Abstract
Germline predisposition plays an important role in breast cancer. Different ethnic populations need respective studies on cancer risks pertinent to germline variants. We aimed to discover the pathogenic and likely pathogenic variants (P/LP-Vs) of germline breast cancer susceptibility genes and to evaluate their correlation with the clinical characteristics in Jakarta populations. The pure DNA was extracted from the blood buffy coat, using reagents from the QIAamp DNA Mini Kit® (Qiagen, Hilden, Germany). The DNA libraries were prepared using the TargetRich™ Hereditary Cancer Panel (Kailos Genetics®, Huntsville, AL, USA). The barcoded DNA libraries were sequenced using the Illumina NextSeq 500 platform. In-house bioinformatics pipelines were used to analyze the gene variants. We identified 35 pathogenic and likely pathogenic (P/LP-Vs) variants (28 frameshift, 5 nonsense, and 2 splice-site variants). The P/LP-Vs group was statistically significantly different in luminal B status (p < 0.05) compared with the non-P/LP-Vs group. The P/LP-Vs found both in BRCA1/2 genes and non-BRCA genes may increase the risk of breast cancer and alter drug responses. The screening of multigene variants is suggested, rather than BRCA testing only. Prior knowledge of the germline variants status is important for optimal breast cancer diagnosis and optimal therapy.
1. Introduction
Among developing nations, breast cancer is the leading cause of mortality and it is the most prevalent type of cancer in women globally. Globally, 2.3 million cases were recorded by GLOBOCAN in 2020, representing the fifth cause of cancer-related mortality, with cases in Asia being higher than for any other continent. By 2020, breast cancer continued to be the most common new case of cancer in women (30.8%) and was the leading cause of death in Indonesia (15.3%) [1].
A new method of genetic study has emerged with the development of next-generation sequencing (NGS) technology. For screening individual genes and small gene panels in clinical settings, NGS has established itself as an alternative, cost-effective tool. The finding of the susceptibility genes linked to cancer risk was accelerated by NGS in the late 2000s [2]. Compared with BRCA1/2 testing alone, multi-gene panel testing could significantly uncover the cancer risk gene variations in more patients [3].
Breast cancer is the most prevalent type of cancer in women, and inherited predisposition is a significant factor. Better information and comprehension of the underlying genetic factors would thus be beneficial for the management and prevention of breast cancer [4]. Practice recommendations for breast cancer will be influenced by knowledge of the prognosis following a diagnosis and the risk assessment of genetic mutations of cancer susceptibility. Many clinical trials are currently evaluating the use of poly (ADP-ribose) polymerase (PARP) inhibitors in the treatment of breast cancer because of their effectiveness in controlling BRCA mutation positive tumors. It is anticipated that these agents will be incorporated into systemic therapy in clinical practice. As a result, systemic therapy for BRCA mutation-associated breast cancer has been made based on the presence of BRCA mutations rather than the disease features only [5].
The genes BRCA1, BRCA2, PALB2, TP53, ATM, CHEK2, STK11, and NBN have all been identified as breast cancer risk genes [6]. BRCA1, BRCA2, CDH1, PTEN, STK11, and TP53 are examples of high-penetrance genes, while ATM, CHEK2, BARD1, and RAD51D are examples of moderate-penetrance genes [7]. Other research has shown that harmful variations in RAD51C, which is crucial for HR repair, increase the chance of developing breast and ovarian cancer [8]. The DNA mismatch repair genes (MLH1, MSH2, MSH6, and PMS2), which are germline variants that are mostly linked to Lynch syndrome, have been found to be correlated with breast cancer risk or survival [5]. It is interesting to note that Chinese and Japanese populations have pathogenic BRIP1 variants that have not been found in Western populations. The carrier frequency of the detrimental PALB2 mutation is different from that in Western populations, similar to the BRIP1 mutation. This variation might be caused by the ethnicity of the genome [8]. We investigated the germline variant linked to breast cancer risk and outcomes in our cohort using gene panels in order to better understand these variants.
It is necessary to conduct individual research in various ethnic populations in order to determine cancer risks related to germline variations. The autosomal dominant conditions most frequently linked to a high risk of breast cancer are hereditary breast and ovarian malignancies brought on by mutations in BRCA1 or BRCA2. Numerous research in Asian groups revealed findings that were distinct from those found in Caucasian people [5,9,10]. In contrast with Western countries, where the incidence of breast cancer rises among postmenopausal women in their 60s, the incidence of breast cancer peaks in premenopausal women in their 40s in Asian countries [11]. Asian patients may have breast cancer at a younger age than their Caucasian peers because their BRCA1/2 mutations are thought to have different contributions than those of Caucasians [12]. Enhancing the database of variants in various ethnic communities is essential for improving the interpretation of ethnically specific germline variants and the management of cancer risks in ethnically appropriate populations. In this work, we sought to identify pathogenic and likely pathogenic variants (P/LP-Vs) of germline genes associated with breast cancer susceptibility and to assess their association with clinical traits in the Jakarta population.
