Abstract
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), causing a disease called COVID-19, is a class of acute respiratory syndrome that has considerably affected the global economy and healthcare system. This virus is diagnosed using a traditional technique known as the Reverse Transcription Polymerase Chain Reaction (RT-PCR) test. However, RT-PCR customarily outputs a lot of false-negative and incorrect results. Current works indicate that COVID-19 can also be diagnosed using imaging resolutions, including CT scans, X-rays, and blood tests. Nevertheless, X-rays and CT scans cannot always be used for patient screening because of high costs, radiation doses, and an insufficient number of devices. Therefore, there is a requirement for a less expensive and faster diagnostic model to recognize the positive and negative cases of COVID-19. Blood tests are easily performed and cost less than RT-PCR and imaging tests. Since biochemical parameters in routine blood tests vary during the COVID-19 infection, they may supply physicians with exact information about the diagnosis of COVID-19. This study reviewed some newly emerging artificial intelligence (AI)-based methods to diagnose COVID-19 using routine blood tests. We gathered information about research resources and inspected 92 articles that were carefully chosen from a variety of publishers, such as IEEE, Springer, Elsevier, and MDPI. Then, these 92 studies are classified into two tables which contain articles that use machine Learning and deep Learning models to diagnose COVID-19 while using routine blood test datasets. In these studies, for diagnosing COVID-19, Random Forest and logistic regression are the most widely used machine learning methods and the most widely used performance metrics are accuracy, sensitivity, specificity, and AUC. Finally, we conclude by discussing and analyzing these studies which use machine learning and deep learning models and routine blood test datasets for COVID-19 detection. This survey can be the starting point for a novice-/beginner-level researcher to perform on COVID-19 classification.
1. Introduction
SARS-CoV-2 was first recognized in China, after which the severe pneumonia yielded by the virus, called COVID-19, rapidly circulated worldwide [1,2]. COVID-19 has different clinical symptoms, such as dyspnea, fever, cough, myalgia, fatigue, gastrointestinal complications, and headache [3,4]. This virus is risky and affects the mortality of individuals with compromised immune systems. Medical professionals and infectious disease experts from the entire globe are seeking a solution for the disease. COVID-19 has been the primary source of death in many countries around the world, with the United States, Italy, and Spain having one of the highest number of deaths [5]. Figure 1 demonstrates the global 14-day COVID-19 case notification rate per 100,000 as of 15 July 2020.
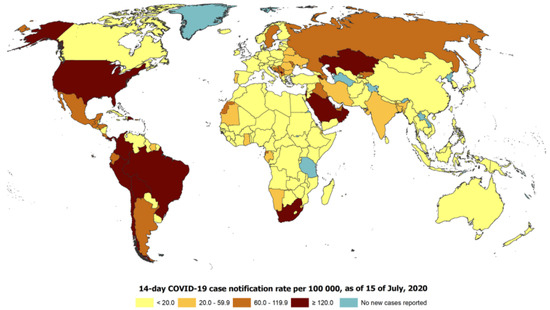
Figure 1.
The global 14-day COVID-19 case notification rate per 100,000 as of 15 July 2020.
To date, a couple of diagnostic techniques have been adopted by physicians, such as RT-PCR [6,7], imaging resolutions and blood checks [8]. RT-PCR, which is the best measure for the analysis of COVID-19 [9,10], tolerates a low sensitiveness (60–71%), more extended waiting duration for the results [11], and poses extra responsibilities to the healthcare system, demanding pricey devices [12,13,14]. Additionally, there is a shortage of testing kits, reagents, and trained personnel for analysis, particularly in less-developed nations [12]. Thus, scientists have searched for extra reachable techniques of diagnosis, among which imaging techniques have obtained remarkable attraction [15]. Chest CT and chest X-rays are mainstream imaging alternatives for diagnosing COVID-19 infection. Chest CT presents a better sensitivity [16], similar to RT-PCR [12]; however, it also has numerous downsides, such as hospital-acquired infection, radiation safety, and lower access rates to CT devices [17,18,19]. Chest X-ray is a less costly imaging choice and uses a lower radiation dose than CT, while virtually every health center and most clinics will have access to X-ray equipment [20,21,22]. Chest-X-ray, similar to CT, affords medical physicians with imaging symptoms of SARS-CoV-2 contamination, e.g., ground-glass opacity, but suffers from an increased rate of false-negative results [23,24,25]. Blood tests are broadly accessible and have much lower costs than RT-PCR and imaging tests. Since biochemical parameters contained in ordinary blood exams, for example, lactate dehydrogenase (LDH), C-reactive protein (CRP), etc. [26], change over the course of the COVID-19 contamination, blood tests can provide physicians with data about the diagnosis of COVID-19 [12]. Consequently, blood tests may additionally supply a potentially precious instrument for the quick screening of infected patients and compensate for the deficiency of RT-PCR and CT scans by providing an initial step of detection [27].
The health industry is impatiently following new techniques and technologies to address the growth of the COVID-19 epidemic in the international health crisis. AI is known to be one of the grandest uses of global technology that can follow the speed and detect the growth rate of COVID-19 and determine the risk and severity of COVID-19 patients. AI can also predict cases of death by adequately analyzing previous patient data. Moreover, AI can help us battle this virus by testing people and providing medical assistance, data and information, and recommendations regarding disease control [28].
Machine learning (ML) [29,30,31], as a class of AI, includes the algorithmic modeling structures of statistical models and only requires a small amount of knowledge to learn how to handle problems [32,33,34]. On the other hand, deep learning (DL) [35] is a class of ML that concentrates on making deep structural neural-network-based models that learn from data utilizing feed-forward and back-propagation. DL has improved significantly in the last two decades in several activities [36,37,38]. However, it needs a vast amount of data to learn. Exceptional cases of DL, where large-scale datasets are not required to train, have been generative models and transfer learning [39,40,41].
This survey introduces a series of main works of AI containing ML and DL research articles on COVID-19 diagnosis using routine blood tests. In total, we investigated 92, of which 82 articles are ML-based, and the rest are based on DL models. Because of the rapid growth of COVID-19 cases, we have quoted many published articles before conducting a thorough investigation, so these articles should be analyzed for their accuracy and quality in peer review.These articles are classified in two tables, in which, by comparing different works, it is determined which AI algorithms or performance metrics are used more to detect COVID-19, as well as which blood test datasets are used.
There are four main sections in this study. Section 2 discusses the procedure followed for selecting the research articles. A summary of contemporary machine learning and deep learning research is given in Section 3. Section 4 presents the blood features used in previous articles. The approaches are examined, and the outcomes of various models are discussed in Section 5. In Section 6, the article is concluded.
2. Protocol for Choosing COVID-19-Related Research Articles
To choose the research articles, the most pertinent keywords, such as COVID-19, routine blood tests, machine learning, and deep learning, were used. Moreover, we employed digital databases, including IEEE Xplore, Elsevier, Springer, and MDPI, to collect only English-language literature. Note that research articles about blood tests and rapid antigen tests were not reviewed. Figure 2 shows the details of the statistics of ML and DL publications on COVID-19. The search strategy was adjusted to obtain the maximum number of documents. Table 1 lists the number of documents retrieved by the queries referenced. The results were thoroughly scrutinized to seek relevant results. The queries yielded 4785 documents.
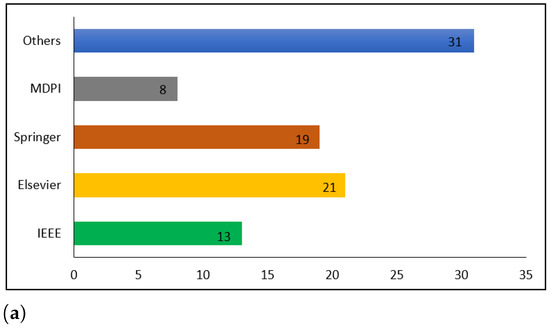
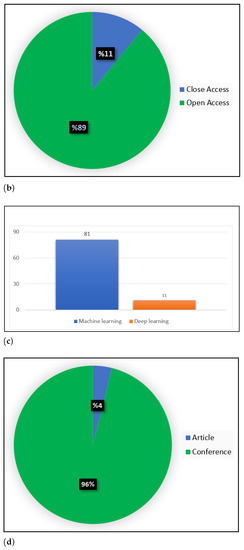
Figure 2.
Statistics on ML and DL publications in COVID-19. (a) Publication per database, (b) COVID-19 publication, (c) Distribution of the published COVID-19 research articles, (d) COVID-19 publication.

Table 1.
Research articles identified in the databases under investigation.
The IEEE database was subject to a review of 620 studies that were evaluated according to their titles and abstracts. This screening led to the exclusion of 459 documents, as their material was not related to the analysis, and the remaining 161 were chosen for a full-text review. After a full-text review of the studies, 148 were rejected, and the remaining 13 were included in the present review. In total, the titles and abstracts of 1123 studies from the Springer database were scanned through. After the screening process, 992 documents were deemed unsuitable and only 131 were qualified for a full-text review. In total, 112 of the studies were disregarded and 19 were ultimately included in this study. The titles and abstracts of 1201 studies in the Elsevier database were examined. After the screening was finished, 1102 documents were disregarded because they had no association with the investigation, leaving 99 documents to be analyzed for a full-text review. From these studies, 21 were accepted and 78 rejected. A review of the MDPI database was conducted, resulting in 337 studies, with the evaluation based on their titles and abstracts. The screening process resulted in the elimination of 301 documents, as they were not relevant to the analysis, while 36 were selected for a full-text review. A complete textual analysis of the studies led to the rejection of 28, while the remaining 8 were included in the current review. Finally, the titles and abstracts of 1504 studies in other databases were evaluated. Following the screening, 178 documents were kept for a full-text review, since the other 1326 had no association to the topic being studied. Out of these studies, 147 were rejected and 31 were used for the final analysis.
This section delineates the content of the 92 studies found in the databases investigated. The results of the research article search are summarized in Figure 3.
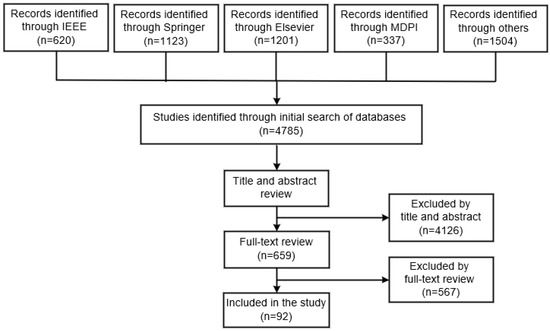
Figure 3.
Summary of the research article selection process.
3. Overview of Machine Learning and Deep Learning Methods
Routine blood tests can be used to quickly diagnose COVID-19 infection utilizing AI-based techniques such as ML and DL. These models can uncover potential connections between different qualities in blood test results and provide information to guide decisions.
This section contains research articles that use routine blood tests to diagnose COVID-19 while taking into account ML and DL models. The majority of COVID-19 detection techniques used are shown in Figure 4.
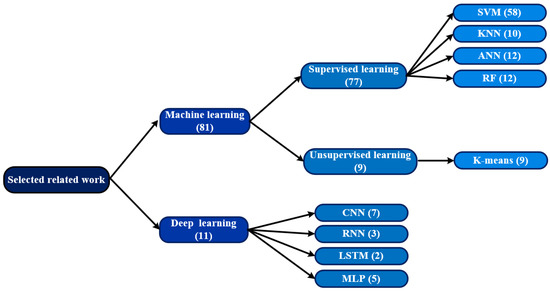
Figure 4.
The most frequently used techniques for COVID-19 detection among the studies included in this survey. The numbers in parentheses indicate the number of research articles.
3.1. Machine Learning
More focus has been placed on the potential use of ML methods to address COVID-19 diagnosis using routine blood testing. The following are some well-known ML algorithms that have been applied for COVID-19 diagnosis [42].
