Characterization of Exosomes and Exosomal RNAs Isolated from Post-Mortem Body Fluids for Molecular Forensic Diagnosis
Abstract
:1. Introduction
2. Materials and methods
2.1. Sample Collection and Preparation
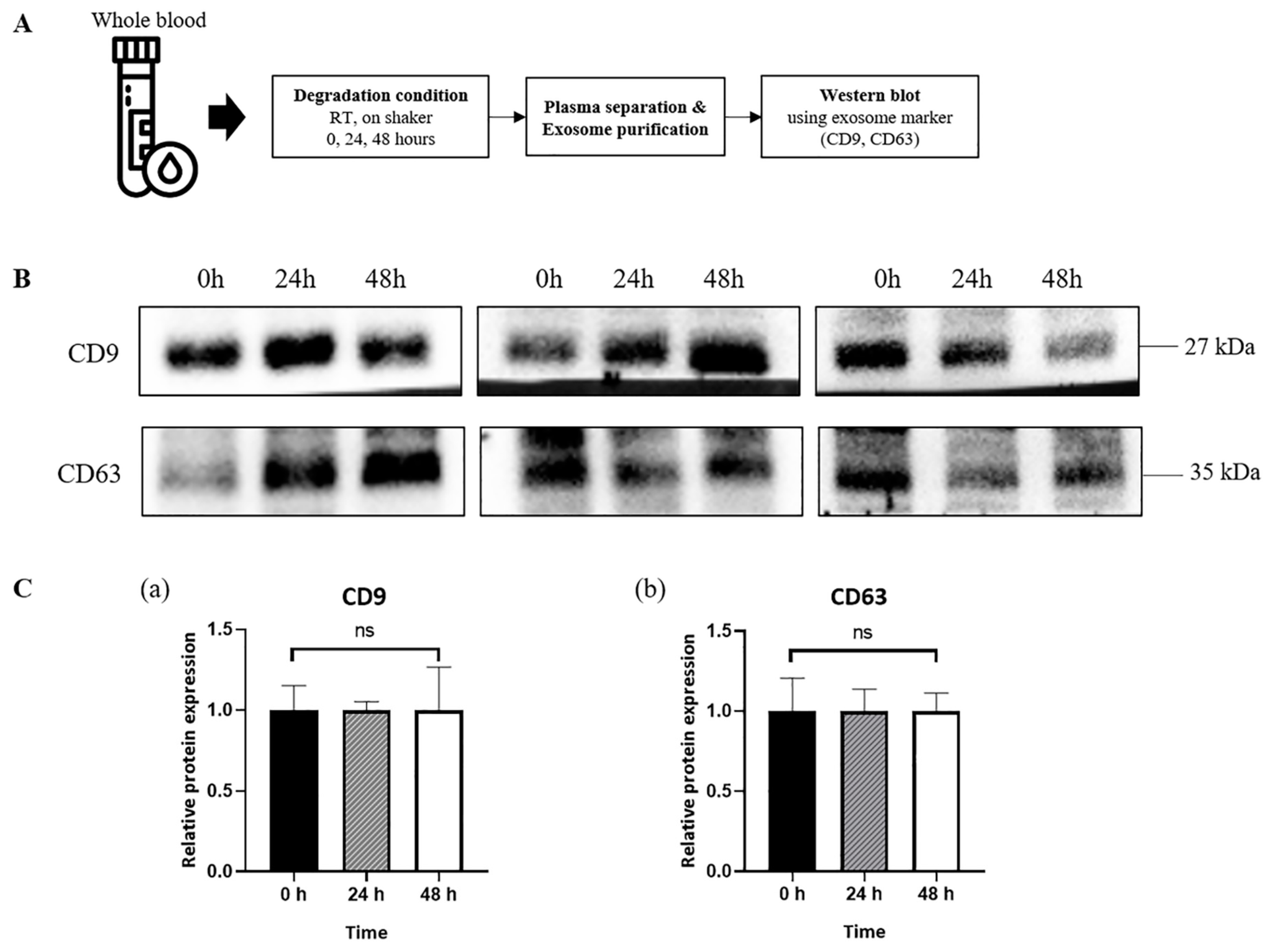
2.2. Simulation of Post-Mortem Changes in Blood Samples
2.3. Exosome and Exosomal RNA Isolation and Quantification
2.4. Western Blot Analysis
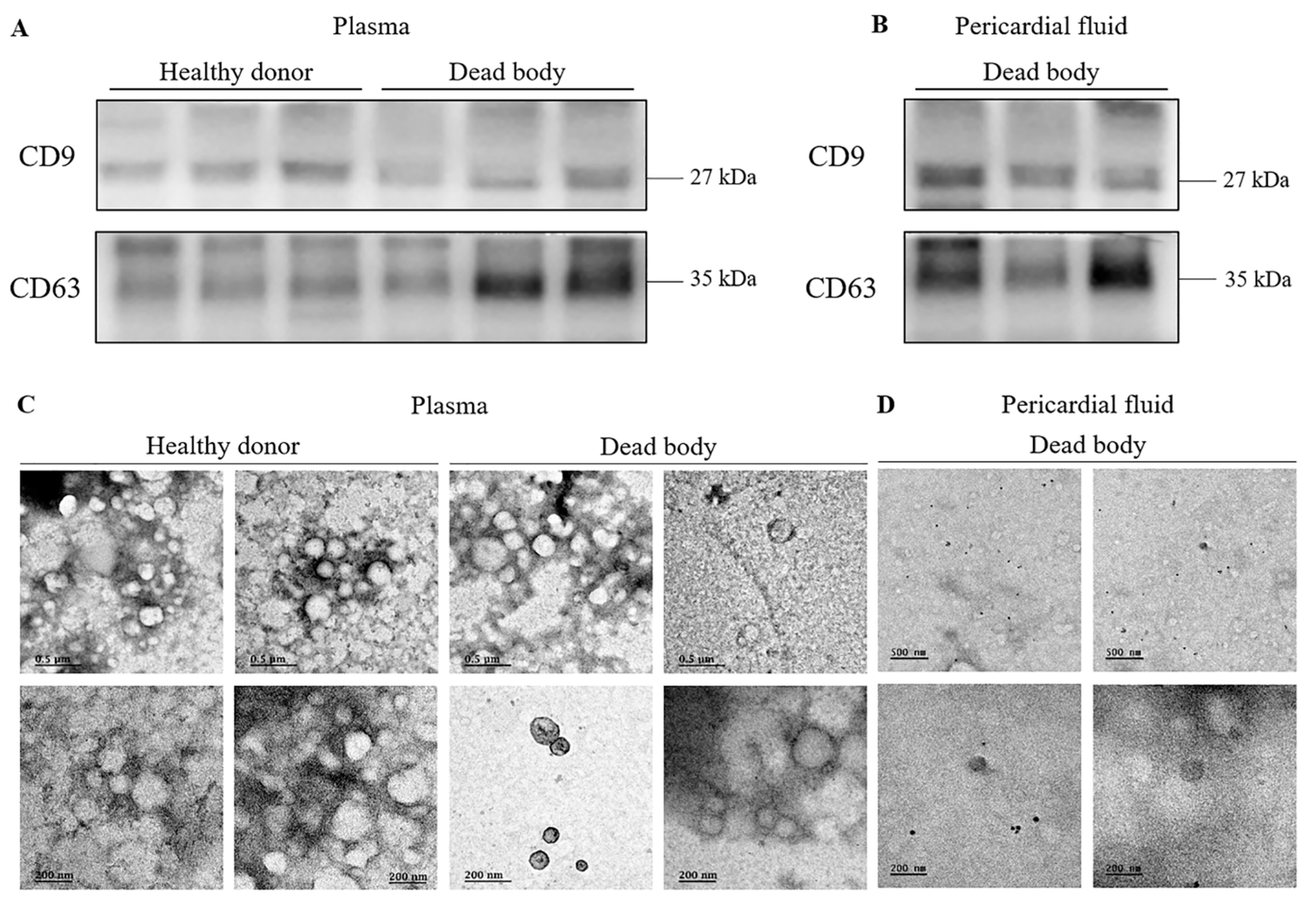
2.5. Transmission Electron Microscopy (TEM)
2.6. Statistical Analysis
3. Results
3.1. Identification and Evaluation of Exosomes Isolated from Post-Mortem Body Fluids
3.2. Sustained Expression of Exosome Surface Markers in Exosomes Derived from Plasma of Healthy Donors
3.3. Bioanalyzer Electropherogram Analysis of Exosomal RNA Derived from Plasma from Dead Bodies and Healthy Donors
3.4. Comparison of Exosome Isolation Kits for Exosomal RNA Extraction from Post-Mortem Body Fluids
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kurata, A. Strategy for postmortem diagnosis of myocardial infarction. Virchows Arch. 2020, 476, 177–178. [Google Scholar] [CrossRef]
- Chen, J.-H.; Inamori-Kawamoto, O.; Michiue, T.; Ikeda, S.; Ishikawa, T.; Maeda, H. Cardiac biomarkers in blood, and pericardial and cerebrospinal fluids of forensic autopsy cases: A reassessment with special regard to postmortem interval. Leg. Med. 2015, 17, 343–350. [Google Scholar] [CrossRef]
