Rapid Evaluation of the Xpert® Xpress CoV-2 plus and Xpert® Xpress CoV-2/Flu/RSV plus Tests
Abstract
1. Introduction
2. Materials and Methods
2.1. Overview
2.2. Ethics
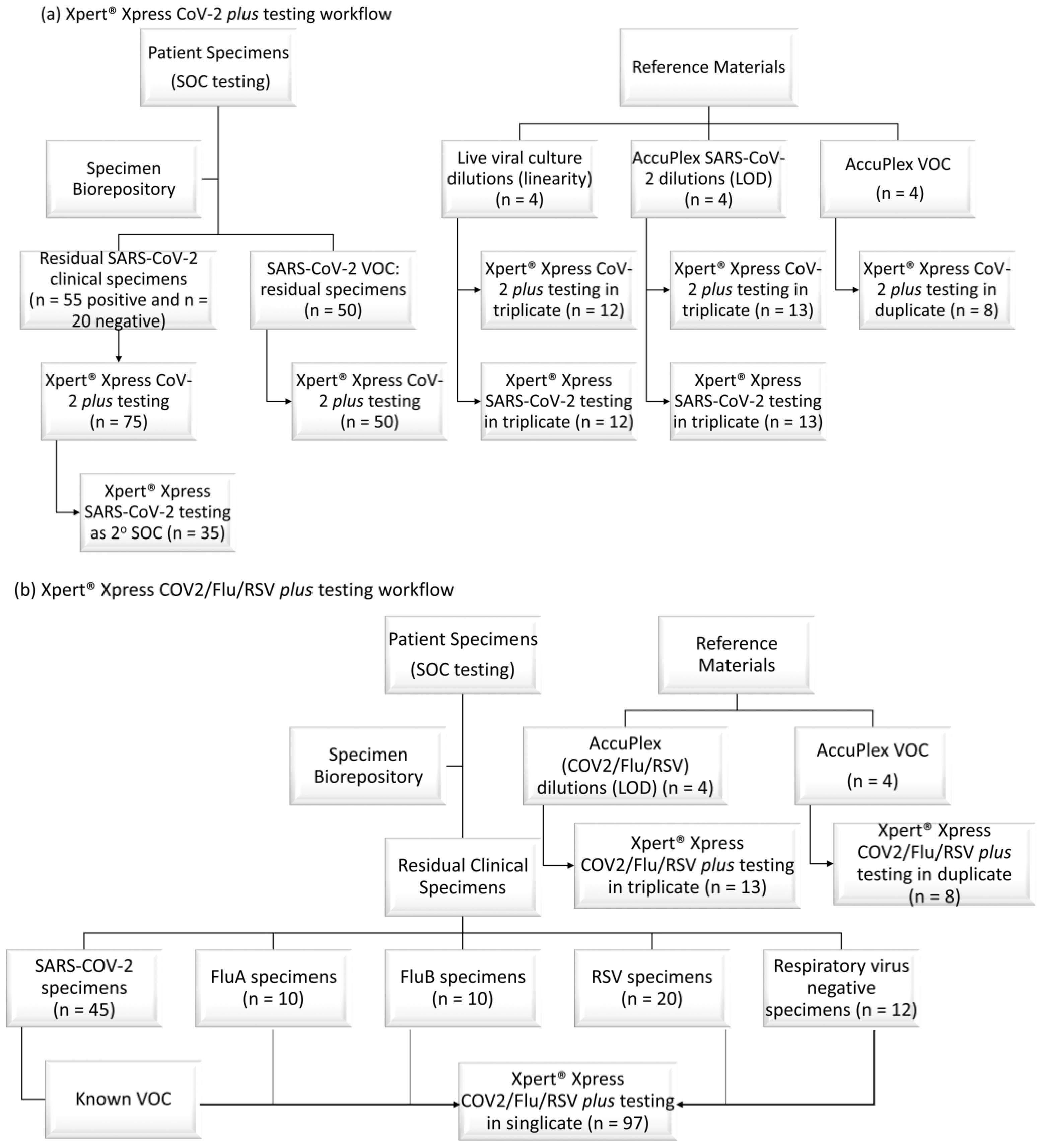
2.3. Xpert® Xpress CoV-2 plus Specimen Selection
2.4. Xpert® Xpress CoV-2/Flu/RSV plus Test Evaluation
2.5. Xpert® Test Overview
2.6. Data Analysis
3. Results
3.1. Xpert® Xpress CoV-2 plus Evaluation
3.1.1. Xpert® Xpress CoV-2 plus Specimen Description
3.1.2. Xpert® Xpress CoV-2 plus Accuracy Analysis
3.1.3. Xpert® Xpress CoV-2 plus Precision Analysis
3.2. Xpert® Xpress CoV-2/Flu/RSV plus Evaluation
3.2.1. Xpert® Xpress CoV-2/Flu/RSV plus Specimen Description
3.2.2. Xpert® Xpress CoV-2/Flu/RSV plus Accuracy Analysis
3.2.3. Xpert® Xpress CoV-2/Flu/RSV plus Precision Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- World Health Organisation. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. 2020. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 2 April 2020).
- United States of America Food and Drug Administation. In Vitro Diagnostics EUAs—Molecular Diagnostic Tests for SARS-CoV-2. Available online: https://www.fda.gov/medical-devices/coronavirus-disease-2019-COVID-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-molecular-diagnostic-tests-SARS-CoV-2 (accessed on 11 August 2022).
- Stevens, W.S.; Scott, L.; Noble, L.; Gous, N.; Dheda, K. Impact of the GeneXpert MTB/RIF Technology on Tuberculosis Control. Microbiol. Spectr. 2017, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Moran, A.; Beavis, K.G.; Matushek, S.M.; Ciaglia, C.; Francois, N.; Tesic, V.; Love, N. The Detection of SARS-CoV-2 using the Cepheid Xpert Xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 Assays. J. Clin. Microbiol. 2020, 58, e00772-20. [Google Scholar] [CrossRef] [PubMed]
