of vascular junctions in fundus images. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), IEEE, Melbourne, VIC, Australia, 18–21 April 2017. [Google Scholar]
- Leeza, M.; Farooq, H. Detection of Severity Level of Diabetic Retinopathy Using Bag of Features Model. IET Comput. Vis. 2019, 13, 523–530. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Q.; You, J.; Zhang, D. A modified matched filter with double-sided thresholding for screening proliferative diabetic retinopathy. IEEE Trans. Inf. Technol. Biomed. 2009, 13, 528–534. [Google Scholar] [CrossRef]
- Qummar, S.; Khan, F.G.; Shah, S.; Khan, A.; Shamshirb, S.; Rehman, Z.U.; Khan, I.A.; Jadoon, W. A Deep Learning Ensemble Approach for Diabetic Retinopathy Detection. IEEE Access 2019, 7, 150530–150539. [Google Scholar] [CrossRef]
- Elgafi, M.; Sharafeldeen, A.; Elnakib, A.; Elgarayhi, A.; Alghamdi, N.S.; Sallah, M.; El-Baz, A. Detection of Diabetic Retinopathy Using Extracted 3D Features from OCT Images. Sensors 2022, 22, 7833. [Google Scholar] [CrossRef]
- Mehboob, A.; Akram, M.U.; Alghamdi, N.S.; Abdul Salam, A. A Deep Learning Based Approach for Grading of Diabetic Retinopathy Using Large Fundus Image Dataset. Diagnostics 2022, 12, 3084. [Google Scholar] [CrossRef]
- Tang, M.C.; Teoh, S.S.; Ibrahim, H.; Embong, Z. Neovascularization detection and localization in fundus images using deep learning. Sensors 2021, 21, 5327. [Google Scholar] [CrossRef]
- Tang, M.C.; Teoh, S.S.; Ibrahim, H.; Embong, Z. A deep learning approach for the detection of neovascularization in fundus images using transfer learning. IEEE Access 2022, 10, 20247–20258. [Google Scholar] [CrossRef]
- Lavanya, S.; Naveen, P. Detection of Retinal Neovascularization Using Optimized Deep Convolutional Neural Networks. J. Trends Comput. Sci. Smart Technol. 2022, 4, 38–49. [Google Scholar] [CrossRef]
- Tariq, A.; Akram, M.U. An Automated System for Colored Retinal Image Background and Noise Segmentation. In Proceedings of the 2010 IEEE Symposium on Industrial Electronics and Applications (ISIEA), Penang, Malaysia, 3–5 October 2010. [Google Scholar]
- Antoine, J.P.; Carette, P.; Murenzi, R.; Piette, B. Image analysis with two-dimensional continuous wavelet transform. Signal Process. 1993, 31, 241–272. [Google Scholar] [CrossRef]
- Akram, M.U.; Khan, S.A. Multilayered thresholding-based blood vessel segmentation for screening of diabetic retinopathy. Eng. Comput. 2011, 29, 165–173. [Google Scholar] [CrossRef]
- Usman, A.; Abbas, S.; Akram, M.U.; Nadeem, Y. A Robust Algorithm for Optic Disc Segmentation from Colored Fundus Images: Image analysis and Recognition; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Nelson, L.S. Evaluating Overlapping Confidence Intervals. J. Qual. Technol. 1989, 21, 140141. [Google Scholar] [CrossRef]
- MESSIDOR. Available online: https://www.adcis.net/en/third-party/messidor/ (accessed on 29 January 2023).







| Parameters | Value for Normal | Value for Abnormal |
|---|---|---|
| Blood Vessels | Blood Vessels | |
| Dilation (a) | 11 | 1.8 |
| Elongation () | 5 | 1 |
| Rotation Angle () | 10° | 10° |
| [0,2.5] | [0,2] |
| Parameters | Value for Normal | Value for Abnormal |
|---|---|---|
| Blood Vessels | Blood Vessels | |
| Dilation(a) | 7 | 2 |
| Elongation () | 10 | 1 |
| Rotation angle () | 10° | 10° |
| [0,2.5] | [0,2.5] |
| Grade | Condition | Class |
|---|---|---|
| 0 | No abnormal blood vessels present | Healthy |
| 1 | A few abnormal blood vessels present 1dd away from the OD | NVE |
| 2 | Abnormal blood vessels present within 1dd of OD | NVD |
| Database | Images | Normal | PDR | NVD | NVE |
|---|---|---|---|---|---|
| MESSIDOR | 1200 | 397 | 37 | 27 | 18 |
| AFIO | 20 | 13 | 7 | 4 | 3 |
| Sr. | Method | Number of Images | Sen | Spec | Acc/F1 Score |
|---|---|---|---|---|---|
| 1 | Jelinek et al. [13] | 27 images | – | – | 0.90 |
| 2 | Saranya et al. [15] | 50 images from MESSIDOR and DRIVE | 0.96 | 0.89 | 0.96 |
| 3 | Lee et al. [17] | 137 images from MESSIDOR | 0.96 | 0.99 | 0.98 |
| 4 | Welikala et al. [18] | 60 images | 0.91 | 0.92 | 0.96 |
| 5 | Garima et al. [15] | 799 images | 0.95 | 0.83 | 0.96 |
| 8 | Proposed | 1200 images from MESIDOR | 0.90 | 1 | 0.98 |
| – | 20 images from AFIO | 0.80 | 1 | 0.95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huda, N.u.; Salam, A.A.; Alghamdi, N.S.; Zeb, J.; Akram, M.U. Proliferative Diabetic Retinopathy Diagnosis Using Varying-Scales Filter Banks and Double-Layered Thresholding. Diagnostics 2023, 13, 2231. https://doi.org/10.3390/diagnostics13132231
Huda Nu, Salam AA, Alghamdi NS, Zeb J, Akram MU. Proliferative Diabetic Retinopathy Diagnosis Using Varying-Scales Filter Banks and Double-Layered Thresholding. Diagnostics. 2023; 13(13):2231. https://doi.org/10.3390/diagnostics13132231
Chicago/Turabian StyleHuda, Noor ul, Anum Abdul Salam, Norah Saleh Alghamdi, Jahan Zeb, and Muhammad Usman Akram. 2023. "Proliferative Diabetic Retinopathy Diagnosis Using Varying-Scales Filter Banks and Double-Layered Thresholding" Diagnostics 13, no. 13: 2231. https://doi.org/10.3390/diagnostics13132231
APA StyleHuda, N. u., Salam, A. A., Alghamdi, N. S., Zeb, J., & Akram, M. U. (2023). Proliferative Diabetic Retinopathy Diagnosis Using Varying-Scales Filter Banks and Double-Layered Thresholding. Diagnostics, 13(13), 2231. https://doi.org/10.3390/diagnostics13132231