Support vector machine (SVM) [43] is used for classification. The goal of SVM is to identify the best hyperplane for differentiating the features. There are numerous ways to draw the hyperplane and an ideal one has been discovered that best separates the dataset. SVM is a highly accurate model, and it is very unlikely that overfitting will occur.
Random Forest (RF) [44] is another ensemble technique using decision tree that utilizes a re-sampling process called the bagging method that creates multiple trees and handles weak classifiers in a different way. RF is effective for highly complex problems and can handle missing data and unbalanced datasets.
K-nearest-neighbor (KNN) [45] is a classification algorithm with which a sample’s label can be predicted using the labels of its closest neighbors. It is necessary to choose the parameter K and use the attribute-distance computation metric to determine which other data points are the closest neighbors.
Logistic regression (LR) [46] is a classification algorithm. Based on the value of independent features, LR models the probability of samples belonging to a particular class. Then, the model can be used for predicting the probability that a given sample belongs to a certain class. LR has simple calculations and can support continuous numerical values, while non-linear data cannot be handled by LR.
Decision tree (DT) [47] is a Supervised ML algorithm. To characterize the connections between attributes and a class label, DT generates a tree-structured model. It divides observations recursively based on the property with the highest gain ratio value that is the most informative. The data are divided in the nodes and decisions are made in the leaves.
Naive Bayes (NB) [48] is a simple probabilistic classifier based on Bayes’ theorem. It cannot handle missing data.
Extreme Gradient Boosting (XGBoost) [49] is a machine learning algorithm based on decision trees, which employ a gradient boosting structure. XGBoost is a library of machine learning which uses a scalable and distributed form of Gradient-Boosted Decision Tree (GBDT). There are many advantages to using this machine learning library, such as parallel tree-boosting, and it is the leading machine learning library for regression, classification, and ranking.
LGBM (LightGBM) [50] is an efficient, distributed, and high-performing gradient boosting framework based on decision tree algorithms, used for ranking, classifying, and other machine learning processes. LGBM is an application of the gradient boosting framework that is based on tree-structured algorithms.
Table 2 displays the most recent machine learning models for early detection of COVID-19 or assessing the disease severity level of COVID-19 patients based on laboratory and clinical data.
3.2. Deep Learning
The performance enhancements of hardware components, such as graphics cards, and the drop in unit costs are two reasons DL has grown in popularity. DL has also been aided by machine learning and information processing studies [51,52,53], as well as an increase in training data. Numerous domains, including computer vision [36,54,55,56], natural language processing [57,58,59,60,61,62], and speech recognition [63], have extensively used various deep learning architectures, such as ANN, CNN, and RNN. ANN is a method of processing information that draws inspiration from the organic nervous system of humans. This structure is made up of neurons, activation processes, and input, output, and hidden layers. In an ANN, each layer comprises a hierarchy of neurons. The input for the following layer is the layer’s output before it. From the incoming data, each layer learns increasingly intricate relationships. A deep learning system called CNN was created to analyze visual data, such as photographs and movies. Different layer types perform various functions on CNN. The names of these components are the convolution layer, pooling layer, fully connected layer, and activation function layer. In RNN structures, the outcome is influenced by both the other and current inputs. These networks produce their results by combining data from the past and present.
In this study, we selected 11 deep-learning-based studies, shown in Table 3. Overall, the number of works presented based on deep models is small; however, their accuracy in datasets with a large number of data is superior to ML methods.

Table 3.
List of DL models that diagnose COVID-19.

Table 2.
List of ML models that diagnose COVID-19.
Table 2.
List of ML models that diagnose COVID-19.
| Ref. | Dataset Source | Dataset Size (COVID-19) | Total Features (Selected) | Model Used | Metric Results |
|---|---|---|---|---|---|
| [64] | Lanzhou Pulmonary Hospital | 253 (105) | 49 (11) | RF | Acc: 95.95%, Sen: 95.12%, Spe: 96.97% |
| [65] | Hospital Sirio Libanes | 1945 (545) | 40 (40) | RF, DT, XGBoost, GBM, LGBM, SVM, ANN, KNN | Acc: 90% |
| [66] | Hospital Israelita Albert Einstein, Brazil | 5644 (1128) | 50 (17) | SVM, LR, RF, DT, KNN | Acc: 89.78% |
| [9] | San Raphael Hospital in Italy | 1624 (844) | 72 (34) | NB, LR, RF, KNN, SVM | Acc: 74%, Sen: 70%, Spe: 79%, AUC: 74% |
| [67] | Premier Healthcare Database | 2183 (1020) | 29 (15) | XGBoost | Sen: 95.9%, AUC: 91% |
| [16] | Taizhou hospital in China | 114 (32) | 14 (14) | LR, NB, IBk, DT | Acc: 84.21% |
| [68] | - | 55,676 (1564) | 12 (10) | GBM, MLP, RF, DT, KNN, LR, SVM, XGBoost | Sen: 93%, AUC: 91% |
| [69] | Rennes academic hospital | 536 (106) | - (-) | RF, LR, ANN | AUC: 93% |
| [70] | San Raffaele Hospital in Italy | 279 (177) | 15 (15) | CNN and 15 supervised ML algorithms | Acc: 99.28%, AUC: 98.80% |
| [71] | - | 300 (137) | 8 (8) | SVM, LR, XGBoost | Acc: 87% |
| [72] | Hospital Israelita Albert
Einstein, Brazil | 5644 (1128) | 13 (12) | RF, LR, XGBoost, SVM, KNN | Acc: 91%, Sen: 94%, Spe: 71%, AUC: 91% |
| [73] | University Medical Center, Ljubljana, Slovenia | 5333 (160) | 117 (35) | RF, ANN, XGBoost, SVM | Sen: 81.90%, Spe: 97.90%, AUC: 97% |
| [74] | Routinely collected laboratory, clinical, and demographic data | 1040 (-) | 5 (-) | ANN, extra trees, RF, XGBoost, catboost | Sen:92%, Spe: 82%, AUC: 92% |
| [75] | Hospital Israelita Albert Einstein, Brazil | 608 (84) | 108 (16) | RF, DT | Acc: 88%, Sen: 66%, Spe: 91%, AUC: 86% |
| [76] | Hospital Israelita Albert Einstein, Brazil | 5644 (559) | 108 (24) | NB, MLP, DT, BN, SVM | Acc: 95.15%, Sen: 96.80%, Spe: 93.60% |
| [77] | Three datasets of routine laboratory blood tests | 2503 (1043) | Many (-) | SHAP, ELI5, LIME | Sen: 92%, Spe: 82%, AUC: 92% |
| [78] | Hospitals in Zhejiang, China | 912 (361) | 31 (10) | SVM, LR, RF, DT | Acc: 91%, Sen: 87%, Spe: 95%, AUC: 95% |
| [79] | - | 398 (-) | 42 (19) | XGBoost, SOM | Sen: 92.50%, Spe: 97.90% |
| [80] | West China Hospital, China | 620 (211) | 19 (19) | Multivariate LR | AUC: 87.2% |
| [81] | 11 regions in China | 659 (-) | Many (-) | DT | Acc: 89%, AUC: 88% |
| [82] | SMART hospitals | - (-) | - (-) | SVM, RF, NB | Acc: 93.33% |
| [83] | Hospital Israelita Albert Einstein, Brazil | 5644 (2210) | Many (-) | LDA | Acc: 99.60%, Sen: 98.72% Spe: 98.99%, AUC: 99.38% |
| [84] | Hospital Israelita Albert Einstein, Brazil | 598 (81) | 108 (14) | RF, LR, ANN | Acc: 81%-87%, Sen: 43%-65%, Spe: 81%-91% |
| [85] | Hospital Israelita Albert Einstein, Brazil | 5644 (558) | Many (-) | ANN, RF, Shallow learning | AUC: 95% |
| [86] | Hospital Israelita Albert Einstein, Brazil | 5644 (65) | Many (-) | Er-CoV | Sen: 70%, Spe: 85%, AUC: 86% |
| [87] | Kepler University Hospital | 1357 (653) | 28 (-) | RF | Acc: 86%, AUC: 74% |
| [88] | Three Brazilian hospitals | 1521 (-) | 130 (-) | Adaboost, SVM, XGBoost, RF | Sen 96%, Spe: 93% |
| [89] | Three Brazilian hospitals | 1521 (-) | 130 (-) | HUST-19 | Acc 94% |
| [90] | Oxford University hospitals | 1157 (349) | - (-) | Different ML classifiers | Sen: 77%, Spe: 95% AUC: 93% |
| [91] | Five hospitals in New York | 4098 (-) | Many (-) | XGBoost | AUC: 89% |
| [92] | Hospital Israelita Albert Einstein, Brazil | 5644 (559) | 108 (18) | RF, KNN, SVM, ET | Acc: 99.88%, Sen: 98.72%, Spe: 99.99%, AUC: 99.38% |
| [20] | San Raffaele Hospital in Italy | 279 (177) | -(15) | KNN, ET, LR, DT, NB, RF, SVM | Acc: 82–86%, Sen: 92–95% |
| [93] | The RT-PCR COVID-19 test on the basis of routinely acquired blood tests | 127,115 (1573) | Many (100) | SNN, KNN, LR, SVM, RF, XGBoost | AUC: 95% |
| [94] | New York Presbyterian Hospital | 3346 (1394) | 685 (33) | DT, LR, RF, GBDT, DT | Sen: 75.80%, Spe: 80.20%, AUC: 85.30% |
| [95] | Stanford Health Care, CA, USA | 390 (31) | -(4) | LR | Sen: 86%-96%, Spe: 35%-55% |
| [96] | Hospital Israelita Albert Einstein, Brazil | 5644 (558) | 106 (9) | LR, ANN, SVM, RF, GB | Sen: 95% Spe: 95%, AUC: 95% |
| [97] | Hospital Israelita Albert Einstein, Brazil | 253 (102) | 108 (15) | ANN, RF, GBT, Albert LR, SVM | Sen: 68%, Spe: 85% |
| [98] | Hospital Israelita Albert Einstein, Brazil | 599 (81) | 108 (16) | SVM | Sen: 70.25%, Spe: 85.98% |
| [99] | A case series from Wenzhou, Zhejiang, China | 53 (-) | 23 (-) | LR, KNN, RF, DT, SVM | AUC: 80% |
| [100] | West London Hospital | 398 (-) | Many (-) | CRM, ANN | AUC: 88.1% |
| [101] | Hospital Sirio Libanes | 1945 (545) | 40 (40) | DT, RF, GBM, XGBoost, SVM, LGBM, KNN, ANN | Acc: 90% |
| [102] | Tongji Hospital Affiliated to Huazhong University of Science and Technology | 137 (-) | 100 (28) | SVM, LR | Acc: 99% |
| [103] | 3000 examples collected at a hospital in Poland | 3114 (1941) | - (-) | LR, XGBoost | Sen: 97%, Spe: 11% |
| [104] | - | 3819 (-) | Many (-) | LR, SKLearn, RF, LGBM | - |
| [105] | Hospital Israelita Albert Einstein, Brazil | 5644 (558) | 18 (18) | XGBoost, LR | |
| [106] | UCLA Health System in Los Angeles, California | 1455 (182) | Many (27) | RF, LR, SVM, ANN, SGD, XGBoost, ADABoost | Sen: 93%, Spe: 64% |
| [107] | Three hospitals in the United States, Iran, and Italy | 295 (117) | Many (1691) | RF | Sen: 96.1%, AUC: 88.4% |
| [108] | NYU Langone Health (NYULH) | 206,67 (12,47) | Many (-) | LR, XGBoost, MLP, RNN, GRU, LSTM | Sen: 85%, Spe: 86%, AUC: 92% |
| [109] | Hospitals in the Mount Sinai Health System in New York City | 4098 (4098) | Many | XGBoost | AUC: 89% |
| [110] | A large acute care healthcare system the Mount Sinai Health System | 567 (567) | 1360 338 | RF | AUC: 85.5% |
| [111] | HM hospitals’ network in Madrid (Spain) | 2307 (1696) | 29 (25) | LR, RF | Sen: 81.69%, Spe: 81.46%, AUC: 89% |
| [112] | Patients from eight centers in China, Italy, and Belgium | 725 (-) | 29 (13) | LR | Acc: 87.5%, Sen: 96.9%, Spe: 88%, AUC: 93% |
| [113] | Hospitals of Tongji Medical Colleg, Huazhong University of Science and Technology | 2520 (-) | 53 (34) | LR, SVM, GBDT, ANN | AUC: 97.6% |
| [114] | Korea Centers for Disease Control and Prevention | 3524 (3524) | 44 (44) | LR, SVM, KNN, RF, GBDT | AUC: 83% |
| [115] | Electronic Health Records | 86,355 (4759) | Many (-) | LR, XGBoost | AUC: 83.8% |
| [116] | The ED of an urban multicenter health system | 300 (300) | 50 (50) | RF | - |
| [117] | UK Biobank (UKBB data) | 465,728 (7846) | 97 (15) | XGboost | AUC: 81% |
| [118] | Hospital Israelita Albert Einstein, Brazil | 5644 (279) | 106 (97) | RF, LR, XGBoost | Sen: 80%, Spe: 98% |
| [119] | Hospital Israelita Albert Einstein, Brazil | 510 (73) | 108 (15) | NB | - |