- Lee, D.-G.; Yang, K.E.; Hwang, J.W.; Kang, H.-S.; Lee, S.-Y.; Choi, S.; Shin, J.; Jang, I.-S.; An, H.J.; Chung, H.; et al. Degradation of Kidney and Psoas Muscle Proteins as Indicators of Post-Mortem Interval in a Rat Model, with Use of Lateral Flow Technology. PLoS ONE 2016, 11, e0160557. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Doyle, L.; Wang, M. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Bang, C.; Thum, T. Exosomes: New players in cell–cell communication. Int. J. Biochem. Cell Biol. 2012, 44, 2060–2064. [Google Scholar] [CrossRef]
- Zhu, Q.; Heon, M.; Zhao, Z.; He, M. Microfluidic engineering of exosomes: Editing cellular messages for precision therapeutics. Lab A Chip 2018, 18, 1690–1703. [Google Scholar] [CrossRef]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2012, 1820, 940–948. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef]
- Otero-Ortega, L.; Alonso-López, E.; Pérez-Mato, M.; Laso-García, F.; Frutos, M.C.G.-D.; Diekhorst, L.; García-Bermejo, M.L.; Conde-Moreno, E.; Fuentes, B.; De Leciñana, M.A.; et al. Similarities and Differences in Extracellular Vesicle Profiles between Ischaemic Stroke and Myocardial Infarction. Biomedicines 2020, 9, 8. [Google Scholar] [CrossRef]
- Saha, P.; Sharma, S.; Korutla, L.; Datla, S.R.; Shoja-Taheri, F.; Mishra, R.; Bigham, G.E.; Sarkar, M.; Morales, D.; Bittle, G.; et al. Circulating exosomes derived from transplanted progenitor cells aid the functional recovery of ischemic myocardium. Sci. Transl. Med. 2019, 11, eaau1168. [Google Scholar] [CrossRef]