2. Materials and Methods
2.1. Patients
The study was approved by the Ethical Committee at Faculty of Medicine Universitas Indonesia on 16 October 2017 (approval number: 958/UN2.F1/ETIK/2017). All procedures were carried out in conformity with the applicable rules and regulations. Before the patients’ samples were taken, all of the patients gave their informed consent for this study. Through participant surveys and computerized medical records, clinical data were gathered.
A total of 75 female breast cancer patients under the age of 45 who were receiving treatment at the Surgical Oncology Division, Department of Surgery, Cipto Mangunkusumo National Hospital Jakarta were included in this study. For the purposes of this analysis, all patients who had breast cancer as determined by histology and immunohistochemistry between January 2014 and December 2017 were eligible. Patients with incomplete histology and medical record information were disqualified from this investigation. None of the participants knew their gene variations associated with cancer risk prior to being recruited. Distant metastases and stage grouping classification were defined according to the American Joint Committee for Cancer (AJCC) 7th Edition [13]. Molecular subtypes (Luminal A, Luminal B, HER2, and triple-negative) were defined based on hormone receptors and HER2 status.
2.2. DNA Extraction, Library Preparation, and Sequencing
Pure DNA was extracted from the blood buffy coat, using reagents from the QIAamp DNA Mini Kit® (Qiagen, Hilden, Germany). TargetRichTM Hereditary Cancer Panel (Kailos Genetics®, Huntsville, AL, USA) was used to create the DNA libraries. This panel is 22 genes targeted-sequencing panel (Table S1). TargetRichTM UMI/Index Adapter Plate (Kailos Genetics®, Huntsville, AL, USA) was combined with DNA libraries to enable sample barcoding, multiplex sequencing, and tagging of individual collected DNA molecules. The Illumina NextSeq 500 platform was used for the sequencing of the barcoded DNA libraries. All of the isolation and genome sequencing methods of 75 samples were described in Panigoro et al., 2020 [14].
2.3. Bioinformatics Analysis
Using FastQC 0.11.9 [15], the paired-end raw readings from each sample were examined for quality. After quality control, using the Trimmomatic [16] program, all of the low-quality reads were eliminated before the alignment phase. The BWA algorithm was used to align the fastq files with the hg38 human reference genome (Genome Reference Consortium GRCh38), which was retrieved from ENSEMBL (http://asia.ensembl.org/Homo_sapiens/Info/Index (accessed on 5 January 2022)) [17]. With the use of the Genome Analysis Toolkit (GATK, version 4.1.9) [18], the base quality score was recalculated. HaplotypeCaller from GATK (4.1.9 version) was used for the variant calling methods. The variant calling file output then went to the hard-filtering process using specific thresholds and throwing out any variants that had annotation values above or below the set threshold. The thresholds for SNV were QualByDepth (QD) > 2.0, FisherStrand(FS) < 60, StrandOddsRatio (SOR) < 3, and RMSMappingQuality (MQ) > 40.0. The thresholds for MNV were QualByDepth (QD) > 2.0, FisherStrand(FS) < 200.0 and StrandOddsRatio (SOR) < 10.0.
All of the variants were annotated using snpEff [19] (GRCh38.101 annotation database version), and snpSift (dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/ (accessed on 10 January 2022)) annotated the variants with synonymous, missense, nonsense, frameshift, and splicing variants; ClinVar databases (http://www.ncbi.nlm.nih.gov/clinvar/ (accessed on 10 January 2022)) classified the variants as follows: 0—uncertain significance, 1—not provided, 2—benign, 3—likely benign, 4—likely pathogenic, 5—pathogenic, 6—drug response, 7—histocompatibility, and 255—other. The impact of novel variants on the protein function or structure was analyzed using VarSome [20], an integrated search engine that allows to access multiple databases, prediction tools, and publications at a single site. Nonsense, frameshift, and splice-site variants that resulted in a truncated protein product were classified as pathogenic.
SnpSift from the SnpEff package was used to filter the raw variants. Filtered variants were exported from variant files into tab-delimited files using SnpSift and were concatenated into a single tab-delimited file including all variants of all patients. Variants were visualized using Lollipop plot by MutationMapper (https://www.cbioportal.org/mutation_mapper (accessed on 2 February 2022)) [21,22]. The raw data of all samples were deposited in the SRA under BioProject number: PRJNA606794 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA606794 (accessed on 25 December 2021)).