| [120] | - | 689 (362) | 43 (-) | KNN, RF, SVM | ACC: 97.7% |
| [121] | Painel COVID-19 Estado Do ESPÍRITO Santo | 8443 (4826) | Many (-) | LR, LDA, NB, KNN, DT, XGboost, SVM | Sen: 88%, Spe: 82%, AUC: 92% |
| [122] | Tongji Hospital Affiliated to Huazhong University of Science and Technology | 362 (362) | - (-) | RF | Acc: 95% |
| [123] | Hospital Israelita Albert Einstein, Brazil | 5644 (674 ) | Many (-) | - | AUC: 94% |
| [124] | Mass General Brigham (MGB) Healthcare | 10,826 (3713) | Many (-) | ANN, SVM | - |
| [125] | San Raffaele Hospital (Milan, Italy) | 207 (105) | Many (-) | LR | - |
| [126] | 24 hospitals in Hong Kong | 5148 (-) | Many (-) | - | Sen: 77.8%, Spe:98.3%, AUC: 96.8% |
| [127] | Tumor Center of Union Hospital affiliated with Tongji Medical College, China | 99 (11) | 33 (33) | - | AUC: 95.3% |
| [128] | Tongji Hospital of Wuhan, China | 110 (59) | 47 (7) | LR | Sen: 98%, Spe: 91% |
| [129] | World Health Organization | 601 (-) | Many (-) | XGBoost | Acc: 100%, Sen: 84.6%, Spe: 84.6%, AUC: 95.3% |
| [130] | Hospitals in Wuhan, China | 294 (208) | 15 (-) | RF, SVM | Acc: 84%, Sen: 88%, Spe: 80% |
| [131] | First Medical Center, Beijing, China | 132 (26) | 46 (18) | LR, DT, Adaboost | Sen: 100%, Spe: 77.80% |
| [132] | Hospital Israelita Albert Einstein, Brazil | 5644 (520) | 17 (17) | DES | Acc: 99.81%, AUC: 99.81% |
| [133] | National Center for Biotechnology Information Gene Expression Omnibus and European Bioinformatics Institute ArrayExpress | 705 (100) | - (-) | LR | AUC: 90% |
| [134] | Ethics Committee of the Affiliated Yueqing Hospital of Wenzhou Medical University | 51 (51) | 22 (22) | KELM | Acc: 100%, Sen: 100%, Spe: 100% |
| [135] | Seven different hospital clinical biomarker datasets from Italy, Brazil, and Ethiopia. | 1624 (844) | 21 (21) | LDA, XGBoost, RF, LR, KNN | Acc: 91.45%, Sen: 91.44% , Spe: 91.44% |
| [136] | Keio University Hospital | 312 (300) | 35 (35) | LR | - |
| [137] | Wuhan, China | 485 (-) | - (-) | XGBoost | Acc: 97% |
| [138] | San Raffaele Hospital Milan Italia and Hospital Israelita Albert Einstein | 5644 (560) | 111 (111) | SVM | Acc: 99.29%, Sen: 92.79%, Spe: 100% |
| [139] | Tongji Hospital of Wuhan, China | 375 (201) | 300 (3) | XGBoost | Sen: 83% |
| [140] | Veterans Health Administration Sites, USA | 5002 (1079) | 68 (54) | RF | Acc: 83.30%, Sen: 83.40% Spe: 89.80% |
| [141] | Oxford University Hospitals, UK | 40,732 (437) | 74 (-) | RF, LR, XGBoost | Acc: 92.30%, Sen: 77.40%, Spe: 95.70% |
4. Features
The importance of features in routine blood tests for COVID-19 diagnosis is significant, because in machine learning and deep learning, these features can be used to build predictive models that can identify patterns in data, allowing for earlier detection and diagnosis of COVID-19. Additionally, routine blood tests can be used to monitor disease progression and treatment response in patients with COVID-19. Changes in these features over time can indicate the severity of the disease and the effectiveness of treatment. Moreover, the identification of the most informative features from routine blood tests can help develop models that can predict disease outcomes and mortality rates. This information can be used to allocate resources and prioritize treatment for patients most at risk of severe illness.
The consequences of a wrong selection of features in medical diagnosis using routine blood tests, especially for COVID-19, can have severe consequences for patient health and treatment outcomes. Importantly, machine learning and deep learning algorithms rely heavily on the selection of appropriate features to make accurate predictions. If the wrong features are chosen, the accuracy and reliability of the diagnostic results can be significantly impacted. In the case of COVID-19 diagnosis, a wrong selection of features could result in misdiagnosis or delayed diagnosis, leading to delayed treatment and potentially worse outcomes for patients. For example, if important features, such as inflammatory markers or lymphocyte counts, are excluded from the model, patients with mild or asymptomatic cases of COVID-19 may be missed, leading to the spread of the disease. Moreover, a wrong selection of features can lead to false positives or false negatives, which can have significant implications for patient care. False positives can lead to unnecessary medical interventions or treatments, which can be costly, time-consuming, and may cause patient harm. On the other hand, false negatives can result in delayed treatment, which can lead to the progression of the disease and worse outcomes for the patient. Additionally, a wrong selection of features can result in a lack of generalizability and poor performance of the model. The model may not perform well on new data or may be specific to a particular population or dataset, limiting its usefulness and applicability in real-world settings.
The blood features that were used by previous studies (see Table 4) are as follows [152]:

Table 4.
List of blood features selected by previous research articles.
- Hematocrit: The computation of the ratio of erythrocytes (commonly referred to as red blood cells) in the blood is carried out. When this percentage is low, it could signify respiratory difficulties and possibly reveal the severity of COVID-19 cases [153].
- Hemoglobin: A particular material found in red blood cells carries oxygen in the bloodstream. When someone is diagnosed with pneumonia due to COVID-19, a drop in the level of this material (known as Hb) shortly after the diagnosis could suggest that the pneumonia is getting worse. It is worth noting that anemia is a frequent occurrence in COVID-19 cases as well [154].
- Red blood cell distribution width (RDW): Another term used to describe this is the RDW coefficient of variation. It offers a way to quantify the variability in the dimensions of erythrocytes; while initially utilized as a diagnostic tool for anemia, it has since evolved into an indicator of infections and more severe ailments, including cardiovascular and cancer. Although it is not a reliable indicator of the presence of COVID-19, it has been recognized as a sign of the disease’s severity, as elevated RDW levels have been associated with mortality in cases of COVID-19 [155].
- Mean corpuscular hemoglobin (MCH): It pertains to the mean amount of hemoglobin, a protein that carries oxygen throughout the body, present in every individual red blood cell [75]. Sarkar et al. [156] suggested that changes in MCH levels could indicate the presence of COVID-19. A decrease in the MCH value typically signifies a lack of iron in the body, known as iron deficiency anemia. In general, people with COVID-19 tend to display MCH values that are slightly below the normal range, falling within one standard deviation.
- Mean corpuscular hemoglobin concentration (MCHC): It is a measurement that determines the average hemoglobin concentration inside an individual red blood cell, similar to MCH [75]. A low MCHC value suggests that a person’s red blood cells have insufficient hemoglobin, indicating anemia. According to [157], this metric aided in the differentiation of cases of COVID-19 from pneumonia contracted within the general community. MCV, which stands for Mean Corpuscular Volume, is an indicator of the typical size or volume of erythrocytes [75]. Changes in the mean size of red blood cells, whether an increase or a decrease, can signal underlying health concerns, and research has associated such alterations with the severity of COVID-19 [158].
- Lymphocytes absolute: A reduced level can be an indication of serious COVID-19, which can prompt early treatment or suggest a negative outcome [154]. This metric serves as a marker for infectious processes.
- Leukocytes: The immune system’s defensive cells are called white blood cells. Research has demonstrated that COVID-19 has the ability to attack these cells, causing them to discharge pro-inflammatory cytokines that result in an increase in inflammation within the affected individual [159]. Furthermore, it is possible to utilize indicators present in the genetic composition of leukocytes to detect the existence of COVID-19 [160].
- Basophils absolute: They are crucial cells of the immune system, and their levels tend to rise during prolonged inflammation or allergic reactions. However, research has shown that individuals infected with COVID-19, particularly those with severe cases, experience a significant decrease in basophil counts [161]. Similarly, eosinophils, which play a role in defending the body against parasites and infections, also exhibit reduced levels in COVID-19 patients [162].
- Platelets: The bone marrow produces these cells that aid in the process of blood coagulation. Keeping a close eye on this measurement is crucial because a rise in its levels may not always be indicative of COVID-19, but can instead point to complications related to the disease, including thrombosis [163].
- Monocytes absolute: The protection against different microorganisms and viruses is provided by monocytes and macrophages, which are essential constituents of the immune system [164]. Macrophages exist in bodily tissues, whereas monocytes can be found in the bloodstream and are identifiable through blood counts. Despite their beneficial characteristics, these cells can have harmful effects on those with COVID-19, leading to lung infections and lesions. Several studies have revealed a reduction in the number of monocytes in people with COVID-19 [164]. According to other research, such as Meidaninikjeh et al. [165], it is proposed to create novel methods to detect the migration of these cells towards the lungs as a potential sign of COVID-19, and to utilize suitable treatments to reduce lung harm.
- SARS_CoV2_PCR: It is dependable in verifying the existence of a COVID-19 infection because it detects the virus’s genetic material. The variable under consideration will be allocated a value of 0, which indicates negative instances, and 1, which signifies positive instances.
5. Discussion and Analysis
It is challenging to diagnose coronavirus using routine blood testing. Researchers have employed numerous preprocessing strategies, feature extraction approaches, and classification models [166]. Identifying a single strategy or set of methodologies that produce the best outcomes for detecting COVID-19 from regular blood tests is challenging. Most research articles showed accuracy rates of more than 90%, which may be extremely high. Nevertheless, the goal would be to raise the accuracy to about 100%, as inaccurate disease classification, even in a small number of cases, is wholly unacceptable. On the other hand, generalization capacity poses a serious issue for all learning-based methodologies. It results from both the procedures themselves and the diversity of the training dataset. As a result, deep learning has had a significant impact on how routine blood tests are applied, and we anticipate that it will become a more effective methodology in the future [167].
Different techniques of ML and DL have been used in 92 reviewed studies. The ML or DL methods utilized are displayed in Figure 4. Figure 5 shows that with a percentage of 16%, Random Forest is the most used machine learning method, followed by LR (14%), SVM and XGBOOST (11%), KNN and ANN (7%), DT (6%), etc.
