Abstract
We conducted this Systematic Review to create an overview of the currently existing Artificial Intelligence (AI) methods for Magnetic Resonance Diffusion-Weighted Imaging (DWI)/Fluid-Attenuated Inversion Recovery (FLAIR)—mismatch assessment and to determine how well DWI/FLAIR mismatch algorithms perform compared to domain experts. We searched PubMed Medline, Ovid Embase, Scopus, Web of Science, Cochrane, and IEEE Xplore literature databases for relevant studies published between 1 January 2017 and 20 November 2022, following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines. We assessed the included studies using the Quality Assessment of Diagnostic Accuracy Studies 2 tool. Five studies fit the scope of this review. The area under the curve ranged from 0.74 to 0.90. The sensitivity and specificity ranged from 0.70 to 0.85 and 0.74 to 0.84, respectively. Negative predictive value, positive predictive value, and accuracy ranged from 0.55 to 0.82, 0.74 to 0.91, and 0.73 to 0.83, respectively. In a binary classification of ±4.5 h from stroke onset, the surveyed AI methods performed equivalent to or even better than domain experts. However, using the relation between time since stroke onset (TSS) and increasing visibility of FLAIR hyperintensity lesions is not recommended for the determination of TSS within the first 4.5 h. An AI algorithm on DWI/FLAIR mismatch assessment focused on treatment eligibility, outcome prediction, and consideration of patient-specific data could potentially increase the proportion of stroke patients with unknown onset who could be treated with thrombolysis.
1. Introduction
Magnetic resonance imaging (MRI) is the primary imaging modality for stroke detection and classification in patients with an unknown onset and wake-up stroke (WUS), using Diffusion-weighted imaging (DWI) and T2-Weighted Fluid-Attenuated Inversion Recovery (FLAIR) sequences [1]. DWI sequences are useful for the detection of early signs of infarction and can be used to outroot stroke mimics [2,3]. The visibility of hyperintense FLAIR sequence lesions compatible with acute infarction increases with time since stroke onset (TSS). However, the intensity of FLAIR hyperintensity lesions cannot provide an exact time of stroke onset [4,5,6,7,8].
Based on results from randomized clinical trials, alteplase or recombinant tissue plasminogen activator (rtPA) can be administered up to 4.5 h after stroke symptoms onset, as the risk of hemorrhage and poor outcomes increases with time [6,7,8,9]. For patients with unknown stroke onset, which accounts for 20% to 27% of all ischemic strokes, the DWI/FLAIR lesion mismatch assessment is used to assess whether the patient can be treated with rtPA [8,10,11]. Presently, only Wake-up stroke patients/patients with unknown TSS without visible FLAIR lesions and within 4.5 h from recognition of stroke symptoms are considered eligible for treatment [8,9].
Over recent years, the number of artificial intelligence (AI) algorithms applicable for radiological purposes has increased markedly [12,13,14]. Multiple studies have shown the ability of algorithms to detect whether ischemic lesions are present on DWI and FLAIR sequences with deep learning (DL) algorithms [15,16].
Field experts, e.g., neuroradiologists, perform the current clinically used binary mismatch assessment, but it has been shown to have poor inter- and intra-observer agreement [6,7,8]. An automated assessment of DWI/FLAIR mismatch could create a more standardized method and assist the stroke progression analysis and treatment selection process.
We conducted this Systematic Review to discover the currently existing AI algorithms for automated DWI/FLAIR mismatch assessment and determine how well DWI/FLAIR mismatch algorithms perform as compared to experienced radiologists and neurologists.
2. Materials and Methods
This systematic review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [17]. The study protocol was registered in the Prospective Register of Systematic Reviews (PROSPERO) under number CRD42022377938 during the research process.
2.1. Literature Search
Literature was searched between 26 October 2022, and 20 November 2022, in PubMed MEDLINE, Ovid Embase, IEEE Xplore, Web of Science, Elsevier Scopus, and Cochrane. We searched articles published between 1 January 2017, and 20 November 2022, within the clinical and technical scope of the review.
The preliminary search string was done in PubMed MEDLINE. Afterward, it was converted and edited with the polyglot tool Systematic Review-accelerator (Bond University, Australia) [18]. This made the search strings suitable for the rest of the databases. Search strings for each database can be found in Appendix A.
2.2. Study Selection
The literature was extracted and collected for the reference tool Endnote 20 (Clarivate, London, UK). Duplicates were removed with Endnote’s internal duplicate search function.
The references were exported to Covidence (Melbourne, Australia) to screen and review articles. The internal duplicate tool in Covidence removed duplicates not caught by Endnote 20.
Only studies focusing on machine learning methods to automatically identify DWI and FLAIR lesions on structural brain MRI were included. The studies had to be peer-reviewed and in English. Editorials, case series, letters, conference proceedings, reviews, and inaccessible papers were also excluded. The main inclusion and exclusion criteria for the screening process are shown in Table 1.

Table 1.
Main in- and exclusion criteria.
The two reviewers, a medical doctor (C.M.O.) and a medical student (J.S.), independently screened all records based on titles and abstracts, followed by a full-text review of the potentially relevant papers for final inclusion. Any conflicts during the screening process were resolved by consulting a 3rd and 4th reviewer (K.S. and J.F.C.).
2.3. Data Extraction and Analysis
Three reviewers (J.S., C.M.O., and K.S.) extracted data from the included articles. The extracted study characteristics composed of the following: The number of patients, size of training and test data set, population demographics, National Institutes of Health Stroke Scale (NIHSS) score upon admission, time from onset to MRI, and the percentage of the dataset with an onset to MRI within 4.5 h from symptom onset. In addition, scanner characteristics, the type of scanner, and the used artificial intelligence methods were extracted. Performance data for the algorithms and used comparator was extracted and composed of; Area under the curve (AUC), Sensitivity (Sens), Specificity (Spec), Accuracy (Acc), Precision or positive predictive value (PPV), and negative predictive value (NPV).