- Thery, C.; Clayton, A.; Amigorena, S.; Raposo, G. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar]
- Serrano-Pertierra, E.; Oliveira-Rodríguez, M.; Rivas, M.; Oliva, P.; Villafani, J.; Navarro, A.; Blanco-López, M.C.; Cernuda-Morollón, E. Characterization of Plasma-Derived Extracellular Vesicles Isolated by Different Methods: A Comparison Study. Bioengineering 2019, 6, 8. [Google Scholar] [CrossRef]
- Ding, M.; Wang, C.; Lu, X.; Zhang, C.; Zhou, Z.; Chen, X.; Zhang, C.-Y.; Zen, K.; Zhang, C. Comparison of commercial exosome isolation kits for circulating exosomal microRNA profiling. Anal. Bioanal. Chem. 2018, 410, 3805–3814. [Google Scholar] [CrossRef]
- Théry, C.; Regnault, A.; Garin, J.; Wolfers, J.; Zitvogel, L.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Molecular Characterization of Dendritic Cell-Derived Exosomes. Selective accumulation of the heat shock protein hsc73. J. Cell Biol. 1999, 147, 599–610. [Google Scholar] [CrossRef]
- Escola, J.-M.; Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective Enrichment of Tetraspan Proteins on the Internal Vesicles of Multivesicular Endosomes and on Exosomes Secreted by Human B-lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. [Google Scholar] [CrossRef]
- Rikkert, L.G.; Nieuwland, R.; Terstappen, L.; Coumans, F.A.W. Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J. Extracell. Vesicles 2019, 8, 1555419. [Google Scholar] [CrossRef]