2.4. Clinical Correlation and Statistical Analysis
For the study participants who previously had breast cancer, age of onset and familial history were taken from participant survey. Using the Chi-square test or t test, statistical correlations between the clinical features and P/LP-Vs or non-P/LP-Vs status were assessed. p-values and 95% confidence intervals were used to show statistical significance. To reject the null hypothesis, an alpha level of 0.05 was deemed statistically significant different. Prism 9.0.0 software was used for all of the analyses (trial version).
3. Results
3.1. Patients’ Characteristics
There were 75 patients aged ≤45 years at diagnosis, among whom 18 patients were included with familial breast cancer (mother or aunt or grandmother had breast cancer), and the remaining 57 patients were women without a familial history of breast cancer. The clinical characteristics of the patients are shown in Table 1. The mean age of the breast cancer group was 34 years. Regarding tumor histopathology, patients with familial breast cancer exhibited stage I (4; 22.2%), II (5; 27.8%), III (3; 16.7%), and IV (6; 33.3%), whether in sporadic patients exhibited stage I (9; 15.8%), II (22; 38.6%), III (16; 28.113%), and IV (10; 17.5%).

Table 1.
Demographic characteristics of the breast cancer patients.
3.2. Characteristics of the Germline Variants
In this study, we sequenced the established breast cancer susceptibility genes in 75 breast cancer patients. We annotated the variants based on the dbSNP and ClinVar databases. Novel variants that had not yet been found in the public databases were predicted using VarSome software. Pathogenic mutations included nonsense, frameshift, and splice-site variations that lead to shortened protein products. A flow chart of germline variant characteristics is shown in Figure 1.
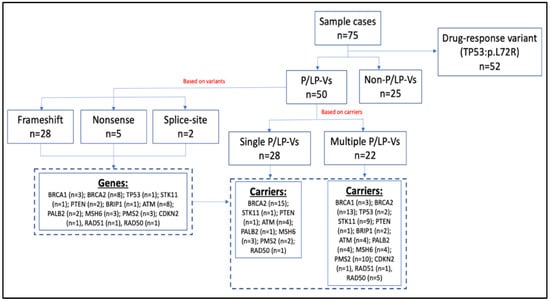
Figure 1.
Flow chart of the germline variants characteristics.
From Figure 1, we could identify 50 patients (67%) that carried P/LP-Vs. Based on the carriers, there were 28 (56%) of P/LP-Vs patients carried single P/LP-Vs and the remaining 22 patients (44%) carried multiple P/LP-Vs (Table S2). Based on the gene variants, we found 35 P/LP-Vs (Table 2) that contained 28 frameshift, 5 nonsense, and 2 splice-site variants.

Table 2.
Pathogenic and likely pathogenic germline variants (P/LP-Vs).
In the distribution of genes in single and multiple P/LP-Vs diagram (Figure 2), BRCA2 was the most predominant in both of the groups, with 53% and 22% in single P/LP-Vs carriers and multiple P/LP-Vs carriers (Table S2), respectively. Potentially pathogenic VUS missense variants were also found in this study. We identified 31 variants of uncertain significance (VUS), of which most of the variants had been recorded in the dbSNP database (Table S3).
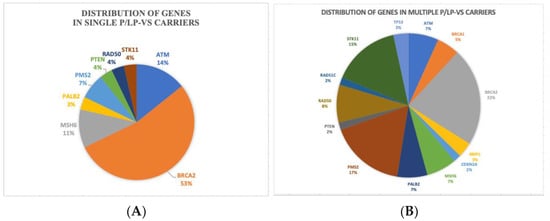
Figure 2.
Distribution of genes in: (A) single P/LP-Vs carriers, (B) multiple P/LP-Vs carriers.
The positions of the pathogenic variants for high-penetrance genes, such as BRCA1, BRCA2, PTEN, STK11, and TP53, were visualized in Lollipop plot, as shown in Figure 3. Most of the P/LP-Vs had not been recorded yet in the dbSNP or CinVar databases, and were designated as novel variants with their clinical significance predicted by VarSome.