If multiple performance data were mentioned for different algorithms, only the performance data from the best algorithm was selected unless otherwise stated.
2.4. Quality Assessment
The two reviewers used the questionnaire Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool to perform the quality assessment of the included studies [19]. The QUADAS-2 assess the risk of bias and concern for applicability in the domains of patient selection, index test, reference standard, and flow and timing. The domains were categorized as having a high, unclear, or low risk of bias or concern for applicability.
3. Results
3.1. Study Selection
The literature search resulted in 3273 articles. Duplicates and conference abstracts accounted for 1595. The total number of titles and abstracts to screen was then 1678. Eighteen articles were selected relevant for full-text assessment. In five articles, the AI algorithm did not perform a DWI-FLAIR mismatch assessment. One was not in English. Two had outcomes not relevant for this study, i.e., a focus on prediction accuracy for upper extremity motor outcomes 90 days post-stroke and a focus on detecting FLAIR and DWI lesions independently and not as a mismatch [20,21]. One article was a preprint of an already-included article. Ultimately, this led to a total number of five articles included in the review. This study’s inclusion process is presented in Figure 1.
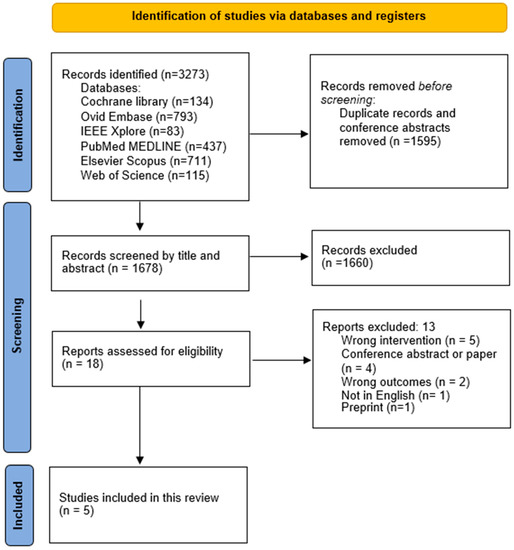
Figure 1.
PRISMA workflow. The study inclusion process.
3.2. Study Characteristics
All five included studies were retrospective. They used image data from different hospitals or university databases from the period between 2011 and 2021. Study characteristics and scanner specifications are summarized in Table 2.

Table 2.
Study characteristics and scanner specifications.
The study population sizes varied from 268 to 587 patients diagnosed with stroke. The study conducted by Polson et al. had two study populations, one internal and one external [24]. The external population was the same as the one used in the study by Lee et al. [25]. Therefore, the internal study population from Polson et al. was chosen for reporting in this review. All five studies divided their dataset into training populations and test populations. In Lee et al., Polson et al., and Zhang et al. the distribution of patients in the two groups was very similar, ranging from 16% to 19% [23,24,25]. Zhu et al. and Jiang et al. had a much higher proportion of the test population; the distributions were 30% and 40%, respectively [22,26].
The median population age covering all five studies ranged from 63 to 70 years. The maximal difference in NIHSS scores upon admission between training and test populations in the included articles was minor. Polson et al. and Zhang et al. had a higher NIHSS score upon admission in the training population compared to the test population [23,24]. Lee et al. and Jiang et al. had a higher score in the test population [25,26]. Zhu et al. only reported an average score for the whole study population [22].
Zhu et al. had a very high average time from onset to MRI of 10.3 h [22]. The median time from stroke onset to a performed MRI in the four other studies ranged from 3.5 h to 4.5 h across training and test populations [23,24,25,26]. Polson et al., Jiang et al., and Zhu et al. stated the percentage of patients who had a stroke onset time to MRI within 4.5 h [22,24,26]. The variation across training and test population was 64.55% to 50%.
Four out of five studies used stroke neuroradiologists or neurologists for mismatch assessment as the comparator for their AI algorithm [22,23,24,25]. Polson et al. and Zhang et al. used the independent mismatch assessments of three neuroradiologists and aggregated their results [23,24]. Lee et al. used two neurologists’ mismatch assessments, potential disagreements were resolved, and a consensus was obtained [25]. Zhu et al. used an unspecified number of neuroradiologists for the mismatch assessment [22]. Jiang et al. did not have a human comparator in the study, but due to the scope of this review, it was relevant for inclusion [26].
3.3. Automated Assessment of Time since Stroke
Lee et al., Jiang et al., and Zhu et al. used a DL algorithm in conjunction with conventional machine learning (ML) algorithms in a two-step model for the assessment of TSS [22,25,26]. The first step for all three studies was the segmentation of the stroke lesion on the DWI sequence to determine the region of interest (ROI) and volume of interest (VOI) using DL algorithms derived from the U-net architecture. The specific segmentation methods are seen in Table 2.
After the segmentation, all three studies used different ML techniques for the TSS classification. All three studies tested and compared three to seven various ML models to find the best-performing ML model. In Lee et al. and Jiang et al., the best-performing ML models were Random Forest and a support vector machine with a radial kernel (svmRadial), respectively [25,26]. Zhu et al. used an ensemble approach with one ML model integrating the outputs of the top five best-performing ML algorithms to produce a final prediction [22]. Polson et al. and Zhang et al. only used an end-to-end convolutional neural networks model (CNN) for stroke segmentation and mismatch assessment [23,24]. Both studies trained the CNN to segment the lesion on undefined sequences and afterward trained it to classify TSS using one unified architecture.
3.4. Study Performance
For this review, the performance measures for the AI models were registered, and missing values were calculated from the existing data when possible. The best performance metrics and preferentially from out-of-distribution external validation datasets are the performance results for the TSS classification reported in Table 3.

Table 3.
Performance results of TSS classification.