- Kakimoto, Y.; Kamiguchi, H.; Ochiai, E.; Satoh, F.; Osawa, M. MicroRNA Stability in Postmortem FFPE Tissues: Quantitative Analysis Using Autoptic Samples from Acute Myocardial Infarction Patients. PLoS ONE 2015, 10, e0129338. [Google Scholar] [CrossRef]
- Yu, S.; Na, J.-Y.; Lee, Y.-J.; Kim, K.-T.; Park, J.-T.; Kim, H.-S. Forensic application of microRNA-706 as a biomarker for drowning pattern identification. Forensic Sci. Int. 2015, 255, 96–101. [Google Scholar] [CrossRef]
- Vuokila, N.; Aronica, E.; Korotkov, A.; van Vliet, E.A.; Nuzhat, S.; Puhakka, N.; Pitkänen, A. Chronic Regulation of miR-124-3p in the Perilesional Cortex after Experimental and Human TBI. Int. J. Mol. Sci. 2020, 21, 2418. [Google Scholar] [CrossRef]
- Park, J.-P.; Choi, M.; Yang, K.-M.; Lee, K.; Kim, J.H.; Kim, M.; Yu, M.; Yang, H.W. Evaluation of the Usefulness of Cardiac Marker Analysis for Postmortem Diagnosis of Acute Myocardial Infarction. Korean J. Leg. Med. 2021, 45, 7–13. [Google Scholar] [CrossRef]
- Xu, M.; Ji, J.; Jin, D.; Wu, Y.; Wu, T.; Lin, R.; Zhu, S.; Jiang, F.; Ji, Y.; Bao, B.; et al. The biogenesis and secretion of exosomes and multivesicular bodies (MVBs): Intercellular shuttles and implications in human diseases. Genes Dis. 2022. [Google Scholar] [CrossRef]
- Prendergast, E.N.; de Souza Fonseca, M.A.; Dezem, F.S.; Lester, J.; Karlan, B.Y.; Noushmehr, H.; Lin, X.; Lawrenson, K. Optimizing exosomal RNA isolation for RNA-Seq analyses of archival sera specimens. PLoS ONE 2018, 13, e0196913. [Google Scholar] [CrossRef]