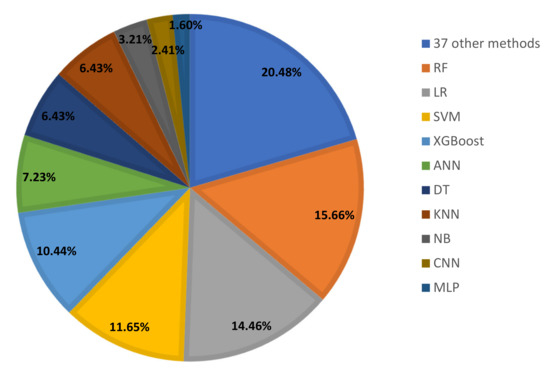
Figure 5.
ML and DL techniques used for COVID-19 diagnosis.
Four metrics, including accuracy, sensitivity (recall), specificity, and AUC, were used to diagnose COVID-19 and to evaluate and compare the performance of the suggested methods quantitatively. These four performance metrics used in the literature of COVID-19 diagnosis are shown in Table 5. Accuracy indicates that how many samples are classified properly (ratio of true predictions over all predictions ). Sensitivity or recall refers to the rate of the number of correctly classified COVID-19-positive samples to the total number of suspected samples. Specificity refers to the rate of identifying negative samples correctly. The area under the ROC curve is represented by AUC, from (0, 0) to (1, 1).

Table 5.
Performance metrics used in the research articles.
A lot of AI knowledge is needed to implement a standard ML-based diagnosis. The most crucial step is identifying the distinctive features that can help with COVID-19 diagnosis. Because of the absence of adequate hardware resources and data availability, the development of DL systems for the diagnosis of COVID-19 has been a difficult endeavor. Although websites such as Google Colab already provide researchers with powerful computing processors, applying and employing these approaches in the real world presents many challenges. While DL and ML models hold great potential for improving COVID-19 diagnosis using routine blood tests, there are still several limitations that must be addressed before they can be used widely in clinical settings: (1) DL and ML models rely on large amounts of diverse data to train and learn from. However, data on COVID-19 are still relatively limited, especially in terms of routine blood tests. This can make it difficult to develop accurate models that apply to different populations and settings. (2) Routine blood tests can be affected by many factors, such as age, sex, underlying medical conditions, and medications. ML models may struggle to account for these factors and may generate inaccurate predictions as a result. (3) ML models are only as good as the data they are trained on and must be carefully validated and tested before they are used in clinical settings. This requires extensive testing and validation of the model on large and diverse datasets. (4) DL models are often described as “black boxes”, because it is difficult to interpret how they arrive at their predictions. This can make it difficult for healthcare providers to understand and trust the predictions generated by these models. (5) ML models used in healthcare are subject to regulatory approval, and there are strict requirements for validation and testing. This can slow down the adoption of these models in clinical settings.
Although we conducted a search in four of the most major databases and found over 4785 documents, there are some drawbacks to this research. The first one is the possibility of selection bias because only English-language research articles were chosen. The keywords used for the queries also influence the results that are obtained. Another limitation of this research is the time frame in which the search was conducted. As with any rapidly evolving field, new research is constantly being published, and the results of this study may not reflect the most recent findings. Additionally, the scope of the search may have been too narrow, focusing only on research articles related to a specific aspect of the topic. Moreover, the quality of the research articles selected in the review may vary, as not all research articles undergo the same level of peer review or scrutiny. It is possible that some of the research articles chosen may have had limitations in their methodology or analysis, which could impact the validity of the conclusions drawn from the review.
We want to take note that the primary benefit of the current study is that it gives the reader a comprehensive list of recent research articles that use various forecasting approaches based on routine blood tests. The reader gets access to a method-based categorization of publications (ML and DL). For anyone interested in this subject, the research articles given in this study would be a good place to start and would hasten their learning in this area of study. For everyone involved in a literature review, the flowchart shown in Figure 3 would be useful. There are two primary drawbacks to this study. We have only considered studies published within the last one to two years. This survey has not covered the material from other captivating databases. This is because of the overwhelming volume of publications that authors would have had to manage.
6. Conclusions
Millions of people’s lives have been gravely threatened by the ongoing COVID-19 pandemic in a short amount of time. As the CT scan technique is more expensive and time-consuming than routine blood tests, it is apparent that routine blood tests are more broadly accessible than the CT image dataset. As a result, the majority of researchers used standard blood testing to identify COVID-19. After reviewing the literature in this field, we discover that there is a dearth of annotated data on those impacted by COVID-19. The performance of the aforementioned data-hungry models can be significantly improved by enhancing high-quality datasets of COVID-19 patients. ML and DL can detect the coronavirus using AI techniques when applied to routine blood testing. This study compares some recent research utilizing ML and DL algorithms to detect coronaviruses from routine blood tests obtained from multiple open-source datasets. This study only covers the 92 studies examined, while numerous studies have recently been undertaken based on these findings.
After reviewing 4785 research articles, only 92 were deemed pertinent to the topic under investigation for this study. This can provide the reader with a sense of how uncommon this topic is in the studied field. The application of ML and DL to the prediction of COVID-19 using routine blood tests remains unexplored. It should be noted that 559 full-text research articles were examined for this revision work. In order to appeal to readers, the authors suggest that any upcoming research should take into consideration other databases in the literature review, such as Emerald, Scopus, and Web of Science. In addition, the authors suggest researchers consider using open-source datasets to train and test ML and DL models for predicting COVID-19 using routine blood tests. Open-source datasets can provide a standardized and accessible platform for researchers to develop and test their models. This can also facilitate collaboration and sharing of data and results among researchers in the field. The authors also recommend that future research should explore the potential of using ML and DL in combination with other medical technologies, such as imaging and genomics, to develop more accurate and comprehensive diagnostic tools for COVID-19. By leveraging multiple sources of data, researchers can develop more holistic approaches to predicting and treating the disease.
Author Contributions
Conceptualization, S.A.H.; methodology, S.A.H. and M.K.; software, S.A.H.; validation, R.A.; formal analysis, M.K.; investigation, R.A.; resources, S.A.H.; data curation, R.A.; writing—original draft preparation, S.A.H.; writing—review and editing, S.A.H., M.K., and R.A.; visualization, S.A.H. and R.A.; supervision, M.K.; project administration, M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data will be made available on request.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this study:
| ML | machine learning |
| DL | deep learning |
| Acc | accuracy |
| Sen | sensitivity |
| Spe | specificity |
| AUC | area under the curve |
| RF | random forest |
| DT | decision tree |
| XGBoost | gradient boosting |
| GBM | gradient boosting machine |
| LGBM | light gradient boosting machine |
| SVM | support vector machine |
| ANN | artificial neural network |
| KNN | k-nearest neighbor |
| LR | logistic regression |
| NB | naive Bayes |
| IBk | lazy-classifier |
| MLP | multilayer perceptron |
| BN | Bayesian network |
| SOM | self-organizing-map |
| LDA | linear discriminant analysis |
| ET | extra trees |
| GBDT | gradient boosted decision trees |
| GB | gradient boosting |
| GBT | gradient boosting trees |
| CRM | Cox regression model |
| SGD | stochastic gradient descent |
| RNN | recurrent neural network |
| GRU | gated recurrent units |
| LSTM | long short-term memory |
| DES | dynamic ensemble selection |
| KELM | kernel extreme learning machine |
| VAE | variational autoencoder |
| CNN | convolutional neural network |
| IEEE | institute of electrical and electronics engineers |
| MDPI | multidisciplinary digital publishing institute |
References
- Xie, X.; Xie, B.; Xiong, D.; Hou, M.; Zuo, J.; Wei, G.; Chevallier, J. New theoretical ISM-K2 Bayesian network model for evaluating vaccination effectiveness. J. Ambient. Intell. Humaniz. Comput. 2022, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Chen, D.; Lv, H. Smart city construction and management by digital twins and BIM big data in COVID-19 scenario. ACM Trans. Multimed. Comput. Commun. Furth. Appl. (TOMM) 2022, 18, 1–21. [Google Scholar] [CrossRef]
- Afaghi, S.; Tarki, F.E.; Rahimi, F.S.; Besharat, S.; Mirhaidari, S.; Karimi, A.; Alamdari, N.M. Prevalence and clinical outcomes of vitamin D deficiency in COVID-19 hospitalized patients: A retrospective single-center analysis. Tohoku J. Exp. Med. 2021, 255, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zheng, S.; Li, L.; Zhang, X.; Zhang, X.; Huang, Z.; Chen, J.; Wang, R.; Zhao, H.; Chong, Y.; et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans. Comput. Biol. Furth. Bioinform. 2021, 18, 2775–2780. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Qiu, L.; Xia, W.; Liu, C.F.; Xi, X.; Zhao, S.; Yu, J.; Wei, S.; Hu, X.; Su, N.; et al. Spatiotemporal evolution of online attention to vaccines since 2011: An empirical study in China. Front. Public Health 2022, 2310. [Google Scholar] [CrossRef]
- Tahamtan, A.; Ardebili, A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev. Mol. Diagn. 2020, 20, 453–454. [Google Scholar] [CrossRef]
- Arevalo-Rodriguez, I.; Buitrago-Garcia, D.; Simancas-Racines, D.; Zambrano-Achig, P.; Del Campo, R.; Ciapponi, A.; Sued, O.; Martinez-Garcia, L.; Rutjes, A.W.; Low, N.; et al. False-negative results of initial RT-PCR assays for COVID-19: A systematic review. PLoS ONE 2020, 15, e0242958. [Google Scholar] [CrossRef]