The area under the curve (AUC) was reported in all five studies. It ranged from 0.74 to 0.90 for the AI models. The AUC was not reported for the comparators, as these assessments were dichotomous. The sensitivity, also known as the true positive rate, and the specificity, also known as the true negative rate, were reported in all five studies. It ranged from 0.70 to 0.82 and from 0.74 to 0.84, respectively. Evaluation of the best performances showed that the specificity was a little higher than the sensitivity in all five studies. Results are shown in Table 3.
For Lee et al., Jiang et al., Polson et al., and Zhu et al., the positive predictive value (PPV) and negative predictive value (NPV) were reported or calculated, ranging from 0.74 to 0.91 and 0.65 to 0.80, respectively. In the same four studies, the accuracy ranged from 0.73 to 0.83 [22,24,25,26]. In Zhu et al., precision, also known as PPV, was registered for the machine learning methods, but neither precision nor PPV was registered for the comparators. PPV was therefore calculated for both the AI method and the comparators in order to align with the other studies.
Performance measures for the comparators were obtained in all studies except for Jiang et al. The sensitivity and specificity ranged from 0.49 to 0.82 and 0.59 to 0.91, respectively. The accuracy ranged from 0.61 to 0.74 [22,23,24,25]. For Zhu et al., Polson et al., and Lee et al., PPV and NPV for the comparators were either reported or calculated. It ranged from 0.73 to 0.89 and 0.55 to 0.72, respectively [22,24,25].
These results suggest that AI sensitivity, NPV, and accuracy in the determination of TSS were better than for the radiologists’ but the AI specificity and PPV were comparable to that of the radiologists’ in a binary classification of TSS ±4.5 h.
3.5. Quality Assessment
The Quality Assessment of Diagnostic Accuracy Studies 2 tool was applied to all included studies of this review. The results of the risk of bias/concern for applicability analysis are presented in Table 4.

Table 4.
Tabular presentation of the QUADAS-2 assessments in each study.
Cohort sizes were reasonably large in all five included studies and comparable to the cohort sizes in studies included in another systematic review with a focus on the potential of automated segmentation of stroke lesions in MRI images [27].
Sampled study cohorts, description of inclusion and exclusion criteria, and processing of the images for optimization of the AI algorithm performances were assessed.
The studies description of the comparator process and methods used for the segmentations also affected the evaluation.
None of the studies were assessed with a low risk of bias and concern for applicability in all domains.
No meta-analysis was conducted due to the small number of included articles.
4. Discussion
The currently existing AI-based DWI/FLAIR mismatch assessment uses FLAIR lesion visibility as a surrogate marker of TSS with a binary decision, i.e., ±4.5 h since stroke onset. Because of the non-trivial correlation between FLAIR changes and TSS, it is not recommended that FLAIR lesion visibility is used for the determination of time since stroke onset within the first 4.5 h [5]. However, an AI-assisted DWI/FLAIR mismatch assessment could be useful in patients of unknown onset time/wake-up stroke if a mismatch is identified and the time from recognized symptoms is less than 4.5 h. Then the patient could receive rtPA treatment.
4.1. Artificial Intelligence vs. Human Readings
Ebinger et al. have shown that AI algorithms can register several features in the images, such as size, homogeneity, gradient, and intensity [5]. This creates an opportunity for a more nuanced analysis than the current binary decision, i.e., mismatch or no-mismatch.
In the five included studies, we found that the used AI algorithms performed equivalent to or even better than neuroradiologists and neurologists in the binary classification of time since onset ±4.5 h from DWI/FLAIR images. The automated registration of several features could explain these results.
Thomalla et al. and Ebinger et al. have found that even though there is a relationship between the time clock and tissue progression, there is no significant correlation between relative signal intensity on FLAIR imaging and TSS [5,8]. The FLAIR hyperintensity lesions can occur before 4.5 h but may also occur later [5,6,8]. The automated analysis of DWI/FLAIR lesion mismatch could be useful assistance in the daily acute setting concerning lesion visibility, but the determination of TSS from these identified lesions should be considered with caution.
The sensitivities and specificities reported for the AI algorithms were more consistent across the different studies than for the neuroradiologists and neurologists, with the exception of one study, showing the opposite. This supports the idea that automated segmentations could provide a standardized assessment of lesions and potentially decrease inter-rater variability. In a study on fully automatic acute ischemic lesion segmentation on DWI, they argue that domain experts could save time and effort if the manual segmentations were based on automated segmentation [28].
In a study from 2022 on automated segmentation, they tested whether the combination of T2w-FLAIR and DWI sequences could lead to a better segmentation of stroke lesions than single modal modality approaches, i.e., only DWI [20]. Jiang et al. also compared the performance of their model with different modalities, i.e., DWI vs. FLAIR vs. DWI + FLAIR. They found that DWI + FLAIR had the best performance [26].
Similarly, in a study on infarct segmentation on DWI, ADC, and low b-value-weighted images from 2019, they found that the combination of sequences led to an improved segmentation of lesions compared to only diffusion maps and could produce results comparable with manual lesions segmented by domain experts [29].
These studies suggest that automated segmentations could assist neuroradiologists in the assessment of infarction lesions and maybe neurologists in the treatment decision of patients with acute ischemic stroke.
4.2. FLAIR Hyperintensity Segmentation
To conduct the FLAIR segmentation, three of the included articles, Zhu et al., Jiang et al., and Lee et al., used the DWI segmentation data and features, such as location, to improve their FLAIR segmentation [22,25,26]. Lee et al. used the apparent diffusion coefficient (ADC) maps.
Other studies on FLAIR segmentation have highlighted the difficulties in segmenting FLAIR lesions in stroke and mixed pathology cases, yielding mediocre DICE values ranging from 0.58 to 0.79 [30,31]. This could indicate that the segmentation of FLAIR lesions is not only difficult for the human eye but also a challenge for an AI algorithm.
To improve FLAIR segmentation, the intensity correction by an algorithm, the use of an automated filter to remove inhomogeneity, and the negative influence of high-intensity regions were tested in a study from 2019. The FLAIR lesion segmentation method showed promising results in a follow-up FLAIR imaging dataset on lesion volume estimations [30].