- Tham, J.W.M.; Ng, S.C.; Chai, C.N.; Png, S.; Tan, E.J.M.; Ng, L.J.; Chua, R.P.; Sani, M.; Chiang, D.; Tan, K.X.; et al. Parallel testing of 241 clinical nasopharyngeal swabs for the detection of SARS-CoV-2 virus on the Cepheid Xpert Xpress SARS-CoV-2 and the Roche cobas SARS-CoV-2 assays. Clin. Chem. Lab. Med. 2020, 59, e45–e48. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Chen, J.; Wang, Y.; Lu, Y.; Zhu, Y.; Zhang, B.; Wang, F.; Mao, L.; Tang, Y.W.; Hu, B.; et al. Multicenter Evaluation of the Cepheid Xpert Xpress SARS-CoV-2 Assay for the Detection of SARS-CoV-2 in Oropharyngeal Swab Specimens. J. Clin. Microbiol. 2020, 58, e01288-20. [Google Scholar] [CrossRef] [PubMed]
- Smithgall, M.C.; Scherberkova, I.; Whittier, S.; Green, D.A. Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche cobas for the Rapid Detection of SARS-CoV-2. J. Clin. Virol. 2020, 128, 104428. [Google Scholar] [CrossRef]
- Wolters, F.; van de Bovenkamp, J.; van den Bosch, B.; van den Brink, S.; Broeders, M.; Chung, N.H.; Favie, B.; Goderski, G.; Kuijpers, J.; Overdevest, I.; et al. Multi-center evaluation of cepheid xpert(R) xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J. Clin. Virol. 2020, 128, 104426. [Google Scholar] [CrossRef]
- Loeffelholz, M.J.; Alland, D.; Butler-Wu, S.M.; Pandey, U.; Perno, C.F.; Nava, A.; Carroll, K.C.; Mostafa, H.; Davies, E.; McEwan, A.; et al. Multicenter Evaluation of the Cepheid Xpert Xpress SARS-CoV-2 Test. J. Clin. Microbiol. 2020, 58, e00926-20. [Google Scholar] [CrossRef]
- Cao, X.J.; Fang, K.Y.; Li, Y.P.; Zhou, J.; Guo, X.G. The Diagnostic Accuracy of Xpert Xpress to SARS-CoV-2: A systematic review. J. Virol. Methods 2022, 301, 114460. [Google Scholar] [CrossRef]
- European Centres for Disease Prevention and Control. Rapid Increase of a SARS-CoV-2 Variant with Multiple Spike Protein Mutations Observed in the United Kingdom (20 December 2020). 2020. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/SARS-CoV-2-variant-multiple-spike-protein-mutations-United-Kingdom.pdf (accessed on 12 January 2021).