- Luan, D.; Liu, A.; Wang, X.; Xie, Y.; Wu, Z. Robust two-stage location allocation for emergency temporary blood supply in postdisaster. Discret. Dyn. Nat. Soc. 2022, 2022, 1–20. [Google Scholar] [CrossRef]
- Cabitza, F.; Campagner, A.; Ferrari, D.; Di Resta, C.; Ceriotti, D.; Sabetta, E.; Colombini, A.; De Vecchi, E.; Banfi, G.; Locatelli, M.; et al. Development, evaluation, and validation of machine learning models for COVID-19 detection based on routine blood tests. Clin. Chem. Lab. Med. (CCLM) 2021, 59, 421–431. [Google Scholar] [CrossRef]
- Karimi, A.; Hashemian, S.M.R. Cytokine storm in COVID-19 and the treatment simulacrum. Biomed. Biotechnol. Res. J. (BBRJ) 2020, 4, 41. [Google Scholar]
- Alsharif, W.; Qurashi, A. Effectiveness of COVID-19 diagnosis and management tools: A review. Radiography 2021, 27, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Kalane, P.; Patil, S.; Patil, B.; Sharma, D.P. Automatic detection of COVID-19 disease using U-Net architecture based fully convolutional network. Biomed. Signal Process. Control 2021, 67, 102518. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lin, L.; Zhang, L.; Xu, R.; Chen, X.; Ji, J.; Li, Y. Long noncoding RNA p21 enhances autophagy to alleviate endothelial progenitor cells damage and promote endothelial repair in hypertension through SESN2/AMPK/TSC2 pathway. Pharmacol. Res. 2021, 173, 105920. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Qu, Y.Y.; Liu, L.; Qiao, Y.N.; Geng, H.R.; Lin, Y.; Xu, W.; Cao, J.; Zhao, J.Y. Homocysteine inhibits pro-insulin receptor cleavage and causes insulin resistance via protein cysteine-homocysteinylation. Cell Rep. 2021, 37, 109821. [Google Scholar] [CrossRef] [PubMed]
- Sarbazi, F.; Akbari, E.; Karimi, A.; Nouri, B.; Ardebili, S.N. The Clinical Outcome of Laparoscopic Surgery for Endometriosis on Pain, Ovarian Reserve, and Cancer Antigen 125 (CA-125): A Cohort Study. Int. J. Fertil. Steril. 2021, 15, 275. [Google Scholar] [CrossRef]
- Arpaci, I.; Huang, S.; Al-Emran, M.; Al-Kabi, M.N.; Peng, M. Predicting the COVID-19 infection with fourteen clinical features using machine learning classification algorithms. Multimed. Tools Appl. 2021, 80, 11943–11957. [Google Scholar] [CrossRef]
- Azar, A.S.; Ghafari, A.; Najar, M.O.; Rikan, S.B.; Ghafari, R.; Khamene, M.F.; Sheikhzadeh, P. Covidense: Providing a suitable solution for diagnosing COVID-19 lung infection based on Deep Learning from chest X-ray images of patients. Front. Biomed. Technol. 2021, 8, 131–142. [Google Scholar] [CrossRef]
- Wang, Y.; Zhai, W.; Cheng, S.; Li, J.; Zhang, H. Surface-functionalized design of blood-contacting biomaterials for preventing coagulation and promoting hemostasis. Friction 2023, 1–24. [Google Scholar] [CrossRef]
- Zheng, J.; Yue, R.; Yang, R.; Wu, Q.; Wu, Y.; Huang, M.; Chen, X.; Lin, W.; Huang, J.; Chen, X.; et al. Visualization of Zika virus infection via a light-initiated bio-orthogonal cycloaddition labeling strategy. Front. Bioeng. Biotechnol. 2022, 10. [Google Scholar] [CrossRef]
- Brinati, D.; Campagner, A.; Ferrari, D.; Locatelli, M.; Banfi, G.; Cabitza, F. Detection of COVID-19 infection from routine blood exams with machine learning: A feasibility study. J. Med. Syst. 2020, 44, 1–12. [Google Scholar] [CrossRef]
- Wang, Y.; Zhai, W.; Zhang, H.; Cheng, S.; Li, J. Injectable Polyzwitterionic Lubricant for Complete Prevention of Cardiac Adhesion. Macromol. Biosci. 2023, 2200554. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, L.; Ni, S.; Li, D.; Liu, J.; Chu, H.Y.; Zhang, N.; Sun, M.; Li, N.; Ren, Q.; et al. Targeting loop3 of sclerostin preserves its cardiovascular protective action and promotes bone formation. Nat. Commun. 2022, 13, 4241. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Chen, D.; Feng, H.; Zhu, H.; Lv, H. Digital twins in unmanned aerial vehicles for rapid medical resource delivery in epidemics. IEEE Trans. Intell. Transp. Syst. 2021, 23, 25106–25114. [Google Scholar] [CrossRef] [PubMed]
- Golpayegani, G.; Jafari, M.; Karimi, A. Arhinia and Bilateral Anophthalmia: Report of a Rare Case and Review of Literature. Acta Medica Iran. 2021, 59, 621. [Google Scholar] [CrossRef]
- Wang, L.; Yu, Y.; Ni, S.; Li, D.; Liu, J.; Xie, D.; Chu, H.Y.; Ren, Q.; Zhong, C.; Zhang, N.; et al. Therapeutic aptamer targeting sclerostin loop3 for promoting bone formation without increasing cardiovascular risk in osteogenesis imperfecta mice. Theranostics 2022, 12, 5645. [Google Scholar] [CrossRef]
- Mobarakeh, Z.T.; Hasanzadeh, E.; Farzin, A.; Goodarzi, A.; Farahani, M.S.; Shirian, S.; Mahmoodi, N.; Zamani, N.; Karimi, A.; Ai, J. Enhanced sciatic nerve regeneration with fibrin scaffold containing human endometrial stem cells and insulin encapsulated chitosan particles: An in vivo study. Injury 2023, in press. [Google Scholar] [CrossRef]
- Tahermanesh, K.; Hanjani, S.; Saadat Mostafavi, S.R.; Shahriyari, R.; Fazel Anvari-Yazdi, A.; Karimi-Zarchi, M.; Samimi, M.; Karimi, A.; Allahqoli, L.; Alkatout, I. Hourglass cesarean scar: A neglected external niche in association with the internal niche. Int. J. Gynecol. Obstet. 2022, 157, 478–480. [Google Scholar] [CrossRef]
- Sartakhti, M.S.; Kahaki, M.J.M.; Moravvej, S.V.; javadi Joortani, M.; Bagheri, A. Persian language model based on BiLSTM model on COVID-19 corpus. In Proceedings of the 2021 5th International Conference on Pattern Recognition and Image Analysis (IPRIA), Kashan, Iran, 28–29 April 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Wang, F.; Wang, H.; Zhou, X.; Fu, R. A Driving Fatigue Feature Detection Method Based on Multifractal Theory. IEEE Sensors J. 2022, 22, 19046–19059. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, R.; Heidari, A.A.; Wang, X.; Chen, Y.; Wang, M.; Chen, H. Towards augmented kernel extreme learning models for bankruptcy prediction: Algorithmic behavior and comprehensive analysis. Neurocomputing 2021, 430, 185–212. [Google Scholar] [CrossRef]
- Yuan, Q.; Kato, B.; Fan, K.; Wang, Y. Phased array guided wave propagation in curved plates. Mech. Syst. Signal Process. 2023, 185, 109821. [Google Scholar] [CrossRef]
- Vakilian, S.; Moravvej, S.V.; Fanian, A. Using the cuckoo algorithm to optimizing the response time and energy consumption cost of fog nodes by considering collaboration in the fog layer. In Proceedings of the 2021 5th International Conference on Internet of Things and Applications (IoT), Isfahan, Iran, 19–20 May 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1–5. [Google Scholar]
- Liu, Y.; Tian, J.; Hu, R.; Yang, B.; Liu, S.; Yin, L.; Zheng, W. Improved feature point pair purification algorithm based on SIFT during endoscope image stitching. Front. Neurorobotics 2022, 16. [Google Scholar] [CrossRef] [PubMed]
- Vakilian, S.; Moravvej, S.V.; Fanian, A. Using the artificial bee colony (ABC) algorithm in collaboration with the fog nodes in the Internet of Things three-layer architecture. In Proceedings of the 2021 29th Iranian Conference on Electrical Engineering (ICEE), Tehran, Iran, 18–20 May 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 509–513. [Google Scholar] [CrossRef]
- Ban, Y.; Liu, M.; Wu, P.; Yang, B.; Liu, S.; Yin, L.; Zheng, W. Depth estimation method for monocular camera defocus images in microscopic scenes. Electronics 2022, 11, 2012. [Google Scholar] [CrossRef]
- Moravvej, S.V.; Alizadehsani, R.; Khanam, S.; Sobhaninia, Z.; Shoeibi, A.; Khozeimeh, F.; Sani, Z.A.; Tan, R.S.; Khosravi, A.; Nahavandi, S.; et al. RLMD-PA: A reinforcement learning-based myocarditis diagnosis combined with a population-based algorithm for pretraining weights. Contrast Media Mol. Imaging 2022, 2022. [Google Scholar] [CrossRef]
- Liu, S.; Yang, B.; Wang, Y.; Tian, J.; Yin, L.; Zheng, W. 2D/3D multimode medical image registration based on normalized cross-correlation. Appl. Sci. 2022, 12, 2828. [Google Scholar] [CrossRef]
- Cao, Z.; Wang, Y.; Zheng, W.; Yin, L.; Tang, Y.; Miao, W.; Liu, S.; Yang, B. The algorithm of stereo vision and shape from shading based on endoscope imaging. Biomed. Signal Process. Control. 2022, 76, 103658. [Google Scholar] [CrossRef]
- Alafif, T.; Tehame, A.M.; Bajaba, S.; Barnawi, A.; Zia, S. Machine and deep learning towards COVID-19 diagnosis and treatment: Survey, challenges, and future directions. Int. J. Environ. Res. Public Health 2021, 18, 1117. [Google Scholar] [CrossRef] [PubMed]
- Haghighi, R.D. Effect of ECAP and extrusion on particle distribution in Al-nano–Al2O3 composite. Bull. Mater. Sci. 2015, 38, 1205–1212. [Google Scholar] [CrossRef]
- Gao, H.; Hsu, P.H.; Li, K.; Zhang, J. The real effect of smoking bans: Evidence from corporate innovation. J. Financ. Quant. Anal. 2020, 55, 387–427. [Google Scholar] [CrossRef]
- Alballa, N.; Al-Turaiki, I. Machine learning approaches in COVID-19 diagnosis, mortality, and severity risk prediction: A review. Informatics Med. Unlocked 2021, 24, 100564. [Google Scholar] [CrossRef]
- Suthaharan, S. Machine learning models and algorithms for big data classification. Integr. Ser. Inf. Syst 2016, 36, 1–12. [Google Scholar] [CrossRef]
- Rigatti, S.J. Random forest. J. Insur. Med. 2017, 47, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.E. K-nearest neighbor. Scholarpedia 2009, 4, 1883. [Google Scholar] [CrossRef]
- Wright, R.E. Logistic regression. In Reading and Understanding Multivariate Statistics; American Psychological Association: Worcester, MA, USA, 1995. [Google Scholar]
- Song, Y.Y.; Ying, L. Decision tree methods: Applications for classification and prediction. Shanghai Arch. Psychiatry 2015, 27, 130. [Google Scholar]
- Webb, G.I.; Keogh, E.; Miikkulainen, R. Naïve Bayes. Encycl. Mach. Learn. 2010, 15, 713–714. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Aziz, R.M.; Baluch, M.F.; Patel, S.; Ganie, A.H. LGBM: A machine learning approach for Ethereum fraud detection. Int. J. Inf. Technol. 2022, 1–11. [Google Scholar] [CrossRef]
- Kan, J.; Zhou, X.; Sun, Y.; Sun, L.; Chu, H.; Qian, Z.; Zhou, J. Molecular engineering and biomedical applications of ultra-sensitive fluorescent probe for Ag+. Chin. Chem. Lett. 2021, 32, 3066–3070. [Google Scholar] [CrossRef]
- Li, R.; Zha, Z.; Miao, Z.; Xu, C.Y. Emerging 2D pnictogens for biomedical applications. Chin. Chem. Lett. 2022, 33, 2345–2353. [Google Scholar] [CrossRef]