In a study on FLAIR visibility and 90-day outcome prediction but no DWI/FLAIR mismatch assessment, they found that patients with an early visible FLAIR lesion within 4.5 h and who were given intravenous (IV) thrombolysis had a poorer 90-day outcome than those who had a later development of visible FLAIR lesions [32].
Even though FLAIR lesion visibility cannot serve as a surrogate marker for TSS, FLAIR changes could resemble a biological marker of stroke progression/tissue salvage-ability, though the influence of the blood-brain barrier (BBB) permeability and vascular recanalization in subacute ischemic stroke is still questioned [5,8,9,33].
4.3. Limitations of the Included Studies
The risks of bias and concern for applicability seen in the four domains of the QUADAS-2 assessment were mainly due to a limited description of the human comparator process, convenience sampled cohorts, insufficient description of inclusion and exclusion criteria, and/or extensive exclusion of images which could limit the use in a daily clinical setting.
Only two of the included studies transparently specified their stroke patient selection with inclusion and exclusion criteria. The exclusion criteria were small or unsegmented lesions, DWI lesions on sites with extensive leucoaraiosis on FLAIR, and heavy artifacts on DWI or FLAIR sequences [25,26]. These exclusion criteria narrow the number of patients suited for the AI algorithms. One study only reported characteristics for the cohort in general, which could raise the risk of an unbalanced training and test cohort selection [22]. Furthermore, another of the five studies did not report the percentage of patients who had a stroke within 4.5 h [23]. This could potentially bias the test data and facilitate better test results, which could introduce an overestimation of model performance and limit integration into clinical practices.
The split size of the training set and test set in all five studies followed the general trend and ranged from 70/30 to 90/10. Consideration of sample size is important as deep-learning-based algorithms have higher recognition accuracy on larger sample data sets [34].
4.4. Limitations of This Review
The search process was performed according to PRISMA guidelines. Given the speed and interest to develop AI algorithms for neuroradiology purposes, there is a probability that newer and relevant studies are not included in this systematic review when published.
One of the inclusion criteria was to only include algorithms on DWI and FLAIR sequences, based on the present process to determine treatment eligibility for patients with unknown symptom onset. In the preliminary search process, we found a study on stroke onset determination with the use of perfusion-weighted imaging [35]. This shows the limitation of our search due to the narrow inclusion criteria. Another limitation of this review is the risk of publication bias. It is not possible to rule out the risk of conducted studies with poor performance not being published. Unpublished work with poor performance would lead to a false view of the potential and performance of AI algorithms for mismatch assessment. The five reviewed methods all focus on TSS from the classification of DWI/FLAIR mismatch. We searched for studies on DWI/FLAIR mismatch assessment without excluding articles with a focus on the prediction of outcomes but did not find any that fit the scope of this review. We did find studies on DWI and FLAIR segmentation, which focused on the outcome, but there were no studies on DWI/FLAIR mismatch, outcome prediction, and AI.
4.5. Perspectives
The continued advancement of radiology AI methods offers new opportunities to improve the detection, diagnosis, and prediction of patient outcomes. Still, even if the results of a study seem promising, a method can sometimes perform poorer than expected when tested on an external dataset with a larger, consecutive cohort. External validation is, therefore, important, and a validation and evaluation framework could be necessary [36].
For remote hospitals and in countries where the MRI scanner density is low, the use of an approved AI algorithm could assist regarding diagnosis, decrease unnecessary transportation to other clinics, and improve treatment decisions and, thereby, patient outcomes [37].
Time is of the essence in the treatment of acute ischemic stroke. A rapid and efficient workflow is, therefore, essential. The exclusion of hemorrhage, stroke mimics, and the estimation of stroke volume on imaging is important to identify patients eligible for rtPA-treatment [38]. The door-to-needle (DTN) time, which is the time from the arrival of the acute ischemic stroke (AIS) patient at the emergency room to the initiation of rtPA-treatment, can be used as a measurement of quality improvement [39]. An automated stroke detection with a reliable imaging method could possibly decrease DTN time [39]. Another method to improve stroke care is discussed in a study from 2021 on the development of a prediction model for daily stroke occurrences. They describe how the prediction could be improved by synchronizing a variety of medical information and have patients perform self-care. Further knowledge of stroke occurrence could also have an influence on the improvement in medical resources [40]. This could lower DTN time, which would improve stroke care and patient outcome.
Another way to improve MRI workflow was tested in a study on a synthetic FLAIR sequence automatically developed from DWI sequences. The synthetic FLAIR sequence was tested against the usual FLAIR sequence in DWI/FLAIR mismatch assessment by neuroradiologists. It showed comparable diagnostic performances [41].
A number of AI algorithms for outcome prediction of stroke patients based on MRI have used modified Rankin scales as a reference to predict stroke outcomes and were able to predict the likelihood of hemorrhagic transformation [42,43,44,45].
In a Meta-analysis on examination of delayed thrombolysis among patients selected according to mismatch criteria, they found that patients who received delayed thrombolysis, i.e., after 3 h, was associated with increased reperfusion/recanalization, but the relation between delayed thrombolysis in mismatch patients and the post-treatment outcome was not established [46]. Several other studies have later shown the importance of early management, which is also included in the guidelines from 2019 by Power et al. [1].
In WAKE-UP, a randomized, placebo-controlled trial of thrombolysis in stroke with an unknown time of symptom onset, they used MRI criteria to determine patients’ eligibility for thrombolysis. In a report from the WAKE–UP study on systematic image interpretation, the physicians went through systematic image training before rating the images. The results showed a high level of consistency amongst the raters in the assessment of the different imaging criteria, e.g., the presence of an acute ischemic lesion and the extent of infarction [47]. The DWI/FLAIR mismatch assessment was described as the most difficult imaging criteria to rate. The interrater agreement showed good consistency, but it was lower than for the other imaging criteria [47]. Several other studies argue that there is a noticeable interrater variability in the DWI/FLAIR mismatch assessment [6,7,8]. A study from 2018 found an insufficient interrater agreement on mismatch assessment and expressed uncertainty about this method to be the center of decisions in patients with unknown TSS [48]. This emphasizes that the use of an automated assessment could assist the interpretation and potentially improve the current standard.