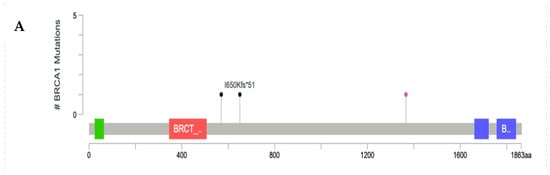
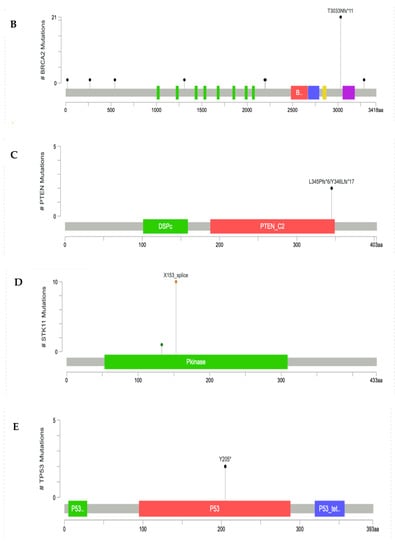
Figure 3.
Lollipop plot of P/LP-Vs in high-penetrance genes: (A) BRCA1 gene, (B) BRCA2 gene, (C) PTEN gene, (D) STK11 gene, and (E) TP53 gene. *: terminated protein.
3.3. Prediction of Drug Response Alteration Associated with Gene Variants
There were some variants that showed alterations to drug therapy, based on the Onco-KB database prediction (Table 2). Patients with certain P/LP-Vs were predicted to benefit from targeted therapy. P/LP-Vs in BRCA1/2, BRIP1, ATM, PALB2, and ATM genes (23 of 36 P/LP-Vs) were predicted to benefit from the PARP inhibitor (Talazoparib or Olaparib). We also found one frameshift variant in CDKN2A that could be sensitive to CDK4/6 inhibitors such as Palbociclib, Ribociclib, and Abemaciclib [23,24]. Related to two variants in the PTEN gene, we found that loss-of-function variants could be sensitive to beta-isoform-selective PI3K-targeted inhibitors, such as the investigational agents GSK2636771 and AZD8186 [25]. Besides P/LP-Vs in several genes, there was a missense variant of TP53:p.P72R (dbSNP ID: rs1042522) in 52 patients (69.3%) that resistance to neoadjuvant chemotherapy, such as 5-fluorouracil and cisplatin.
3.4. Correlation of Variants with Tumor Characteristics
To characterize the clinical features upon cancer diagnosis in the germline variant carriers, we compared clinical characteristics between the pathogenic and likely pathogenic variant (P/LP-Vs) group and non-P/LP-Vs group (Table 3). None of the patients had prior knowledge of cancer susceptibility gene variant status at the time of their breast cancer diagnosis or treatment.

Table 3.
Cancer characteristics of breast cancer patients and their correlation with pathogenic and likely pathogenic variants (P/LP-Vs) (N = 75).
Age of onset for breast cancer patients with P/LP-Vs with P/LP-Vs variants and non-P/LP-Vs was not statistically significantly different, but for the respective mean age of onset, patients with the P/LP-Vs variant showed a younger in age of onset (33.7 years old) than the patients without the P/LP-Vs variant (34.8 years old). This is because all of the samples were young women. The triple negative characteristics were shown to be statistically significant differences in the non-P/LP-Vs variant group. In the P/LP-Vs group, a statistically significant difference was found in the higher percentage of Luminal B status than in the non-P/LP-Vs group. This result could be due to the fact that the number of patients with luminal B was twice as high as the triple negative patients.
4. Discussion
The probability of dying from breast cancer and distant metastases was considerably higher for BRCA1 and BRCA2 mutation carriers [26]. Many breast cancer patients who had a higher inherited risk of developing the disease tested negative for BRCA1 or BRCA2 P/LP-Vs, necessitating additional genetic testing using larger gene panels. Because of sequencing limitations, some individuals who test negative for BRCA1/2 P/LP-Vs may also have P/LP-Vs in other cancer susceptibility genes, such as TP53, PALB2, ATM, PTEN, STK11, and others, in 3–4% cases [3,27]. For this reason, the usage of multi-gene panels encompassing multiple susceptibility genes beyond BRCA1/2 is increasing because of the advancements made by next-generation sequencing (NGS) technology, which have transformed clinical approaches to genetic testing [6,28].
In this study, we identified 35 P/LP-Vs in the 13 established hereditary breast cancer genes (Table 2). Although ClinVar has registered many pathogenic variants in those 13 genes, only 20% (7/35) of P/LP-Vs have been registered. BRCA2 genes were found to be the most predominant genes with P/LP-Vs. In potentially pathogenic VUS variants, we also found 31 variants in 15 genes, and VUS variants in the MSH6 gene were found to be predominant among the others. Unfortunately, we did not have the recurrence data from all of the patients, so we could not analyze the impact of P/LP-Vs in BRCA1/2 genes with recurrence. However, in several previous studies, P/LP-Vs in BRCA1/2 genes could drive recurrence [5,29,30,31].