- Li, C.; Zhang, Y.; Wan, Y.; Wang, J.; Lin, J.; Li, Z.; Huang, P. STING-activating drug delivery systems: Design strategies and biomedical applications. Chin. Chem. Lett. 2021, 32, 1615–1625. [Google Scholar] [CrossRef]
- Danaei, S.; Bostani, A.; Moravvej, S.V.; Mohammadi, F.; Alizadehsani, R.; Shoeibi, A.; Alinejad-Rokny, H.; Nahavandi, S. Myocarditis Diagnosis: A Method using Mutual Learning-Based ABC and Reinforcement Learning. In Proceedings of the 2022 IEEE 22nd International Symposium on Computational Intelligence and Informatics and 8th IEEE International Conference on Recent Achievements in Mechatronics, Automation, Computer Science and Robotics (CINTI-MACRo), Budapest, Hungary, 21–22 November 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 000265–000270. [Google Scholar] [CrossRef]
- Jin, K.; Yan, Y.; Wang, S.; Yang, C.; Chen, M.; Liu, X.; Terasaki, H.; Yeo, T.H.; Singh, N.G.; Wang, Y.; et al. iERM: An Interpretable Deep Learning System to Classify Epiretinal Membrane for Different Optical Coherence Tomography Devices: A Multi-Center Analysis. J. Clin. Med. 2023, 12, 400. [Google Scholar] [CrossRef]
- Xie, B.; Li, S.; Li, M.; Liu, C.H.; Huang, G.; Wang, G. Sepico: Semantic-guided pixel contrast for domain adaptive semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2023, in press. [Google Scholar] [CrossRef]
- Moravvej, S.V.; Mousavirad, S.J.; Oliva, D.; Schaefer, G.; Sobhaninia, Z. An Improved DE Algorithm to Optimise the Learning Process of a BERT-based Plagiarism Detection Model. In Proceedings of the 2022 IEEE Congress on Evolutionary Computation (CEC), Padua, Italy, 18–23 July 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–7. [Google Scholar] [CrossRef]
- Moravvej, S.; Maleki Kahaki, M.; Salimi Sartakhti, M.; Joodaki, M. Efficient GAN-based method for extractive summarization. J. Electr. Comput. Eng. Innov. 2022, 10, 287–298. [Google Scholar] [CrossRef]
- Moravvej, S.V.; Mirzaei, A.; Safayani, M. Biomedical text summarization using conditional generative adversarial network (CGAN). arXiv 2021, arXiv:2110.11870. [Google Scholar] [CrossRef]
- Moravvej, S.V.; Joodaki, M.; Kahaki, M.J.M.; Sartakhti, M.S. A method based on an attention mechanism to measure the similarity of two sentences. In Proceedings of the 2021 7th International Conference on Web Research (ICWR), Tehran, Iran, 19–20 May 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 238–242. [Google Scholar]
- Moravvej, S.V.; Kahaki, M.J.M.; Sartakhti, M.S.; Mirzaei, A. A method based on attention mechanism using bidirectional long-short term memory (BLSTM) for question answering. In Proceedings of the 2021 29th Iranian Conference on Electrical Engineering (ICEE), Tehran, Iran, 18–20 May 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 460–464. [Google Scholar] [CrossRef]
- Moravvej, S.V.; Mousavirad, S.J.; Moghadam, M.H.; Saadatmand, M. An lstm-based plagiarism detection via attention mechanism and a population-based approach for pre-training parameters with imbalanced classes. In Proceedings of the Neural Information Processing: 28th International Conference, ICONIP 2021, Sanur, Bali, Indonesia, 8–12 December 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 690–701. [Google Scholar] [CrossRef]
- Gaikwad, S.K.; Gawali, B.W.; Yannawar, P. A review on speech recognition technique. Int. J. Comput. Appl. 2010, 10, 16–24. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, P.; Zhang, L.; Meng, W.; Li, J.; Tong, C.; Li, Y.; Cai, J.; Yang, Z.; Zhu, J.; et al. Rapid and accurate identification of COVID-19 infection through machine learning based on clinical available blood test results. MedRxiv 2020, arXiv:02.20051136. [Google Scholar] [CrossRef]
- Aktar, S.; Ahamad, M.M.; Rashed-Al-Mahfuz, M.; Azad, A.; Uddin, S.; Kamal, A.; Alyami, S.A.; Lin, P.I.; Islam, S.M.S.; Quinn, J.M.; et al. Machine learning approach to predicting COVID-19 disease severity based on clinical blood test data: Statistical analysis and model development. JMIR Med. Inform. 2021, 9, e25884. [Google Scholar] [CrossRef]
- Tamer, A.; Pester, A. Large Scale COVID-19 Detection with Blood Platelets Using Artificial Neural Network. In Proceedings of the Online Engineering and Society 4.0: Proceedings of the 18th International Conference on Remote Engineering and Virtual Instrumentation; Springer: Berlin/Heidelberg, Germany, 2022; pp. 393–399. [Google Scholar] [CrossRef]
- Plante, T.B.; Blau, A.M.; Berg, A.N.; Weinberg, A.S.; Jun, I.C.; Tapson, V.F.; Kanigan, T.S.; Adib, A.B. Development and external validation of a machine learning tool to rule out COVID-19 among adults in the emergency department using routine blood tests: A large, multicenter, real-world study. J. Med. Internet. Res. 2020, 22, e24048. [Google Scholar] [CrossRef]
- Viana dos Santos Santana, Í.; CM da Silveira, A.; Sobrinho, Á.; Chaves e Silva, L.; Dias da Silva, L.; Santos, D.F.; Gurjao, E.C.; Perkusich, A. Classification models for COVID-19 test prioritization in Brazil: Machine learning approach. J. Med. Internet. Res. 2021, 23, e27293. [Google Scholar] [CrossRef]
- Gangloff, C.; Rafi, S.; Bouzillé, G.; Soulat, L.; Cuggia, M. Machine learning is the key to diagnose COVID-19: A proof-of-concept study. Sci. Rep. 2021, 11, 7166. [Google Scholar] [CrossRef]
- Abayomi-Alli, O.O.; Damaševičius, R.; Maskeliūnas, R.; Misra, S. An ensemble learning model for COVID-19 detection from blood test samples. Sensors 2022, 22, 2224. [Google Scholar] [CrossRef]
- Yousif, A.Y.; Younis, S.M.; Hussein, S.A.; Al-Saidi, N.M.G. An intelligent computing for diagnosing COVID-19 using available blood tests. Int. J. Innov. Comput. Inf. Furth. Control 2022, 18, 57–72. [Google Scholar]
- Chadaga, K.; Chakraborty, C.; Prabhu, S.; Umakanth, S.; Bhat, V.; Sampathila, N. Clinical and laboratory approach to diagnose COVID-19 using machine learning. Interdiscip. Sci. Comput. Life Sci. 2022, 14, 452–470. [Google Scholar] [CrossRef]
- Kukar, M.; Gunčar, G.; Vovko, T.; Podnar, S.; Černelč, P.; Brvar, M.; Zalaznik, M.; Notar, M.; Moškon, S.; Notar, M. COVID-19 diagnosis by routine blood tests using machine learning. Sci. Rep. 2021, 11, 10738. [Google Scholar] [CrossRef]
- Fernandes, F.T.; de Oliveira, T.A.; Teixeira, C.E.; Batista, A.F.d.M.; Dalla Costa, G.; Chiavegatto Filho, A.D.P. A multipurpose machine learning approach to predict COVID-19 negative prognosis in São Paulo, Brazil. Sci. Rep. 2021, 11, 3343. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.A.; Castro, G.Z.; Oliveira, B.A.S.; Ferreira, L.A.; Ramírez, J.A.; Silva, R.; Guimarães, F.G. Explaining machine learning based diagnosis of COVID-19 from routine blood tests with decision trees and criteria graphs. Comput. Biol. Med. 2021, 132, 104335. [Google Scholar] [CrossRef] [PubMed]
- de Freitas Barbosa, V.A.; Gomes, J.C.; de Santana, M.A.; Albuquerque, J.E.d.A.; de Souza, R.G.; de Souza, R.E.; dos Santos, W.P. Heg. IA: An intelligent system to support diagnosis of COVID-19 based on blood tests. Res. Biomed. Eng. 2021, 1–18. [Google Scholar] [CrossRef]
- Rikan, S.B.; Azar, A.S.; Ghafari, A.; Mohasefi, J.B.; Pirnejad, H. COVID-19 diagnosis from routine blood tests using artificial intelligence techniques. Biomed. Signal Process. Control 2022, 72, 103263. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.N.; Yang, Y.; Tang, L.L.; Dai, Y.N.; Gao, H.N.; Pan, H.Y.; Ju, B. A prediction model based on machine learning for diagnosing the early COVID-19 patients. MedRxiv 2020, arXiv:03.20120881. [Google Scholar] [CrossRef]
- Li, W.T.; Ma, J.; Shende, N.; Castaneda, G.; Chakladar, J.; Tsai, J.C.; Apostol, L.; Honda, C.O.; Xu, J.; Wong, L.M.; et al. Using machine learning of clinical data to diagnose COVID-19: A systematic review and meta-analysis. BMC Med. Informatics Decis. Mak. 2020, 20, 1–13. [Google Scholar] [CrossRef]
- Meng, Z.; Wang, M.; Song, H.; Guo, S.; Zhou, Y.; Li, W.; Zhou, Y.; Li, M.; Song, X.; Zhou, Y.; et al. Development and utilization of an intelligent application for aiding COVID-19 diagnosis. MedRxiv 2020, arXiv:18.20035816. [Google Scholar] [CrossRef]
- Xu, W.; Sun, N.N.; Gao, H.N.; Chen, Z.Y.; Yang, Y.; Ju, B.; Tang, L.L. Risk factors analysis of COVID-19 patients with ARDS and prediction based on machine learning. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Abdulkareem, K.H.; Mohammed, M.A.; Salim, A.; Arif, M.; Geman, O.; Gupta, D.; Khanna, A. Realizing an effective COVID-19 diagnosis system based on machine learning and IOT in smart hospital environment. IEEE Internet Things J. 2021, 8, 15919–15928. [Google Scholar] [CrossRef] [PubMed]
- Willette, A.A.; Willette, S.A.; Wang, Q.; Pappas, C.; Klinedinst, B.S.; Le, S.; Larsen, B.; Pollpeter, A.; Li, T.; Mochel, J.P.; et al. Using machine learning to predict COVID-19 infection and severity risk among 4510 aged adults: A UK Biobank cohort study. Sci. Rep. 2022, 12, 7736. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Ray, S.; Vorselaars, B.; Kitson, J.; Mamalakis, M.; Weeks, S.; Baker, M.; Mackenzie, L.S. Use of machine learning and artificial intelligence to predict SARS-CoV-2 infection from full blood counts in a population. Int. Immunopharmacol. 2020, 86, 106705. [Google Scholar] [CrossRef] [PubMed]
- Darapaneni, N.; Gupta, M.; Paduri, A.R.; Agrawal, R.; Padasali, S.; Kumari, A.; Purushothaman, P. A novel machine learning based screening method for high-risk COVID-19 patients based on simple blood exams. In Proceedings of the 2021 IEEE International IOT, Electronics and Mechatronics Conference (IEMTRONICS), Toronto, ON, Canada, 21–24 April 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Tschoellitsch, T.; Dünser, M.; Böck, C.; Schwarzbauer, K.; Meier, J. Machine learning prediction of SARS-CoV-2 polymerase chain reaction results with routine blood tests. Lab. Med. 2021, 52, 146–149. [Google Scholar] [CrossRef] [PubMed]
- Delafiori, J.; Navarro, L.C.; Siciliano, R.F.; De Melo, G.C.; Busanello, E.N.B.; Nicolau, J.C.; Sales, G.M.; De Oliveira, A.N.; Val, F.F.A.; De Oliveira, D.N.; et al. COVID-19 automated diagnosis and risk assessment through metabolomics and machine learning. Anal. Chem. 2021, 93, 2471–2479. [Google Scholar] [CrossRef] [PubMed]
- Ning, W.; Lei, S.; Yang, J.; Cao, Y.; Jiang, P.; Yang, Q.; Zhang, J.; Wang, X.; Chen, F.; Geng, Z.; et al. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nat. Biomed. Eng. 2020, 4, 1197–1207. [Google Scholar] [CrossRef]