- World Health Organisation. SARS-CoV-2 Variants (31 December 2020). 2020. Available online: https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/ (accessed on 17 December 2021).
- World Health Organisation. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 17 December 2021).
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Consortium, C.-G.U.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Public Health England. Confirmed Cases of COVID-19 Variant from South Africa Identified in UK. Published 15 January 2021; 2021. Available online: https://www.gov.uk/government/news/confirmed-cases-of-COVID-19-variants-identified-in-uk (accessed on 18 January 2021).
- Tegally, H.; Wilkinson, E.; Althaus, C.L.; Giovanetti, M.; San, J.E.; Giandhari, J.; Pillay, S.; Naidoo, Y.; Ramphal, U.; Msomi, N.; et al. Rapid replacement of the Beta variant by the Delta variant in South Africa. medRxiv 2021. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Giovanetti, M.; Fonseca, V.; Wilkinson, E.; Tegally, H.; San, E.J.; Althaus, C.L.; Xavier, J.; Nanev Slavov, S.; Viala, V.L.; Ranieri Jeronimo Lima, A.; et al. Replacement of the Gamma by the Delta variant in Brazil: Impact of lineage displacement on the ongoing pandemic. Virus Evol. 2022, 8, veac024. [Google Scholar] [CrossRef] [PubMed]
- Subramoney, K.; Mtileni, N.; Bharuthram, A.; Davis, A.; Kalenga, B.; Rikhotso, M.; Maphahlele, M.; Giandhari, J.; Naidoo, Y.; Pillay, S.; et al. Identification of SARS-CoV-2 Omicron variant using spike gene target failure and genotyping assays, Gauteng, South Africa, 2021. J. Med. Virol. 2022, 94, 3676–3684. [Google Scholar] [CrossRef]
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef]
- Choi, H.; Hwang, M.; Lukey, J.; Jinadatha, C.; Navarathna, D.H. Presumptive positive with the Cepheid Xpert Xpress SARS-CoV-2 assay due to N mutations in the Delta variant. Diagn. Microbiol. Infect. Dis. 2022, 103, 115699. [Google Scholar] [CrossRef]
- Foster, C.S.P.; Madden, M.; Chan, R.; Agapiou, D.; Bull, R.A.; Rawlinson, W.D.; Van Hal, S.J. SARS-CoV-2 N-gene mutation leading to Xpert Xpress SARS-CoV-2 assay instability. Pathology 2022, 54, 499–501. [Google Scholar] [CrossRef]
- US Food and Drug Administation. SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests. Last Updated 27 December 2021; 2021. Available online: https://www.fda.gov/medical-devices/coronavirus-COVID-19-and-medical-devices/SARS-CoV-2-viral-mutations-impact-COVID-19-tests#omicron (accessed on 17 January 2022).