Previous studies held in the Asian region showed that the prevalence of P/LP-Vs of BRCA1/2 genes among women of a young age of onset and with familial history were 8.8% to 26.7% (Table 4). In our result, the variant rate of BRCA1/2 in familial breast cancer (FBC) samples was 44.4% (8/18). Overall, our variant rate of P/LP-Vs of BRCA1/2 in FBC was higher than previous studies findings (Table 4). Howver, the discrepancy regarding the high percentage in these studies may in part result from limitations due to small sample sizes.

Table 4.
Previous studies analyzing BRCA variants using NGS in young breast cancer patients.
In our study, the P/LP-Vs group showed statistically significant difference with the non-P/LP-Vs group in Luminal B (p < 0.05), which might be because of P/LP-Vs that were found to be predominant in the BRCA2 gene. This result was in line with previous studies that showed that approximately 75% of BCs in BRCA2 PV carriers were more often Luminal B. On the other hand, TNBCs were found to make up around 70% of BCs developing in BRCA1 PV carriers [36,37,38].
Patients with P/LP-Vs, such as in BRCA1/2, BRIP1, ATM, PALB2, and ATM genes, were predicted to benefit from the PARP inhibitor (Talazoparib or Olaparib). Cancer cells with harmful mutations in breast cancer susceptibility genes 1 or 2 (BRCA1/2) lack the ability to repair DNA double strand breaks, making these tumors heavily reliant on the single strand break repair pathway. The poly(adenosine diphosphate-ribose) polymerase (PARP) enzyme controls this pathway. Inhibition of PARP results in cell death in BRCA1/2 mutant cells as a result of the accumulation of irreparable DNA damage [39].
The CDKN2A gene is a gene that encodes two proteins, p16INK4A and p14ARF, which regulate cell growth and survival. One of our patients that had P/LP-Vs in the CDKN2A gene may have benefited from CDK4/6 inhibitors. Laboratory data suggest that cancer cells with loss-of-function alterations of CDKN2A may be sensitive to CDK4/6 inhibitors such as Palbociclib, Ribociclib, and Abemaciclib. In ER+ and HER2-amplified cell lines, Palbociclib was shown to be synergistic with Tamoxifen and Trastuzumab, respectively. In cell lines that had conditioned resistance to ER blocking, Palbociclib increased sensitivity to Tamoxifen [23].
One of the genes in cancer that is most frequently altered is PTEN, a lipid and protein phosphatase. Many malignancies, including breast cancer, frequently lose PTEN function. Preclinical research has shown that the essential lipid kinase that predominantly regulates PI3K pathway activation, cell proliferation, and survival in PTEN-deficient tumor cells is the PI3Kβ isoform (containing the p110β catalytic subunit). Related to variants in the PTEN gene, we found that two patients with P/LP-Vs could be sensitive to beta-isoform selective PI3K-targeted inhibitors, such as the investigational agents GSK2636771 and AZD8186 [25].
Besides P/LP-Vs in several genes, there was a missense variant in the TP53 genes (p. P72R) that was found in 52 patients (69.3%). Variants of apoptosis-related genes, such as TP53 codon 72, are associated with the susceptibility or prognosis of solid tumors. The TP53 codon 72 variant was found to be a strong predictor of the pathologic response to neoadjuvant chemotherapy in breast cancer [40,41]. Based on this finding, we suggest that clinicians consider using genomic sequencing to identify the presence of variants in BRCA1/2 and other genes related to breast cancer, such as TP53, ATM, PTEN, and CDKN2A. Genomic sequencing could lead to precision medicine. For example, if there are BRCA1/2 pathogenic variants, clinicians could consider using a PARP inhibitor; if there is a P72R mutation in the TP53 gene, clinicians could avoid using 5-Fluorouracil and Cisplatin because of resistance prediction. The limitations of our study were related to the small number of breast cancer patients. Compared with breast cancer populations, our sample size was small, only around 2.6%. In Indonesia, genomic sequencing research is an expensive research field. We tried to explore genomic aspects in breast cancer, although with limited resources. In addition to this limitation, we realized that we had a bias number in the patient characterization distribution, for example, in familial history distribution (n = 18 of 75) and TNBC subtype (n = 18 of 75). So, this might have resulted in a high variant rate of P/LP-Vs compared with previous studies.