Adding stroke treatment and patient-specific data, such as the location of lesions, symptoms, and comorbidities, will improve the interpretation of the relationship between imaging features and functional outcomes. A reliable algorithm to serve as assistance to determine patient eligibility for rtPA treatment could prove to provide a more standardized evaluation, be less time-consuming, and overall improve the treatment of patients admitted to the hospital under suspicion of stroke.
However, the creation of an AI method that will lower the DTN time, be accurate, and also bring predictive knowledge of the influenced tissues still comprise a challenge [49].
In all of the reviewed studies and in the daily clinical practice, some patients are misclassified in regard to the cut-off time of 4.5 h. In Polson et al., the DL model identified 29/37 (78%), and the radiologists identified 23/37 (62%) of the evaluation set patients with known onset time within 4.5 h [24]. The AI model used imaging features not only from the lesion area but also from the surrounding brain regions. The results could underline the relevance of the tissue status not only in the stroke lesion but also in the surrounding regions.
To be able to improve the analysis of the tissue status and stroke progression and identify those cases currently misclassified, further research is required. The development of AI methods and awareness in daily clinical practice could be amplified through new research in the field with a status of the currently existing AI methods. R. Karthik and R. Menaka et al. propose the development of an end-to-end automatic framework to identify both stroke lesions and prediction of the outcome of the influenced tissues to increase awareness of existing computer-aided detection frameworks [49].
This review elucidates the existing AI methods on DWI/FLAIR mismatch assessment, creates awareness, and focuses on the next potential steps towards an improvement in the current standard analysis and treatment selection for rtPA.
5. Conclusions
In a binary classification of ±4.5 h, the machine learning and deep learning algorithms in this review performed equivalent to or even better than domain experts, but surveyed AI methods are most likely not able to determine TSS only using DWI and FLAIR sequences. Furthermore, the relation between time since stroke onset and increasing visibility of FLAIR hyperintensity lesions is not recommended for the determination of TSS within the first 4.5 h. An AI DWI/FLAIR mismatch assessment focused on treatment eligibility and outcome prediction, adding patient-specific data, could pave the road for improving stroke treatments.
Author Contributions
Conceptualization, C.M.O., J.S., K.S., J.F.C., A.R.L., T.C.T., A.P. and M.B.N.; methodology, C.M.O., J.S., K.S. and J.F.C.; investigation, C.M.O., J.S., J.F.C. and K.S.; Formal analysis, C.M.O., J.S. and K.S.; writing—original draft preparation, C.M.O. and J.S.; writing—review and editing, C.M.O., K.S., J.S., J.F.C., T.C.T. and M.B.N.; supervision, J.F.C., T.C.T., A.R.L., A.P. and M.B.N.; project administration, C.M.O.; funding acquisition, M.B.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
- Search strings for each database
- PubMed MEDLINE
(Stroke[Mesh] OR (Stroke[tw] OR “wake-up stroke”[tw] OR “ischemic stroke”[tw] OR “cerebral infarc*”[tw] OR “brain infarc*”[tw])) AND (“Artificial Intelligence”[Mesh] OR (“Artificial Intelligence”[tw] OR ai[tw] OR “deep learning”[tw] OR “machine learning”[tw])) AND (mri[tw] OR “Magnetic Resonance Imaging”[tw] OR dwi[tw] OR flair*[tw] OR “flair-dwi mismatch”[tw] OR “Magnetic Resonance Imaging”[Mesh])
- OVID Embase
(exp Stroke/ OR (Stroke. mp. OR “wake-up stroke”.mp. OR “ischemic stroke”.mp. OR “cerebral infarc*”.mp. OR “brain infarc*”.mp.)) AND (exp “Artificial Intelligence”/OR (“Artificial Intelligence”.mp. OR ai.mp. OR “deep learning”.mp. OR “machine learning”.mp.)) AND (mri.mp. OR “Magnetic Resonance Imaging”.mp. OR dwi.mp. OR flair*.mp. OR “flair-dwi mismatch”.mp. OR exp “Magnetic Resonance Imaging”/)
- Elsevier Scopus
(INDEXTERMS(Stroke) OR (TITLE-ABS-KEY(Stroke) OR TITLE-ABS-KEY(“wake-up stroke”) OR TITLE-ABS-KEY(“ischemic stroke”) OR TITLE-ABS-KEY(“cerebral infarc*”) OR TITLE-ABS-KEY(“brain infarc*”))) AND (INDEXTERMS(“Artificial Intelligence”) OR (TITLE-ABS-KEY(“Artificial Intelligence”) OR TITLE-ABS-KEY(ai) OR TITLE-ABS-KEY(“deep learning”) OR TITLE-ABS-KEY(“machine learning”))) AND (TITLE-ABS-KEY(mri) OR TITLE-ABS-KEY(“Magnetic Resonance Imaging”) OR TITLE-ABS-KEY(dwi) OR TITLE-ABS-KEY(flair*) OR TITLE-ABS-KEY(“flair-dwi mismatch”) OR INDEXTERMS(“Magnetic Resonance Imaging”))
- Cochrane
(([mh Stroke] OR (Stroke:ti,ab,kw OR “wake-up stroke”:ti,ab,kw OR “ischemic stroke”:ti,ab,kw OR (“cerebral” NEXT infarc*):ti,ab,kw OR (“brain” NEXT infarc*):ti,ab,kw)) AND ([mh “Artificial Intelligence”] OR (“Artificial Intelligence”:ti,ab,kw OR ai:ti,ab,kw OR “deep learning”:ti,ab,kw OR “machine learning”:ti,ab,kw)) AND (mri:ti,ab,kw OR “Magnetic Resonance Imaging”:ti,ab,kw OR dwi:ti,ab,kw OR flair*:ti,ab,kw OR “flair-dwi mismatch”:ti,ab,kw OR [mh “Magnetic Resonance Imaging”])):ti,ab,kw
- IEEE Xplore
((“All Metadata”: Stroke OR “Wake-up stroke” OR “ischemic stroke” OR “brain infrarc*” OR “cerebral infarc*”) AND (“All Metadata”: “Artificial intelligence” OR ai OR “deep learning” OR “machine learning”) AND (“All Metadata”: “magnetic resonance imaging” OR mri OR flair OR dwi OR “flair-dwi mismatch”))