- Burns, B.L.; Moody, D.; Tu, Z.J.; Nakitandwe, J.; Brock, J.E.; Bosler, D.; Mitchell, S.L.; Loeffelholz, M.J.; Rhoads, D.D. Design and Implementation of Improved SARS-CoV-2 Diagnostic Assays to Mitigate the Impact of Genomic Mutations on Target Failure: The Xpert Xpress SARS-CoV-2 Experience. Microbiol. Spectr. 2022, e0135522. [Google Scholar] [CrossRef]
- Johnson, G.; Gregorchuk, B.S.J.; Zubrzycki, A.; Kolsun, K.; Meyers, A.F.A.; Sandstrom, P.A.; Becker, M.G. Clinical Evaluation of the GeneXpert® Xpert® Xpress SARS-CoV-2/Flu/RSV PLUS Combination Test. medRxiv 2022. [Google Scholar] [CrossRef]
- Isles, N.; Badman, S.G.; Ballard, S.; Zhang, B.; Howden, B.P.; Guy, R.; Williamson, D.A. Analytical sensitivity and specificity of the Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV assay. Pathology 2022, 54, 120–122. [Google Scholar] [CrossRef]
- Sluimer, J.; Goderski, G.; van den Brink, S.; Broeders, M.; Rahamat-Langendoen, J.; Then, E.; Wijsman, L.; Wolters, F.; van de Bovenkamp, J.; Melchers, W.J.; et al. Multi-center evaluation of Cepheid Xpert® Xpress SARS-CoV-2/Flu/RSV molecular point-of-care test. J. Clin. Virol. Plus 2021, 1, 100042. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; AlMutawa, F. Tracheal Aspirate and Bronchoalveolar Lavage as Potential Specimen Types for COVID-19 Testing Using the Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV. Microbiol. Spectr. 2022, 10, e0039922. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.; Zubrzycki, A.; Henry, M.; Ranadheera, C.; Corbett, C.; Meyers, A.F.A.; Sandstrom, P.A.; Becker, M.G. Clinical evaluation of the GeneXpert® Xpert® Xpress SARS-CoV-2/Flu/RSV combination test. J. Clin. Virol. Plus 2021, 1, 100014. [Google Scholar] [CrossRef] [PubMed]
- Leung, E.C.; Chow, V.C.; Lee, M.K.; Tang, K.P.; Li, D.K.; Lai, R.W. Evaluation of the Xpert Xpress SARS-CoV-2/Flu/RSV Assay for Simultaneous Detection of SARS-CoV-2, Influenza A and B Viruses, and Respiratory Syncytial Virus in Nasopharyngeal Specimens. J. Clin. Microbiol. 2021, 59, e02965-20. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, H.H.; Carroll, K.C.; Hicken, R.; Berry, G.J.; Manji, R.; Smith, E.; Rakeman, J.L.; Fowler, R.C.; Leelawong, M.; Butler-Wu, S.M.; et al. Multicenter Evaluation of the Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV Test. J. Clin. Microbiol. 2021, 59, e02955-20. [Google Scholar] [CrossRef] [PubMed]
- Cepheid. Xpert® Xpress CoV-2 Plus Instructions for Use 302-7342; Cepheid: Sunnyvale, CA, USA, 2022. [Google Scholar]
- Noble, L.; Scott, L.; Munir, R.; Steegen, K.; du Plessis, M.; Hans, L.; Stevens, W. Guidance for SARS-CoV-2 RNA-Based Molecular Assay Analytical Performance Evaluations. Methods Mol. Biol. 2022, 2511, 99–115. [Google Scholar] [CrossRef]
- Cepheid. Xpert® Xpress CoV-2/Flu/RSV Plus Instructions for Use 302-7085; Cepheid: Sunnyvale, CA, USA, 2021. [Google Scholar]
- Cepheid. Xpert® Xpress SARS-CoV-2 Package Insert. 2020. Available online: https://www.cepheid.com/Package%20Insert%20Files/Xpress-SARS-CoV-2-PI/302-3562-rev%20A-PACKAGE-INSERT-EUA-GX-SARS-CoV2.pdf (accessed on 2 April 2020).