5. Conclusions
The P/LP-Vs found both in BRCA1/2 genes and non-BRCA genes may increase the risk of breast cancer and alter drug responses. The screening of multigene variants is suggested, rather than BRCA testing only. Prior knowledge of the germline variant status is important for optimal breast cancer diagnosis and therapy.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/diagnostics12092241/s1. Table S1: List of genes of TargetRichTM Hereditary Cancer Panel. Table S2: Distribution of P/LP-Vs in multiple P/LP-Vs carriers. Table S3: Variants of uncertain significance (VUS) found among the samples.
Author Contributions
Conceptualization: S.S.P., R.I.P. and F.F.; methodology: S.S.P., K.M.S. and R.I.P.; formal analysis: R.I.P.; writing—original draft preparation and editing: R.I.P.; supervision: S.S.P. and F.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Universitas Indonesia (PUTI 2022 grant; grant number: NKB-213/UN2.RST/HKP.05.00/2022).
Institutional Review Board Statement
The study was approved by the Ethical Committee at the Faculty of Medicine Universitas Indonesia on 16 October 2017 (approval number: 958/UN2.F1/ETIK/2017). All of the methods were performed in accordance with the relevant guidelines and regulations.
Informed Consent Statement
Informed consent for this study was obtained from all of the patients before the patients’ samples were collected. Clinical information was collected through electronic medical records and participant surveys.
Data Availability Statement
The raw data of all samples were deposited in the SRA under BioProject number: PRJNA606794 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA606794 (accessed on 25 December 2021)).
Acknowledgments
We wish to thank the Surgical Oncology Division, Department of Surgery, Cipto Mangunkusumo National Hospital Jakarta; Bioinformatics Core Facilities-IMERI, Faculty of Medicine, Universitas Indonesia; and the Department of Medical Chemistry, Faculty of Medicine, Universitas of Indonesia for supporting this research.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Observatory, G.C. Breast Fact Sheet. 2020, Volume 419. Available online: https://gco.iarc.fr/today/data/factsheets/cancers/20-Breast-fact-sheet.pdf (accessed on 4 March 2022).
- Chan, W.; Lee, M.; Yeo, Z.X.; Ying, D.; Grimaldi, K.A.; Pickering, C.; Yang, M.M.S.; Sundaram, S.K.; Tzang, L.C.H. Development and validation of next generation sequencing based 35-gene hereditary cancer panel. Hered. Cancer Clin. Pract. 2020, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Desmond, A.; Kurian, A.W.; Gabree, M.; Mills, M.A.; Anderson, M.J.; Kobayashi, Y.; Horick, N.; Yang, S.; Shannon, K.M.; Tung, N.; et al. Clinical Actionability of Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Risk Assessment. JAMA Oncol. 2015, 1, 943–951. [Google Scholar] [CrossRef]
- Inagaki-Kawata, Y.; Yoshida, K.; Kawaguchi-Sakita, N.; Kawashima, M.; Nishimura, T.; Senda, N.; Shiozawa, Y.; Takeuchi, Y.; Inoue, Y.; Sato-Otsubo, A.; et al. Genetic and clinical landscape of breast cancers with germline BRCA1/2 variants. Commun Biol. 2020, 3, 578. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.A.; Jian, J.-W.; Hung, C.-F.; Peng, H.-P.; Yang, C.-F.; Cheng, H.-C.S.; Yang, A.-S. Germline breast cancer susceptibility gene mutations and breast cancer outcomes. BMC Cancer 2018, 18, 315. [Google Scholar] [CrossRef] [PubMed]
- Easton, D.F.; Pharoah, P.D.P.; Antoniou, A.C.; Tischkowitz, M.; Tavtigian, S.V.; Nathanson, K.L.; Devilee, P.; Meindl, A.; Couch, F.J.; Southey, M.; et al. Gene-Panel Se-quencing and the Prediction of Breast-Cancer Risk. N. Engl. J. Med. 2015, 372, 2243–2257. [Google Scholar] [CrossRef] [PubMed]
- Couch, F.J.; Shimelis, H.; Hu, C.; Hart, S.N.; Polley, E.C.; Na, J.; Hallberg, E.; Moore, R.; Thomas, A.; Lilyquist, J.; et al. Associations Between Cancer Predisposition Testing Panel Genes and Breast Cancer. JAMA Oncol. 2017, 3, 1190–1196. [Google Scholar] [CrossRef]