- Rocchi, A.; Chiti, E.; Maiese, A.; Turillazzi, E.; Spinetti, I. MicroRNAs: An Update of Applications in Forensic Science. Diagnostics 2020, 11, 32. [Google Scholar] [CrossRef]
- Sidova, M.; Tomankova, S.; Abaffy, P.; Kubista, M.; Sindelka, R. Effects of post-mortem and physical degradation on RNA integrity and quality. Biomol. Detect. Quantif. 2015, 5, 3–9. [Google Scholar] [CrossRef]
- Lobb, R.j.; Becker, M.; Wen, S.W.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Donaldson, A.E.; Lamont, I.L. Biochemistry Changes That Occur after Death: Potential Markers for Determining Post-Mortem Interval. PLoS ONE 2013, 8, e82011. [Google Scholar] [CrossRef]
- He, L.; Zhu, D.; Wang, J.; Wu, X. A highly efficient method for isolating urinary exosomes. Int. J. Mol. Med. 2018, 43, 83–90. [Google Scholar] [CrossRef] [Green Version]
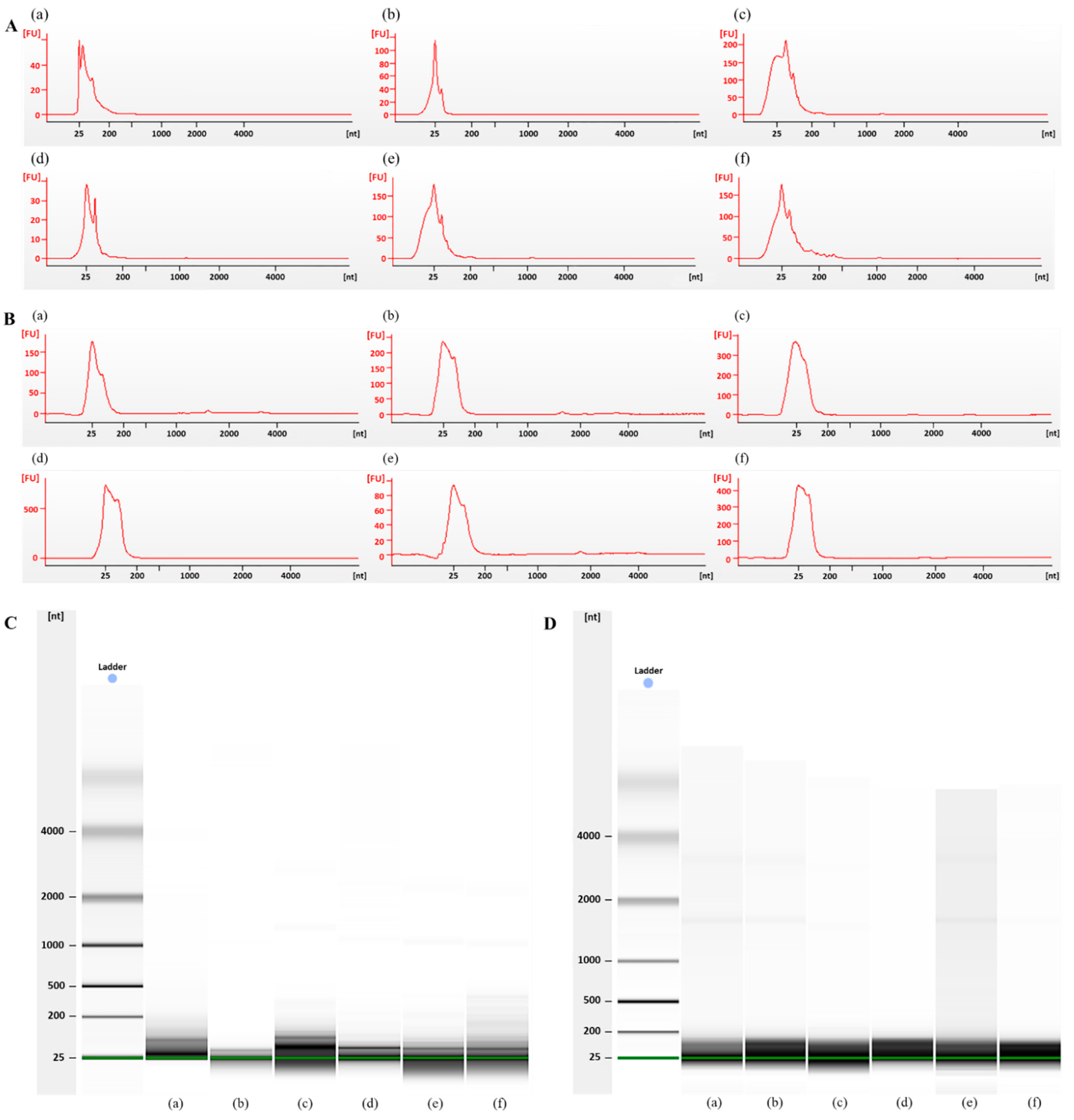
| Group | Sample | Concentration (ng/µL) | OD 260/280 |
|---|---|---|---|
| Healthy donors (Figure 3A,C) | (a) | 19.5 | 1.68 |
| (b) | 32.8 | 2.07 | |
| (c) | 48.8 | 1.91 | |
| (d) | 382.3 | 1.99 | |
| (e) | 10.5 | 1.89 | |
| (f) | 91.3 | 2.03 | |
| Dead bodies (Figure 3B,D) | (a) | 9.1 | 1.42 |
| (b) | 19.8 | 1.82 | |
| (c) | 129.1 | 2.02 | |
| (d) | 25.7 | 1.68 | |
| (e) | 120.3 | 2.02 | |
| (f) | 407.5 | 2.02 |
| Kit (End Product) | Company | Basic Concept | Plasma Volume, μL | Time, min | Complexity | Concentration, ng/μL | Mean ± SD |
|---|---|---|---|---|---|---|---|
| ExoQuick™ (exosome) | System biosciences | Precipitation (polymer) | 250 | 65 | Easy | 134.3 | 151.1 ± 100.7 |