- Obermeier, P.; Muehlhans, S.; Hoppe, C.; Karsch, K.; Tief, F.; Seeber, L.; Chen, X.; Conrad, T.; Boettcher, S.; Diedrich, S.; et al. Enabling Precision Medicine with Digital Case Classification at the Point-of-Care. eBioMedicine 2016, 4, 191–196. [Google Scholar] [CrossRef]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Altman, D. Practical Statistics for Medical Research; Chapman and Hall: London, UK, 1991. [Google Scholar]
- World Health Organisation. COVID-19 Target Product Profiles for Priority Diagnostics to Support Response to the COVID-19 Pandemic v.0.1. 2020. Available online: https://www.who.int/publications/m/item/COVID-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-COVID-19-pandemic-v.0.1 (accessed on 8 August 2020).
- Altman, D.G.; Bland, J.M. Assessing Agreement between Methods of Measurement. Clin. Chem. 2017, 63, 1653–1654. [Google Scholar] [CrossRef]
- Broder, K.; Babiker, A.; Myers, C.; White, T.; Jones, H.; Cardella, J.; Burd, E.M.; Hill, C.E.; Kraft, C.S. Test Agreement between Roche Cobas 6800 and Cepheid GeneXpert Xpress SARS-CoV-2 Assays at High Cycle Threshold Ranges. J. Clin. Microbiol. 2020, 58, e01187-20. [Google Scholar] [CrossRef]
- Wong, R.C.; Wong, A.H.; Ho, Y.I.; Siu, G.K.; Lee, L.K.; Leung, E.C.; Lai, R.W. Application of digital PCR to determine the reliability of Xpert Xpress SARS-CoV-2 assay with envelope (E) gene negative and nucleocapsid (N2) gene positive results. Diagn. Microbiol. Infect. Dis. 2022, 103, 115726. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.L.; Loeffelholz, M.J. Considerations regarding Interpretation of Positive SARS-CoV-2 Molecular Results with Late Cycle Threshold Values. J. Clin. Microbiol. 2022, 60, e0050122. [Google Scholar] [CrossRef] [PubMed]
- Engelmann, I.; Alidjinou, E.K.; Ogiez, J.; Pagneux, Q.; Miloudi, S.; Benhalima, I.; Ouafi, M.; Sane, F.; Hober, D.; Roussel, A.; et al. Preanalytical Issues and Cycle Threshold Values in SARS-CoV-2 Real-Time RT-PCR Testing: Should Test Results Include These? ACS Omega 2021, 6, 6528–6536. [Google Scholar] [CrossRef] [PubMed]


| Type | n | Reference | PPA (95% CI) NPA (95% CI) | Cohen Kappa [38] (95% CI) | Agreement Score [39] |
|---|---|---|---|---|---|
| Clinically relevant specimens | 157 | SOC/expected results | 96.4% (91.7, 98.8) 95.0% (75.1, 99.9) | 0.8416 (0.7185, 0.9647) | Very good |
| Residual clinical specimens | 125 | SOC | 95.2% (89.2, 98.4) 95.0 % (75.1, 99.9) | 0.8348 (0.7070, 0.9626) | Very good |