- Web of Science
(Stroke OR (Stroke OR “wake-up stroke” OR “ischemic stroke” OR “cerebral infarc*” OR “brain infarc*”)) AND (“Artificial Intelligence” OR (“Artificial Intelligence” OR ai OR “deep learning” OR “machine learning”)) AND (mri OR “Magnetic Resonance Imaging” OR dwi OR flair* OR “flair-dwi mismatch” OR “Magnetic Resonance Imaging”)
References
- Powers, W.J.; Rabinstein, A.A.; Ackerson, T.; Adeoye, O.M.; Bambakidis, N.C.; Becker, K.; Biller, J.; Brown, M.; Demaerschalk, B.M.; Hoh, B.; et al. Guidelines for the Early Management of Patients With Acute Ischemic Stroke: 2019 Update to the 2018 Guidelines for the Early Management of Acute Ischemic Stroke: A Guideline for Healthcare Professionals From the American Heart Association/American Stroke Association. Stroke 2019, 50, e344–e418, Erratum in Stroke 2019, 50, e440–e441. [Google Scholar] [CrossRef] [PubMed]
- Radue, E.W.; Weigel, M.; Wiest, R.; Urbach, H. Introduction to Magnetic Resonance Imaging for Neurologists. Contin. Lifelong Learn. Neurol. 2016, 22, 1379–1398. [Google Scholar] [CrossRef] [PubMed]
- Buck, B.H.; Akhtar, N.; Alrohimi, A.; Khan, K.; Shuaib, A. Stroke mimics: Incidence, aetiology, clinical features and treatment. Ann. Med. 2021, 53, 420–436. [Google Scholar] [CrossRef] [PubMed]
- Aoki, J.; Kimura, K.; Iguchi, Y.; Shibazaki, K.; Sakai, K.; Iwanaga, T. FLAIR can estimate the onset time in acute ischemic stroke patients. J. Neurol. Sci. 2010, 293, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Ebinger, M.; Galinovic, I.; Rozanski, M.; Brunecker, P.; Endres, M.; Fiebach, J.B. Fluid-attenuated inversion recovery evolution within 12 hours from stroke onset: A reliable tissue clock? Stroke 2010, 41, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Thomalla, G.; Rossbach, P.; Rosenkranz, M.; Siemonsen, S.; Krützelmann, A.; Fiehler, J.; Gerloff, C. Negative fluid-attenuated inversion recovery imaging identifies acute ischemic stroke at 3 hours or less. Ann. Neurol. 2009, 65, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Emeriau, S.; Serre, I.; Toubas, O.; Pombourcq, F.; Oppenheim, C.; Pierot, L. Can diffusion-weighted imaging-fluid-attenuated inversion recovery mismatch (positive diffusion-weighted imaging/negative fluid-attenuated inversion recovery) at 3 Tesla identify patients with stroke at <4.5 hours? Stroke 2013, 44, 1647–1651. [Google Scholar] [CrossRef] [PubMed]
- Thomalla, G.; Cheng, B.; Ebinger, M.; Hao, Q.; Tourdias, T.; Wu, O.; Kim, J.S.; Breuer, L.; Singer, O.C.; Warach, S.; et al. DWI-FLAIR mismatch for the identification of patients with acute ischaemic stroke within 4·5 h of symptom onset (PRE-FLAIR): A multicentre observational study. Lancet Neurol. 2011, 10, 978–986. [Google Scholar] [CrossRef] [PubMed]
- Thomalla, G.; Simonsen, C.Z.; Boutitie, F.; Andersen, G.; Berthezene, Y.; Cheng, B.; Cheripelli, B.; Cho, T.-H.; Fazekas, F.; Fiehler, J.; et al. MRI-Guided Thrombolysis for Stroke with Unknown Time of Onset. N. Engl. J. Med. 2018, 379, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Fink, J.N.; Kumar, S.; Horkan, C.; Linfante, I.; Selim, M.H.; Caplan, L.R.; Schlaug, G. The stroke patient who woke up: Clinical and radiological features, including diffusion and perfusion MRI. Stroke 2002, 33, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Rimmele, D.L.; Thomalla, G. Wake-up stroke: Clinical characteristics, imaging findings, and treatment option—An update. Front. Neurol. 2014, 5, 35. [Google Scholar] [CrossRef]
- Kelly, B.S.; Judge, C.; Bollard, S.M.; Clifford, S.M.; Healy, G.M.; Aziz, A.; Mathur, P.; Islam, S.; Yeom, K.W.; Lawlor, A.; et al. Radiology artificial intelligence: A systematic review and evaluation of methods (RAISE). Eur. Radiol. 2022, 32, 7998–8007. [Google Scholar] [CrossRef]
- Choy, G.; Khalilzadeh, O.; Michalski, M.; Synho, D.; Samir, A.E.; Pianykh, O.S.; Geis, J.R.; Pandharipande, P.V.; Brink, J.A.; Dreyer, K.J. Current Applications and Future Impact of Machine Learning in Radiology. Radiology 2018, 288, 318–328. [Google Scholar] [CrossRef]
- Letourneau-Guillon, L.; Camirand, D.; Guilbert, F.; Forghani, R. Artificial Intelligence Applications for Workflow, Process Optimization and Predictive Analytics. Neuroimaging Clin. N. Am. 2020, 30, e1–e15. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.M.; Khan, H.; Shafi, U.; Islam, S.U.; Raza, M.; Son, T.T.; Le-Minh, H. 2D-CNN based Segmentation of Ischemic Stroke Lesions in MRI Scans. In Advances in Computational Collective Intelligence; Springer International Publishing: Cham, Switzerland, 2020. [Google Scholar] [CrossRef]
- Liu, L.; Kurgan, L.; Wu, F.X.; Wang, J. Attention convolutional neural network for accurate segmentation and quantification of lesions in ischemic stroke disease. Med. Image Anal. 2020, 65, 101791. [Google Scholar] [CrossRef] [PubMed]
- McInnes, M.D.F.; Moher, D.; Thombs, B.D.; McGrath, T.A.; Bossuyt, P.M.; Clifford, T.; Cohen, J.F.; Deeks, J.J.; Gatsonis, C.; Hooft, L.; et al. Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies: The PRISMA-DTA Statement. JAMA 2018, 319, 388–396, Erratum in JAMA 2019, 322, 2026. [Google Scholar] [CrossRef] [PubMed]
- Systematic Review Accelerator|Institute for Evidence-Based Healthcare|Bond University. Available online: https://sr-accelerator.com/#/polyglot (accessed on 15 November 2022).