- Soltan, A.A.; Kouchaki, S.; Zhu, T.; Kiyasseh, D.; Taylor, T.; Hussain, Z.B.; Peto, T.; Brent, A.J.; Eyre, D.W.; Clifton, D.A. Rapid triage for COVID-19 using routine clinical data for patients attending hospital: Development and prospective validation of an artificial intelligence screening test. Lancet Digit. Health 2021, 3, e78–e87. [Google Scholar] [CrossRef]
- Silveira, E.C. Prediction of COVID-19 from hemogram results and age using machine learning. Front. Health Informatics 2020, 9, 39. [Google Scholar] [CrossRef]
- Singh, R.K.; Sinha, S.; Ramasamy, A.; Kannan, S.; Tambi, G.; Basu, M. COVID–19 AI diagnostic tool using only 13 common blood parameters. Int. J. Inf. Technol. (IJIT) 2020, 6. [Google Scholar]
- AlJame, M.; Ahmad, I.; Imtiaz, A.; Mohammed, A. Ensemble learning model for diagnosing COVID-19 from routine blood tests. Inform. Med. Unlocked 2020, 21, 100449. [Google Scholar] [CrossRef]
- Roland, T.; Böck, C.; Tschoellitsch, T.; Maletzky, A.; Hochreiter, S.; Meier, J.; Klambauer, G. Machine learning based COVID-19 diagnosis from blood tests with robustness to domain shifts. medRxiv 2021, arXiv:06.21254997. [Google Scholar] [CrossRef]
- Yang, H.S.; Hou, Y.; Vasovic, L.V.; Steel, P.A.; Chadburn, A.; Racine-Brzostek, S.E.; Velu, P.; Cushing, M.M.; Loda, M.; Kaushal, R.; et al. Routine laboratory blood tests predict SARS-CoV-2 infection using machine learning. Clin. Chem. 2020, 66, 1396–1404. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.P.; Pejaver, V.; Hammarlund, N.E.; Sung, H.; Lee, S.K.; Furmanchuk, A.; Lee, H.Y.; Scott, G.; Gombar, S.; Shah, N.; et al. A predictive tool for identification of SARS-CoV-2 PCR-negative emergency department patients using routine test results. J. Clin. Virol. 2020, 129, 104502. [Google Scholar] [CrossRef]
- Schwab, P.; DuMont Schütte, A.; Dietz, B.; Bauer, S. Clinical predictive models for COVID-19: Systematic study. J. Med. Internet Res. 2020, 22, e21439. [Google Scholar] [CrossRef] [PubMed]
- de Moraes Batista, A.F.; Miraglia, J.L.; Rizzi Donato, T.H.; Porto Chiavegatto Filho, A.D. COVID-19 diagnosis prediction in emergency care patients: A machine learning approach. MedRxiv 2020, arXiv:04.20052092. [Google Scholar] [CrossRef]
- Soares, F.; Villavicencio, A.; Fogliatto, F.S.; Pitombeira Rigatto, M.H.; José Anzanello, M.; Idiart, M.A.; Stevenson, M. A novel specific artificial intelligence-based method to identify COVID-19 cases using simple blood exams. MedRxiv 2020, arXiv:10.20061036. [Google Scholar] [CrossRef]
- Jiang, X.; Coffee, M.; Bari, A.; Wang, J.; Jiang, X.; Huang, J.; Shi, J.; Dai, J.; Cai, J.; Zhang, T.; et al. Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity. Comput. Mater. Contin. 2020, 63, 537–551. [Google Scholar] [CrossRef]
- Abdulaal, A.; Patel, A.; Charani, E.; Denny, S.; Alqahtani, S.A.; Davies, G.W.; Mughal, N.; Moore, L.S. Comparison of deep learning with regression analysis in creating predictive models for SARS-CoV-2 outcomes. BMC Med. Inform. Decis. Mak. 2020, 20, 1–11. [Google Scholar] [CrossRef]
- Aktar, S.; Ahamad, M.; Rashed-Al-Mahfuz, M.; Azad, A.; Uddin, S.; Kamal, A.; Alyami, S.A.; Lin, P.I.; Islam, S.M.S.; Quinn, J.M.; et al. Predicting patient COVID-19 disease severity by means of statistical and machine learning analysis of blood cell transcriptome data. arXiv 2020, arXiv:2011.10657. [Google Scholar] [CrossRef]
- Yao, H.; Zhang, N.; Zhang, R.; Duan, M.; Xie, T.; Pan, J.; Peng, E.; Huang, J.; Zhang, Y.; Xu, X.; et al. Severity detection for the coronavirus disease 2019 (COVID-19) patients using a machine learning model based on the blood and urine tests. FRontiers Cell Dev. Biol. 2020, 683. [Google Scholar] [CrossRef]
- Henzel, J.; Tobiasz, J.; Kozielski, M.; Bach, M.; Foszner, P.; Gruca, A.; Kania, M.; Mika, J.; Papiez, A.; Werner, A.; et al. Classification supporting COVID-19 diagnostics based on patient survey data. arXiv 2020, arXiv:2011.12247. [Google Scholar] [CrossRef]
- Razavian, N.; Major, V.J.; Sudarshan, M.; Burk-Rafel, J.; Stella, P.; Randhawa, H.; Bilaloglu, S.; Chen, J.; Nguy, V.; Wang, W.; et al. A validated, real-time prediction model for favorable outcomes in hospitalized COVID-19 patients. NPJ Digit. Med. 2020, 3, 130. [Google Scholar] [CrossRef] [PubMed]
- Hallman, R.A.; Chikkula, A.; Prioleau, T. Predicting criticality in COVID-19 patients. In Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Virtual, 21–24 September 2020; pp. 1–7. [Google Scholar] [CrossRef]
- Goodman-Meza, D.; Rudas, A.; Chiang, J.N.; Adamson, P.C.; Ebinger, J.; Sun, N.; Botting, P.; Fulcher, J.A.; Saab, F.G.; Brook, R.; et al. A machine learning algorithm to increase COVID-19 inpatient diagnostic capacity. PLoS ONE 2020, 15, e0239474. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.; Fang, X.; Zhang, J.; Homayounieh, F.; Arru, C.D.; Digumarthy, S.R.; Babaei, R.; Mobin, H.K.; Mohseni, I.; Saba, L.; et al. Integrative analysis for COVID-19 patient outcome prediction. Med. Image Anal. 2021, 67, 101844. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.M.; Liu, W.; Chen, X.; McRae, M.P.; McDevitt, J.T.; Fenyö, D. Predictive modeling of morbidity and mortality in patients hospitalized with COVID-19 and its clinical implications: Algorithm development and interpretation. J. Med. Internet Res. 2021, 23, e29514. [Google Scholar] [CrossRef] [PubMed]
- Vaid, A.; Somani, S.; Russak, A.J.; De Freitas, J.K.; Chaudhry, F.F.; Paranjpe, I.; Johnson, K.W.; Lee, S.J.; Miotto, R.; Richter, F.; et al. Machine learning to predict mortality and critical events in a cohort of patients with COVID-19 in New York City: Model development and validation. J. Med. Internet Res. 2020, 22, e24018. [Google Scholar] [CrossRef]
- Parchure, P.; Joshi, H.; Dharmarajan, K.; Freeman, R.; Reich, D.L.; Mazumdar, M.; Timsina, P.; Kia, A. Development and validation of a machine learning-based prediction model for near-term in-hospital mortality among patients with COVID-19. BMJ Support. Palliat. Care 2022, 12, e424–e431. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Montañés, M.; Rodríguez-Belenguer, P.; Serrano-López, A.J.; Soria-Olivas, E.; Alakhdar-Mohmara, Y. Machine learning for mortality analysis in patients with COVID-19. Int. J. Environ. Res. Public Health 2020, 17, 8386. [Google Scholar] [CrossRef]
- Wu, G.; Yang, P.; Xie, Y.; Woodruff, H.C.; Rao, X.; Guiot, J.; Frix, A.N.; Louis, R.; Moutschen, M.; Li, J.; et al. Development of a clinical decision support system for severity risk prediction and triage of COVID-19 patients at hospital admission: An international multicentre study. Eur. Respir. J. 2020, 56. [Google Scholar] [CrossRef]
- Gao, Y.; Cai, G.Y.; Fang, W.; Li, H.Y.; Wang, S.Y.; Chen, L.; Yu, Y.; Liu, D.; Xu, S.; Cui, P.F.; et al. Machine learning based early warning system enables accurate mortality risk prediction for COVID-19. Nat. Commun. 2020, 11, 5033. [Google Scholar] [CrossRef]
- Das, A.K.; Mishra, S.; Gopalan, S.S. Predicting CoVID-19 community mortality risk using machine learning and development of an online prognostic tool. PeerJ 2020, 8, e10083. [Google Scholar] [CrossRef]
- Davis, C.; Gao, M.; Nichols, M.; Henao, R. Predicting hospital utilization and inpatient mortality of patients tested for COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Casiraghi, E.; Malchiodi, D.; Trucco, G.; Frasca, M.; Cappelletti, L.; Fontana, T.; Esposito, A.A.; Avola, E.; Jachetti, A.; Reese, J.; et al. Explainable machine learning for early assessment of COVID-19 risk prediction in emergency departments. IEEE Access 2020, 8, 196299–196325. [Google Scholar] [CrossRef]
- Wong, K.C.; Xiang, Y.; So, H.C. Uncovering clinical risk factors and prediction of severe COVID-19: A machine learning approach based on UK Biobank data. MedRxiv 2020, arXiv:18.20197319. [Google Scholar] [CrossRef]
- Soltan, A.A.; Kouchaki, S.; Zhu, T.; Kiyasseh, D.; Taylor, T.; Hussain, Z.B.; Peto, T.; Brent, A.J.; Eyre, D.W.; Clifton, D. Artificial intelligence driven assessment of routinely collected healthcare data is an effective screening test for COVID-19 in patients presenting to hospital. MedRxiv 2020, arXiv:07.20148361. [Google Scholar] [CrossRef]
- Avila, E.; Kahmann, A.; Alho, C.; Dorn, M. Hemogram data as a tool for decision-making in COVID-19 management: Applications to resource scarcity scenarios. PeerJ 2020, 8, e9482. [Google Scholar] [CrossRef]
- Xu, M.; Ouyang, L.; Gao, Y.; Chen, Y.; Yu, T.; Li, Q.; Sun, K.; Bao, F.S.; Safarnejad, L.; Wen, J.; et al. Accurately differentiating COVID-19, other viral infection, and healthy individuals using multimodal features via late fusion learning. medRxiv 2020, arXiv:18.20176776. [Google Scholar] [CrossRef]
- De Souza, F.S.H.; Hojo-Souza, N.S.; Dos Santos, E.B.; Da Silva, C.M.; Guidoni, D.L. Predicting the disease outcome in COVID-19 positive patients through Machine Learning: A retrospective cohort study with Brazilian data. Front. Artif. Intell. 2021, 4, 579931. [Google Scholar] [CrossRef]
- Chen, Y.; Ouyang, L.; Bao, F.S.; Li, Q.; Han, L.; Zhu, B.; Xu, M.; Liu, J.; Ge, Y.; Chen, S. An interpretable machine learning framework for accurate severe vs non-severe COVID-19 clinical type classification. medRxiv 2020, arXiv:18.20105841. [Google Scholar] [CrossRef]
- Bezzan, V.P.; Rocco, C.D. Predicting special care during the COVID-19 pandemic: A machine learning approach. Health Inf. Sci. Syst. 2021, 9, 34. [Google Scholar] [CrossRef]
- Subudhi, S.; Verma, A.; Patel, A.B.; Hardin, C.C.; Khandekar, M.J.; Lee, H.; McEvoy, D.; Stylianopoulos, T.; Munn, L.L.; Dutta, S.; et al. Comparing machine learning algorithms for predicting ICU admission and mortality in COVID-19. NPJ Digit. Med. 2021, 4, 87. [Google Scholar] [CrossRef] [PubMed]
- Fakhartousi, A.; Davies, P.; Farahbakhshian, S.F. Effect of feature selection on routine blood tests to diagnose COVID-19 infection. Age 2021, 61, 5–64. [Google Scholar]
- Du, R.; Tsougenis, E.D.; Ho, J.W.; Chan, J.K.; Chiu, K.W.; Fang, B.X.; Ng, M.Y.; Leung, S.T.; Lo, C.S.; Wong, H.Y.F.; et al. Machine learning application for the prediction of SARS-CoV-2 infection using blood tests and chest radiograph. Sci. Rep. 2021, 11, 14250. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Chen, T.; Luo, H.; Luo, Y.; Du, G.; Jiming-Yang, M. Machine learning predictive model for severe COVID-19. Infect. Genet. Evol. 2021, 90, 104737. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhou, S.; Wang, Y.; Li, X. Machine learning: A predication model of outcome of SARS-CoV-2 pneumonia. 2020. Available online: https://assets.researchsquare.com/files/rs-23196/v1/2fca4743-cb6c-4e97-b94e-df7c1949fb2a.pdf?c=1631833056 (accessed on 29 March 2023).