| Clinically relevant specimens | 55 | Xpert® Xpress SARS-CoV-2 | 97.9% (88.9, 99.9) 85.7% (42.1, 99.6) | 0.8363 (0.6153, 1.0573) | Very good |
| SARS-CoV-2 Discordant Specimens | |||||||||||||
| Specimen | Wave | SOC (cobas® SARS-CoV-2) | Xpert® Xpress SARS-CoV-2 | Xpert® Xpress CoV-2 plus | |||||||||
| Ct E | Ct ORF1ab | Result | Ct E | Ct N2 | Ct SPC | Result | Ct E | Ct N2 | Ct RdRp | Ct SPC | Result | ||
| 52 | i2 | 0.0 | 0.0 | SARS-CoV-2 Negative | 36.2 | 37.7 | 27.6 | SARS-CoV-2 Positive | 34.0 | 37.0 | 38.0 | 28.9 | SARS-CoV-2 Positive |
| 110 | 4 | 37.6 | 33.8 | SARS-CoV-2 Positive | 0.0 | 0.0 | 30.5 | SARS-CoV-2 Negative | 0.0 | 0.0 | 0.0 | 28.8 | SARS-CoV-2 Negative |
| 114 | 2 | 36.0 | 34.1 | SARS-CoV-2 Positive | 0.0 | 0.0 | 27.5 | SARS-CoV-2 Negative | 37.5 | 42.3 | 0.0 | 28.6 | SARS-CoV-2 Positive |
| 116 | 4 | 35.7 | 35.1 | SARS-CoV-2 Positive | 0.0 | 0.0 | 27.5 | SARS-CoV-2 Negative | 0.0 | 0.0 | 0.0 | 28.5 | SARS-CoV-2 Negative |
| 119 | 3 | 37.2 | 35.1 | SARS-CoV-2 Positive | 38.4 | 41.4 | 27.9 | SARS-CoV-2 Positive | 0.0 | 0.0 | 0.0 | 28.4 | SARS-CoV-2 Negative |
| 122 | 2 | 36.4 | 35.4 | SARS-CoV-2 Positive | not done | 0.0 | 0.0 | 0.0 | 28.2 | SARS-CoV-2 Negative | |||
| 125 | 2 | 36.9 | 35.6 | SARS-CoV-2 Positive | not done | 0.0 | 0.0 | 0.0 | 29.9 | SARS-CoV-2 Negative | |||
| Respiratory Discordant Specimens | |||||||||||||
| Specimen | Wave | SOC (cobas® SARS-CoV-2) | Xpert® Xpress CoV2/Flu/RSV plus | ||||||||||
| E-gene | ORF1a | Result | CoV2 | Flu A1 | Flu A2 | Flu B | RSV | SPC | Result | ||||
| 82 | i2 | 0.0 | 0.0 | SARS-CoV-2 Negative | 38.5 | 0 | 0 | 0 | 0 | 33.2 | SARS-CoV-2 Positive | FluA, FluB, RSV Negative | |
| 83 | i2 | 0.0 | 0.0 | SARS-CoV-2 Negative | 39.8 | 0 | 0 | 0 | 0 | 30.5 | SARS-CoV-2 Positive | FluA, FluB, RSV Negative | |
| Specimen | Allplex SARS-CoV-2/Flu/RSV | Result | CoV2 | Flu A1 | Flu A2 | Flu B | RSV | SPC | Result | ||||
| 27 | 32.76 | RSVB | 36.1 | 0 | 0 | 0 | 33.2 | 28.9 | SARS-CoV-2, RSV Positive | FluA, FluB Negative | |||
| 30 | 18.79 | RSVA | 40.9 | 0 | 0 | 0 | 24.3 | 29.0 | SARS-CoV-2, RSV Positive | FluA, FluB Negative | |||
| 34 | 24.62 | RSVB | 43.5 | 0 | 0 | 0 | 25.8 | 28.6 | SARS-CoV-2, RSV Positive | FluA, FluB Negative | |||
| AccuPlex | 100 cp/mL | Expected SARS-CoV-2, FluA, FluB, RSV positive | 38.7 | 37.5 | 0 | 35.8 | 38 | 28.9 | SARS-CoV-2, FluA, FluB Positive | RSV Negative | |||
| AccuPlex | 100 cp/mL | 38.2 | 38.4 | 38.7 | 35.4 | 39.7 | 29.5 | SARS-CoV-2, FluA, FluB Positive | FluA2 Negative | ||||