- Sato, K.; Koyasu, M.; Nomura, S.; Sato, Y.; Kita, M.; Ashihara, Y.; Adachi, Y.; Ohno, S.; Iwase, T.; Kitagawa, D.; et al. Mutation status of RAD 51C, PALB 2 and BRIP 1 in 100 Japanese familial breast cancer cases without BRCA 1 and BRCA 2 mutations. Cancer Sci. 2017, 108, 2287–2294. [Google Scholar] [CrossRef]
- Ripperger, T.; Gadzicki, D.; Meindl, A.; Schlegelberger, B. Breast cancer susceptibility: Current knowledge and implications for genetic counselling. Eur. J. Hum. Genet. 2009, 17, 722–731. [Google Scholar] [CrossRef]
- Dong, H.; Chandratre, K.; Qin, Y.; Zhang, J.; Tian, X.; Rong, C.; Wang, N.; Guo, M.; Zhao, G.; Wang, S.M. Prevalence of BRCA1/BRCA2 pathogenic variation in Chinese Han population. J. Med. Genet. 2020, 58, 565–569. [Google Scholar] [CrossRef]
- Kharel, S.; Shrestha, S.; Yadav, S.; Shakya, P.; Baidya, S.; Hirachan, S. BRCA1/BRCA2 mutation spectrum analysis in South Asia: A systematic review. J. Int. Med. Res. 2022, 50, 3000605211070757. [Google Scholar] [CrossRef]
- Kim, H.; Choi, D.H. Distribution of BRCA1 and BRCA2 mutations in asian patients with breast cancer. J. Breast Cancer 2013, 16, 357–365. [Google Scholar] [CrossRef]
- Edge, S.B.; Byrd, D.R.; Compton, C.C.; Fritz, A.G.; Greene, F.L.; Trotti, A. Breast. In AJCC Cancer Staging Handbook, 7th ed; Springer: New York, NY, USA, 2010; pp. 345–376. Available online: http://link.springer.com/10.1007/978-0-387-88443-1_29 (accessed on 4 March 2022).
- Panigoro, S.S.; Siswiandari, K.M.; Paramita, R.I.; Fadilah, F.; Erlina, L. Targeted genome sequencing data of young women breast cancer patients in Cipto Mangunkusumo national hospital, Jakarta. Data Brief 2020, 32, 106138. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 25 January 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Poplin, R.; Ruano-Rubio, V.; DePristo, M.A.; Fennell, T.J.; Carneiro, M.O.; Van der Auwera, G.A.; Kling, D.E.; Gauthier, L.D.; Levy-Moonshine, A.; Roazen, D.; et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 2017. preprint. [Google Scholar]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Aguilera, M.A.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Ge-nomics and Clinical Profiles Using the cBioPortal. Sci Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Finn, R.S.; Dering, J.; Conklin, D.; Kalous, O.; Cohen, D.J.; Desai, A.J.; Ginther, C.; Atefi, M.; Chen, I.; Fowst, C.; et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009, 11, R77. [Google Scholar] [CrossRef]
- Van Arsdale, T.; Boshoff, C.; Arndt, K.T.; Abraham, R.T. Molecular pathways: Targeting the cyclin D-CDK4/6 axis for cancer treatment. Clin. Cancer Res. 2015, 21, 2905–2910. [Google Scholar]
- Mateo, J.; Ganji, G.; Lemech, C.; Burris, H.A.; Han, S.-W.; Swales, K.; Decordova, S.; Deyoung, M.P.; Smith, D.A.; Kalyana-Sundaram, S.; et al. A First-Time-in-Human Study of GSK2636771, a Phosphoinositide 3 Kinase Beta-Selective Inhibitor, in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2017, 23, 5981–5992. [Google Scholar] [CrossRef] [PubMed]
- Baretta, Z.; Mocellin, S.; Goldin, E.; Olopade, O.I.; Huo, D. Effect of BRCA germline mutations on breast cancer prognosis. Medicine 2016, 95, e4975. [Google Scholar] [CrossRef] [PubMed]
- Tung, N.; Battelli, C.; Allen, B.; Kaldate, R.; Bhatnagar, S.; Bowles, K.; Timms, K.; Garber, J.E.; Herold, C.; Ellisen, L.; et al. Frequency of mutations in individuals with breast cancer referred for BRCA 1 and BRCA 2 testing using next-generation sequencing with a 25-gene panel. Cancer 2015, 121, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.C.; Lee, H.B.; Yoo, T.K.; Lee, E.S.; Kim, R.N.; Park, B.; Yoon, K.A.; Park, C.; Lee, E.S.; Moon, H.G.; et al. Detection of Germline Mutations in Breast Cancer Pa-tients with Clinical Features of Hereditary Cancer Syndrome Using a Multi-Gene Panel Test. Cancer Res. Treat. 2020, 52, 697–713. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.A.; Clifford, J.; Li, S.; LaDuca, H.; Hulick, P.; Gutierrez, S.; Black, M.H. Prevalence of Germline Pathogenic and Likely Pathogenic Variants in Patients with Second Breast Cancers. JNCI Cancer Spectr. 2020, 4, pkaa094. [Google Scholar] [CrossRef] [PubMed]