- QUADAS-2|Bristol Medical School: Population Health Sciences|University of Bristol. Available online: https://www.bristol.ac.uk/population-health-sciences/projects/quadas/quadas-2/ (accessed on 20 February 2023).
- Moon, H.S.; Heffron, L.; Mahzarnia, A.; Obeng-Gyasi, B.; Holbrook, M.; Badea, C.T.; Feng, W.; Badea, A. Automated multimodal segmentation of acute ischemic stroke lesions on clinical MR images. Magn. Reson. Imaging 2022, 92, 45–57. [Google Scholar] [CrossRef]
- Rajinikanth, V.; Aslam, S.M.; Kadry, S. Deep Learning Framework to Detect Ischemic Stroke Lesion in Brain MRI Slices of Flair/DW/T1 Modalities. Symmetry 2021, 13, 2080. [Google Scholar] [CrossRef]
- Zhu, H.; Jiang, L.; Zhang, H.; Luo, L.; Chen, Y.; Chen, Y. An automatic machine learning approach for ischemic stroke onset time identification based on DWI and FLAIR imaging. NeuroImage Clin. 2021, 31, 102744. [Google Scholar] [CrossRef]
- Zhang, H.; Polson, J.S.; Nael, K.; Salamon, N.; Yoo, B.; El-Saden, S.; Scalzo, F.; Speier, W.; Arnold, C.W. Intra-domain task-adaptive transfer learning to determine acute ischemic stroke onset time. Comput. Med. Imaging Graph. 2021, 90, 101926. [Google Scholar] [CrossRef] [PubMed]
- Polson, J.S.; Zhang, H.; Nael, K.; Salamon, N.; Yoo, B.Y.; El-Saden, S.; Starkman, S.; Kim, N.; Kang, D.; Speier, W.F.; et al. Identifying acute ischemic stroke patients within the thrombolytic treatment window using deep learning. J. Neuroimaging 2022, 32, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, E.J.; Ham, S.; Lee, H.B.; Lee, J.S.; Kwon, S.U.; Kim, J.S.; Kim, N.; Kang, D.W. Machine Learning Approach to Identify Stroke Within 4.5 Hours. Stroke 2020, 51, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, S.; Ai, Z.; Shen, T.; Zhang, H.; Duan, S.; Chen, Y.C.; Yin, X.; Sun, J. Development and external validation of a stability machine learning model to identify wake-up stroke onset time from MRI. Eur. Radiol. 2022, 32, 3661–3669. [Google Scholar] [CrossRef] [PubMed]
- Subudhi, A.; Dash, P.; Mohapatra, M.; Tan, R.-S.; Acharya, U.R.; Sabut, S. Application of Machine Learning Techniques for Characterization of Ischemic Stroke with MRI Images: A Review. Diagnostics 2022, 12, 2535. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Bentley, P.; Rueckert, D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. Neuroimage Clin. 2017, 15, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Winzeck, S.; Mocking, S.; Bezerra, R.; Bouts, M.; McIntosh, E.; Diwan, I.; Garg, P.; Chutinet, A.; Kimberly, W.; Copen, W.; et al. Ensemble of Convolutional Neural Networks Improves Automated Segmentation of Acute Ischemic Lesions Using Multiparametric Diffusion-Weighted MRI. Am. J. Neuroradiol. 2019, 40, 938–945. [Google Scholar] [CrossRef]
- Subbanna, N.K.; Rajashekar, D.; Cheng, B.; Thomalla, G.; Fiehler, J.; Arbel, T.; Forkert, N.D. Stroke Lesion Segmentation in FLAIR MRI Datasets Using Customized Markov Random Fields. Front. Neurol. 2019, 10, 541. [Google Scholar] [CrossRef]
- Duong, M.T.; Rudie, J.D.; Wang, J.; Xie, L.; Mohan, S.; Gee, J.C.; Rauschecker, A.M. Convolutional Neural Network for Automated FLAIR Lesion Segmentation on Clinical Brain MR Imaging. Am. J. Neuroradiol. 2019, 40, 1282–1290. [Google Scholar] [CrossRef]
- Kim, Y.; Luby, M.; Burkett, N.S.; Norato, G.; Leigh, R.; Wright, C.B.; Kern, K.C.; Hsia, A.W.; Lynch, J.K.; Adil, M.M.; et al. Fluid-Attenuated Inversion Recovery Hyperintense Ischemic Stroke Predicts Less Favorable 90-Day Outcome after Intravenous Thrombolysis. Cerebrovasc. Dis. 2021, 50, 738–745. [Google Scholar] [CrossRef]
- Müller, S.; Kufner, A.; Dell’Orco, A.; Rackoll, T.; Mekle, R.; Piper, S.K.; Fiebach, J.B.; Villringer, K.; Flöel, A.; Endres, M.; et al. Evolution of Blood-Brain Barrier Permeability in Subacute Ischemic Stroke and Associations With Serum Biomarkers and Functional Outcome. Front. Neurol. 2021, 12, 730923. [Google Scholar] [CrossRef]
- Wang, P.; Fan, E.; Wang, P. Comparative analysis of image classification algorithms based on traditional machine learning and deep learning. Pattern Recognit. Lett. 2021, 141, 61–67. [Google Scholar] [CrossRef]
- Ho, K.C.; Speier, W.; Zhang, H.; Scalzo, F.; El-Saden, S.; Arnold, C.W. A Machine Learning Approach for Classifying Ischemic Stroke Onset Time from Imaging. IEEE Trans. Med. Imaging 2019, 38, 1666–1676. [Google Scholar] [CrossRef]
- Tanguay, W.; Acar, P.; Fine, B.; Abdolell, M.; Gong, B.; Cadrin-Chênevert, A.; Chartrand-Lefebvre, C.; Chalaoui, J.; Gorgos, A.; Chin, A.S.-L.; et al. Assessment of Radiology Artificial Intelligence Software: A Validation and Evaluation Framework. Can. Assoc. Radiol. J. 2023, 74, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Total Density Per Million Population: Magnetic Resonance Imaging. Available online: https://www.who.int/data/gho/data/indicators/indicator-details/GHO/total-density-per-million-population-magnetic-resonance-imaging (accessed on 5 February 2023).