| AccuPlex | 250 cp/mL | 37.2 | 37.3 | 40.6 | 34.4 | 36.8 | 28.9 | SARS-CoV-2, FluA, FluB Positive | FluA2 Negative | ||||
| AccuPlex Dilution (Wildtype) | E-Gene | N2-Gene | RdRp | SPC (Internal Control) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | %CV | Mean | SD | %CV | Mean | SD | %CV | Mean | SD | %CV | ||
| Xpert® Xpress SARS-CoV-2 test | 100 cp/mL | 37.97 | 3.50 | 9.22 | 39.00 | 0.56 | 1.43 | Target not included | 27.93 | 0.25 | 0.90 | ||
| 250 cp/mL | 35.53 | 0.23 | 0.65 | 38.15 | Not calculated | 28.13 | 0.93 | 3.30 | |||||
| 500 cp/mL | 34.47 | 0.46 | 1.34 | 37.27 | 0.23 | 0.62 | 27.87 | 0.25 | 0.90 | ||||
| 1000 cp/mL | 33.23 | 0.40 | 1.22 | 35.80 | 0.40 | 1.12 | 27.90 | 0.26 | 0.95 | ||||
| Overall | 35.30 | 1.15 | 3.11 | 37.55 | 0.40 | 1.05 | 27.96 | 0.42 | 1.51 | ||||
| Xpert® Xpress CoV-2 plus test | 100 cp/mL | 34.63 | 0.15 | 0.44 | 39.37 | 0.91 | 2.30 | 38.17 | 0.15 | 0.40 | 28.43 | 0.29 | 1.02 |
| 250 cp/mL | 33.97 | 0.32 | 0.95 | 37.23 | 0.15 | 0.41 | 37.03 | 0.40 | 1.09 | 28.90 | 0.30 | 1.04 | |
| 1000 cp/mL | 31.60 | 0.17 | 0.55 | 35.17 | 0.50 | 1.43 | 34.67 | 0.46 | 1.33 | 28.50 | 0.36 | 1.27 | |
| 1000 cp/mL | 31.60 | 0.17 | 0.55 | 35.17 | 0.50 | 1.43 | 34.67 | 0.46 | 1.33 | 28.50 | 0.36 | 1.27 | |
| Overall | 33.29 | 0.20 | 0.60 | 37.06 | 0.49 | 1.31 | 36.50 | 0.37 | 1.03 | 28.61 | 0.26 | 0.92 | |
| Type | n | Reference | PPA (95% CI) NPA (95% CI) | Cohen Kappa [38] (95% CI) | Agreement Score [39] |
|---|---|---|---|---|---|
| Combined results any pathogen | |||||
| Residual clinical specimens | 92 | SOC | 100% (95.6, 100) 83.3% (51.6, 97.9) | 0.8972 (0.7569, 1.0374) | Very good |
| Residual clinical specimens | 95 1 | 100% (95.5, 100) 66.7% (38.4, 88.2) | 0.7711 (0.5810, 0.9612) | Good | |
| Results per pathogen (CoV-2, Flu A, Flu B, RSV) and across all diseases Clinically relevant specimens (residual clinical specimens and AccuPlex) | |||||
| CoV-2 | 113 | SOC/expected reference results | 100% (94.1, 100) 90.4% (79.0, 96.8) | 0.9103 (0.8338, 0.9869) | Very Good |
| FluA | 113 | 91.3% (72.0, 98.9) 100% (96.0, 100) | 0.9436 (0.8662, 1.0210) | Very Good | |
| FluB | 113 | 100% (85.2, 100) 100% (96.2, 100) | 1.000 (1.000, 1.000) | Very Good | |
| RSV | 113 | 97.0% (84.2, 99.9) 100% (95.5, 100) | 0.9784 (0.9363, 1.0205) | Very Good | |
| All pathogens | 452 | 97.9% (93.9, 99.6) 98.4% (96.3, 99.5) | 0.9588 (0.9305, 0.9871) | Very Good | |
| Results per pathogen (CoV-2, Flu A, Flu B, RSV) and across all pathogens Residual clinical specimens only | |||||