- Valachis, A.; Nearchou, A.D.; Lind, P. Surgical management of breast cancer in BRCA-mutation carriers: A systematic review and meta-analysis. Breast Cancer Res. Treat. 2014, 144, 443–455. [Google Scholar] [CrossRef]
- Jiang, Y.; Tian, T.; Yu, C.; Zhou, W.; Yang, J.; Wang, Y.; Wen, Y.; Chen, J.; Dai, J.; Jin, G.; et al. Identification of Recurrent Variants in BRCA1 and BRCA2 across Multiple Cancers in the Chinese Population. BioMed Res. Int. 2020, 2020, 6739823. [Google Scholar] [CrossRef] [PubMed]
- Jouali, F.; Laarabi, F.Z.; Marchoudi, N.; Ratbi, I.; Elalaoui, S.C.; Rhaissi, H.; Fekkak, J.; Sefiani, A. First application of next-generation se-quencing in Moroccan breast/ovarian cancer families and report of a novel frameshift mutation of the BRCA1 gene. Oncol Lett. 2016, 12, 1192–1196. [Google Scholar] [CrossRef][Green Version]
- Bakkach, J.; Mansouri, M.; Derkaoui, T.; Loudiyi, A.; El Fahime, E.; Barakat, A.; Nourouti, N.G.; De Villarreal, J.M.; Bringas, C.C.; Mechita, M.B. Contribution of BRCA1 and BRCA2 germline mutations to early onset breast cancer: A series from north of Morocco. BMC Cancer 2020, 20, 859. [Google Scholar] [CrossRef]
- Lolas Hamameh, S.; Renbaum, P.; Kamal, L.; Dweik, D.; Salahat, M.; Jaraysa, T.; Abu Rayyan, A.; Casadei, S.; Mandell, J.B.; Gulsuner, S.; et al. Genomic analysis of inherited breast cancer among Palestinian women: Genetic heterogeneity and a founder mutation in TP53. Int. J. Cancer 2017, 141, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Akcay, I.M.; Celik, E.; Agaoglu, N.B.; Alkurt, G.; Msc, T.K.A.; Yildiz, J.; Enc, F.; Kir, G.; Canbek, S.; Kilic, A.; et al. Germline pathogenic variant spectrum in 25 cancer susceptibility genes in Turkish breast and colorectal cancer patients and elderly controls. Int. J. Cancer 2021, 148, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Incorvaia, L.; Fanale, D.; Bono, M.; Calò, V.; Fiorino, A.; Brando, C.; Corsini, L.R.; Cutaia, S.; Cancelliere, D.; Pivetti, A.; et al. BRCA1/2 pathogenic variants in triple-negative versus luminal-like breast cancers: Genotype–phenotype correlation in a cohort of 531 patients. Ther. Adv. Med. Oncol. 2020, 12, 175883592097532. Available online: http://www.ncbi.nlm.nih.gov/pubmed/28086035 (accessed on 15 March 2022). [CrossRef]
- Larsen, M.J.; Kruse, T.A.; Tan, Q.; Lænkholm, A.V.; Bak, M.; Lykkesfeldt, A.E.; Sørensen, K.P.; Hansen, T.V.O.; Ejlertsen, B.; Gerdes, A.M.; et al. Classifications within Molecular Sub-types Enables Identification of BRCA1/BRCA2 Mutation Carriers by RNA Tumor Profiling. PLoS ONE 2013, 8, e64268. [Google Scholar]
- Shah, P.D.; Patil, S.; Dickler, M.N.; Offit, K.; Hudis, C.A.; Robson, M.E. Twenty-one-gene recurrence score assay in BRCA-associated versus sporadic breast cancers: Differences based on germline mutation status. Cancer 2016, 122, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Gonçalves, A.; Lee, K.-H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018, 379, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Ganji, G.; Lemech, C.; Burris, H.A.; Han, S.W.; Swales, K.; Kim, M.K.; Lee, K.H.; Hyun, M.S.; Lee, W.S.; et al. TP53 codon 72 polymorphism associated with prognosis in patients with advanced gastric cancer treated with paclitaxel and cisplatin. Clin. Cancer Res. 2009, 11, 355–360. [Google Scholar]
- Xu, Y.; Yao, L.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Lin, B.; Lu, Y.; Xie, Y. p53 Codon 72 Polymorphism Predicts the Pathologic Response to Neoadjuvant Chemotherapy in Patients with Breast Cancer. Clin. Cancer Res. 2005, 11, 7328–7333. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).