- Smith, A.G.; Rowland Hill, C. Imaging assessment of acute ischaemic stroke: A review of radiological methods. Br. J. Radiol. 2018, 91, 20170573. [Google Scholar] [CrossRef]
- Kuhrij, L.S.; Marang-van de Mheen, P.J.; van den Berg-Vos, R.M.; de Leeuw, F.E.; Nederkoorn, P.J.; Dutch Acute Stroke Audit consortium. Determinants of extended door-to-needle time in acute ischemic stroke and its influence on in-hospital mortality: Results of a nationwide Dutch clinical audit. BMC Neurol. 2019, 19, 265. [Google Scholar] [CrossRef]
- Katsuki, M.; Narita, N.; Ishida, N.; Watanabe, O.; Cai, S.; Ozaki, D.; Sato, Y.; Kato, Y.; Jia, W.; Nishizawa, T.; et al. Preliminary development of a prediction model for daily stroke occurrences based on meteorological and calendar information using deep learning framework (Prediction One; Sony Network Communications Inc., Japan). Surg. Neurol. Int. 2021, 12, 31. [Google Scholar] [CrossRef]
- Benzakoun, J.; Deslys, M.-A.; Legrand, L.; Hmeydia, G.; Turc, G.; Ben Hassen, W.; Charron, S.; Debacker, C.; Naggara, O.; Baron, J.-C.; et al. Synthetic FLAIR as a Substitute for FLAIR Sequence in Acute Ischemic Stroke. Radiology 2022, 303, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wang, Z.; Sun, Y.; Bo, W.; Duan, K.; Song, C.; Hu, Y.; Zhou, J.; Mu, Z.; Wu, N. Prognosis of ischemic stroke predicted by machine learning based on multi-modal MRI radiomics. Front. Psychiatry 2023, 13, 1105496. [Google Scholar] [CrossRef]
- Elsaid, A.F.; Fahmi, R.M.; Shehta, N.; Ramadan, B.M. Machine learning approach for hemorrhagic transformation prediction: Capturing predictors’ interaction. Front. Neurol. 2022, 13, 951401. [Google Scholar] [CrossRef]
- Jiang, L.; Miao, Z.; Chen, H.; Geng, W.; Yong, W.; Chen, Y.-C.; Zhang, H.; Duan, S.; Yin, X.; Zhang, Z. Radiomics Analysis of Diffusion-Weighted Imaging and Long-Term Unfavorable Outcomes Risk for Acute Stroke. Stroke 2023, 54, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lyu, J.; Meng, Z.; Wu, X.; Chen, W.; Wang, G.; Niu, Q.; Li, X.; Bian, Y.; Han, D.; et al. Small vessel disease burden predicts functional outcomes in patients with acute ischemic stroke using machine learning. CNS Neurosci. Ther. 2023, 29, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.K.; Albers, G.W.; Davis, S.M.; Donnan, G.A.; Furlan, A.J.; Hacke, W.; Lees, K.R. Mismatch-based delayed thrombolysis: A meta-analysis. Stroke 2010, 41, e25–e33. [Google Scholar] [CrossRef] [PubMed]
- Galinovic, I.; Dicken, V.; Heitz, J.; Klein, J.; Puig, J.; Guibernau, J.; Kemmling, A.; Gellissen, S.; Villringer, K.; Neeb, L.; et al. Homogeneous application of imaging criteria in a multicenter trial supported by investigator training: A report from the WAKE-UP study. Eur. J. Radiol. 2018, 104, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Fahed, R.; Lecler, A.; Sabben, C.; Khoury, N.; Ducroux, C.; Chalumeau, V.; Botta, D.; Kalsoum, E.; Boisseau, W.; Duron, L.; et al. DWI-ASPECTS (Diffusion-Weighted Imaging-Alberta Stroke Program Early Computed Tomography Scores) and DWI-FLAIR (Diffusion-Weighted Imaging-Fluid Attenuated Inversion Recovery) Mismatch in Thrombectomy Candidates: An Intrarater and Interrater Agreement Study. Stroke 2018, 49, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Karthik, R.; Menaka, R. Computer-aided detection and characterization of stroke lesion—A short review on the current state-of-the art methods. Imaging Sci. J. 2017, 66, 1–22. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).