- Zheng, Y.; Zhu, Y.; Ji, M.; Wang, R.; Liu, X.; Zhang, M.; Liu, J.; Zhang, X.; Qin, C.H.; Fang, L.; et al. A learning-based model to evaluate hospitalization priority in COVID-19 pandemics. Patterns 2020, 1, 100092. [Google Scholar] [CrossRef]
- Bao, F.S.; He, Y.; Liu, J.; Chen, Y.; Li, Q.; Zhang, C.R.; Han, L.; Zhu, B.; Ge, Y.; Chen, S.; et al. Triaging moderate COVID-19 and other viral pneumonias from routine blood tests. arXiv 2020, arXiv:2005.06546. [Google Scholar] [CrossRef]
- Feng, C.; Wang, L.; Chen, X.; Zhai, Y.; Zhu, F.; Chen, H.; Wang, Y.; Su, X.; Huang, S.; Tian, L.; et al. A Novel triage tool of artificial intelligence-assisted diagnosis aid system for suspected COVID-19 pneumonia in fever clinics. MedRxiv 2020, arXiv:19.20039099. [Google Scholar] [CrossRef]
- Wu, J.; Shen, J.; Xu, M.; Shao, M. A novel combined dynamic ensemble selection model for imbalanced data to detect COVID-19 from complete blood count. Comput. Methods Programs Biomed. 2021, 211, 106444. [Google Scholar] [CrossRef]
- Buturovic, L.; Zheng, H.; Tang, B.; Lai, K.; Kuan, W.S.; Gillett, M.; Santram, R.; Shojaei, M.; Almansa, R.; Nieto, J.Á.; et al. A 6-mRNA host response classifier in whole blood predicts outcomes in COVID-19 and other acute viral infections. Sci. Rep. 2022, 12, 889. [Google Scholar] [CrossRef]
- Hu, J.; Heidari, A.A.; Shou, Y.; Ye, H.; Wang, L.; Huang, X.; Chen, H.; Chen, Y.; Wu, P. Detection of COVID-19 severity using blood gas analysis parameters and Harris hawks optimized extreme learning machine. Comput. Biol. Med. 2022, 142, 105166. [Google Scholar] [CrossRef]
- Rahman, T.; Khandakar, A.; Abir, F.F.; Faisal, M.A.A.; Hossain, M.S.; Podder, K.K.; Abbas, T.O.; Alam, M.F.; Kashem, S.B.; Islam, M.T.; et al. QCovSML: A reliable COVID-19 detection system using CBC biomarkers by a stacking machine learning model. Comput. Biol. Med. 2022, 143, 105284. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Sumali, B.; Lee, H.; Terai, H.; Ishii, M.; Fukunaga, K.; Mitsukura, Y.; Nishimura, T. Finding of the factors affecting the severity of COVID-19 based on mathematical models. Sci. Rep. 2021, 11, 24224. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhang, H.T.; Goncalves, J.; Xiao, Y.; Wang, M.; Guo, Y.; Sun, C.; Tang, X.; Jing, L.; Zhang, M.; et al. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020, 2, 283–288. [Google Scholar] [CrossRef]
- Tchagna Kouanou, A.; Mih Attia, T.; Feudjio, C.; Djeumo, A.F.; Ngo Mouelas, A.; Nzogang, M.P.; Tchito Tchapga, C.; Tchiotsop, D. An overview of supervised machine learning methods and data analysis for COVID-19 detection. J. Healthc. Eng. 2021, 2021. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, H.T.; Xiao, Y.; Wang, M.; Guo, Y.; Sun, C.; Tang, X.; Jing, L.; Li, S.; Zhang, M.; et al. Prediction of criticality in patients with severe COVID-19 infection using three clinical features: A machine learning-based prognostic model with clinical data in Wuhan. MedRxiv 2020, arXiv:27.20028027. [Google Scholar] [CrossRef]
- Bayat, V.; Phelps, S.; Ryono, R.; Lee, C.; Parekh, H.; Mewton, J.; Sedghi, F.; Etminani, P.; Holodniy, M. A COVID-19 prediction model from standard laboratory tests and vital signs. Inform. Med. Unlocked 2020, 21, 100449. [Google Scholar] [CrossRef]
- Langer, T.; Favarato, M.; Giudici, R.; Bassi, G.; Garberi, R.; Villa, F.; Gay, H.; Zeduri, A.; Bragagnolo, S.; Molteni, A.; et al. Use of Machine Learning to Rapidly Predict Positivity to Severe Acute Respiratory Syndrome Coronavirus 2(SARS-COV-2) Using Basic Clinical Data. 2020. Available online: https://www.researchsquare.com/article/rs-38576/v1 (accessed on 29 March 2023). [CrossRef]
- Dairi, A.; Harrou, F.; Sun, Y. Deep generative learning-based 1-svm detectors for unsupervised COVID-19 infection detection using blood tests. IEEE Trans. Instrum. Meas. 2021, 71, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Göreke, V.; Sarı, V.; Kockanat, S. A novel classifier architecture based on deep neural network for COVID-19 detection using laboratory findings. Appl. Soft Comput. 2021, 106, 107329. [Google Scholar] [CrossRef] [PubMed]
- Alakus, T.B.; Turkoglu, I. Comparison of deep learning approaches to predict COVID-19 infection. Chaos, Solitons Fractals 2020, 140, 110120. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.S.; Ge, P.; Jiang, C.; Zhang, Y.; Li, X.; Zhao, Z.; Zhang, L.; Duong, T.Q. Deep-learning artificial intelligence analysis of clinical variables predicts mortality in COVID-19 patients. J. Am. Coll. Emerg. Physicians Open 2020, 1, 1364–1373. [Google Scholar] [CrossRef] [PubMed]
- Kemal, A.; KILIÇARSLAN, S. COVID-19 diagnosis prediction in emergency care patients using convolutional neural network. Afyon Kocatepe Üniversitesi Fen Ve Mühendislik Bilim. 2021, 21, 300–309. [Google Scholar] [CrossRef]
- Bismadhika, F.; Qomariyah, N.N.; Purwita, A.A. Experiment on Deep Learning Models for COVID-19 Detection from Blood Testing. In Proceedings of the 2021 IEEE International Biomedical Instrumentation and Technology Conference (IBITeC), Yogyakarta, Indonesia, 20–21 October 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 136–141. [Google Scholar] [CrossRef]
- Mei, X.; Lee, H.C.; Diao, K.y.; Huang, M.; Lin, B.; Liu, C.; Xie, Z.; Ma, Y.; Robson, P.M.; Chung, M.; et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020, 26, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- AlJame, M.; Imtiaz, A.; Ahmad, I.; Mohammed, A. Deep forest model for diagnosing COVID-19 from routine blood tests. Sci. Rep. 2021, 11, 16682. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; He, L.; Luo, W.; Tse, R.; Pau, G. Deep Learning Hybrid Models for COVID-19 Prediction. J. Glob. Inf. Manag. (JGIM) 2022, 30, 1–20. [Google Scholar] [CrossRef]
- Turabieh, H.; Karaa, W.B.A. Predicting the existence of COVID-19 using machine learning based on laboratory findings. In Proceedings of the 2021 international conference of women in data science at Taif University (WiDSTaif), Taif, Saudi Arabia, 30–31 March 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1–7. [Google Scholar] [CrossRef]
- Sargiani, V.; De Souza, A.A.; De Almeida, D.C.; Barcelos, T.S.; Munoz, R.; Da Silva, L.A. Supporting Clinical COVID-19 Diagnosis with Routine Blood Tests Using Tree-Based Entropy Structured Self-Organizing Maps. Appl. Sci. 2022, 12, 5137. [Google Scholar] [CrossRef]
- O’Connor, T.; Santaniello, S.; Javidi, B. COVID-19 detection from red blood cells using highly comparative time-series analysis (HCTSA) in digital holographic microscopy. Opt. Express 2022, 30, 1723–1736. [Google Scholar] [CrossRef]
- Zhang, H.J.; Qi, G.Q.; Gu, X.; Zhang, X.Y.; Fang, Y.F.; Jiang, H.; Zhao, Y.J. Lymphocyte blood levels that remain low can predict the death of patients with COVID-19. Medicine 2021, 100. [Google Scholar] [CrossRef]
- Lee, J.J.; Montazerin, S.M.; Jamil, A.; Jamil, U.; Marszalek, J.; Chuang, M.L.; Chi, G. Association between red blood cell distribution width and mortality and severity among patients with COVID-19: A systematic review and meta-analysis. J. Med. Virol. 2021, 93, 2513–2522. [Google Scholar] [CrossRef]
- Sarkar, S.; Kannan, S.; Khanna, P.; Singh, A.K. Role of red blood cell distribution width, as a prognostic indicator in COVID-19: A systematic review and meta-analysis. Rev. Med. Virol. 2022, 32, e2264. [Google Scholar] [CrossRef]
- Dai, W.; Ke, P.F.; Li, Z.Z.; Zhuang, Q.Z.; Huang, W.; Wang, Y.; Xiong, Y.; Huang, X.Z. Establishing classifiers with clinical laboratory indicators to distinguish COVID-19 from community-acquired pneumonia: Retrospective cohort study. J. Med. Internet Res. 2021, 23, e23390. [Google Scholar] [CrossRef]
- Henry, B.M.; Benoit, J.L.; Benoit, S.; Pulvino, C.; Berger, B.A.; Olivera, M.H.S.d.; Crutchfield, C.A.; Lippi, G. Red blood cell distribution width (RDW) predicts COVID-19 severity: A prospective, observational study from the cincinnati SARS-CoV-2 emergency department cohort. Diagnostics 2020, 10, 618. [Google Scholar] [CrossRef] [PubMed]
- Kahn, R.; Schmidt, T.; Golestani, K.; Mossberg, A.; Gullstrand, B.; Bengtsson, A.A.; Kahn, F. Mismatch between circulating cytokines and spontaneous cytokine production by leukocytes in hyperinflammatory COVID-19. J. Leucoc. Biol. 2021, 109, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Liu, Y.; Zou, S.; Liu, P.; Zhao, J.; Yang, C.; Liang, M.; Yang, J. Genome-wide screening of SARS-CoV-2 infection-related genes based on the blood leukocytes sequencing data set of patients with COVID-19. J. Med. Virol. 2021, 93, 5544–5554. [Google Scholar] [CrossRef] [PubMed]
- Vitte, J.; Diallo, A.B.; Boumaza, A.; Lopez, A.; Michel, M.; Allardet-Servent, J.; Mezouar, S.; Sereme, Y.; Busnel, J.M.; Miloud, T.; et al. A granulocytic signature identifies COVID-19 and its severity. J. Infect. Dis. 2020, 222, 1985–1996. [Google Scholar] [CrossRef]
- Tan, Y.; Zhou, J.; Zhou, Q.; Hu, L.; Long, Y. Role of eosinophils in the diagnosis and prognostic evaluation of COVID-19. J. Med. Virol. 2021, 93, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Koupenova, M.; Freedman, J.E. Platelets and COVID-19: Inflammation, hyperactivation and additional questions. Circ. Res. 2020, 127, 1419–1421. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Rial, J.; Rivero-Calle, I.; Salas, A.; Martinón-Torres, F. Role of monocytes/macrophages in COVID-19 pathogenesis: Implications for therapy. Infect. Drug Resist. 2020, 2485–2493. [Google Scholar] [CrossRef]
- Meidaninikjeh, S.; Sabouni, N.; Marzouni, H.Z.; Bengar, S.; Khalili, A.; Jafari, R. Monocytes and macrophages in COVID-19: Friends and foes. Life Sci. 2021, 269, 119010. [Google Scholar] [CrossRef]
- Zhang, Z.; Ma, P.; Ahmed, R.; Wang, J.; Akin, D.; Soto, F.; Liu, B.F.; Li, P.; Demirci, U. Advanced point-of-care testing technologies for human acute respiratory virus detection. Adv. Mater. 2022, 34, 2103646. [Google Scholar] [CrossRef]
- Zheng, J.; Long, X.; Chen, H.; Ji, Z.; Shu, B.; Yue, R.; Liao, Y.; Ma, S.; Qiao, K.; Liu, Y.; et al. Photoclick reaction constructs glutathione-responsive theranostic system for anti-tuberculosis. Front. Mol. Biosci. 2022, 9, 39. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).