| CoV-2 | 92 | SOC | 100% (91.2, 100) 90.4% (79.0, 96.8) | 0.8910 (0.7986, 0.9834) | Very Good |
| FluA | 92 | 100% (69.2, 100) 100% (95.6, 100) | 1.000 (1.000, 1.000) | Very Good | |
| FluB | 92 | 100% (69.2, 100) 100% (95.6, 100) | 1.000 (1.000, 1.000) | Very Good | |
| RSV | 92 | 100% (83.2, 100) 100% (95.0, 100) | 1.000 (1.000, 1.000) | Very Good | |
| All pathogens | 368 | 100% (95.5, 100) 98.3% (96.0, 99.4) | 0.9610 (0.9270, 0.9949) | Very Good | |
| AccuPlex Respiratory | CoV-2 | Flu A1 | Flu A2 | Flu B | RSV | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | %CV | Mean | SD | %CV | Mean | SD | %CV | Mean | SD | %CV | Mean | SD | %CV | |
| 100 cp/mL | 38.30 | 0.36 | 0.94 | 37.87 | 0.47 | 1.25 | 38.65 | Not determined | 35.43 | 0.35 | 0.99 | 38.63 | 0.93 | 2.41 | |
| 250 cp/mL | 37.13 | 0.06 | 0.16 | 36.93 | 0.72 | 1.96 | 38.63 | 1.74 | 4.50 | 34.70 | 0.36 | 1.04 | 36.97 | 0.15 | 0.41 |
| 500 cp/mL | 36.03 | 0.31 | 0.85 | 35.80 | 0.17 | 0.48 | 36.70 | 0.17 | 0.47 | 33.50 | 0.10 | 0.30 | 35.77 | 0.12 | 0.32 |
| 1000 cp/mL | 35.20 | 0.10 | 0.28 | 32.83 | 0.10 | 0.30 | 35.63 | 0.06 | 0.16 | 32.83 | 0.71 | 2.16 | 35.10 | 0.66 | 1.87 |
| Overall | 37.72 | 0.21 | 0.55 | 37.40 | 0.60 | 1.60 | 38.64 | 1.74 | 4.50 | 35.07 | 0.36 | 1.02 | 37.80 | 0.54 | 1.41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noble, L.D.; Scott, L.E.; Munir, R.; Du Plessis, M.; Steegen, K.; Hans, L.; Marokane, P.; Da Silva, P.; Stevens, W.S. Rapid Evaluation of the Xpert® Xpress CoV-2 plus and Xpert® Xpress CoV-2/Flu/RSV plus Tests. Diagnostics 2023, 13, 34. https://doi.org/10.3390/diagnostics13010034
Noble LD, Scott LE, Munir R, Du Plessis M, Steegen K, Hans L, Marokane P, Da Silva P, Stevens WS. Rapid Evaluation of the Xpert® Xpress CoV-2 plus and Xpert® Xpress CoV-2/Flu/RSV plus Tests. Diagnostics. 2023; 13(1):34. https://doi.org/10.3390/diagnostics13010034
Chicago/Turabian StyleNoble, Lara Dominique, Lesley Erica Scott, Riffat Munir, Mignon Du Plessis, Kim Steegen, Lucia Hans, Puleng Marokane, Pedro Da Silva, and Wendy Susan Stevens. 2023. "Rapid Evaluation of the Xpert® Xpress CoV-2 plus and Xpert® Xpress CoV-2/Flu/RSV plus Tests" Diagnostics 13, no. 1: 34. https://doi.org/10.3390/diagnostics13010034
APA StyleNoble, L. D., Scott, L. E., Munir, R., Du Plessis, M., Steegen, K., Hans, L., Marokane, P., Da Silva, P., & Stevens, W. S. (2023). Rapid Evaluation of the Xpert® Xpress CoV-2 plus and Xpert® Xpress CoV-2/Flu/RSV plus Tests. Diagnostics, 13(1), 34. https://doi.org/10.3390/diagnostics13010034