| 91.1 | |||||||
| 117.9 | |||||||
| 86.5 | |||||||
| 253.6 | |||||||
| 354.3 | |||||||
| 58.6 | |||||||
| 112.4 | |||||||
| exoRNeasy (exosomal RNA) | Qiagen | Membrane affinity | 250 | 55 | Easy | 112.8 | 159.4 ± 119.2 |
| 98.2 | |||||||
| 117.6 | |||||||
| 48.7 | |||||||
| 370.8 | |||||||
| 316.7 | |||||||
| 58.9 | |||||||
| 151.7 | |||||||
| ExoLute® (exosome) | Rosetta exosome | Precipitation (polymer) and size exclusion chromatography | 250 | 89 | Easy | 3.2 | 3.9 ± 1.4 |
| 1.8 | |||||||
| 3.0 | |||||||
| 6.3 | |||||||
| 4.5 | |||||||
| 3.8 | |||||||
| 3.5 | |||||||
| 5.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-Y.; Jang, S.; Lee, S.; Park, J.-T.; Lee, S.-J.; Kim, H.-S. Characterization of Exosomes and Exosomal RNAs Isolated from Post-Mortem Body Fluids for Molecular Forensic Diagnosis. Diagnostics 2022, 12, 2153. https://doi.org/10.3390/diagnostics12092153
Kim S-Y, Jang S, Lee S, Park J-T, Lee S-J, Kim H-S. Characterization of Exosomes and Exosomal RNAs Isolated from Post-Mortem Body Fluids for Molecular Forensic Diagnosis. Diagnostics. 2022; 12(9):2153. https://doi.org/10.3390/diagnostics12092153
Chicago/Turabian StyleKim, So-Yeon, Sinae Jang, Sookyoung Lee, Jong-Tae Park, Su-Jin Lee, and Hyung-Seok Kim. 2022. "Characterization of Exosomes and Exosomal RNAs Isolated from Post-Mortem Body Fluids for Molecular Forensic Diagnosis" Diagnostics 12, no. 9: 2153. https://doi.org/10.3390/diagnostics12092153
APA StyleKim, S.-Y., Jang, S., Lee, S., Park, J.-T., Lee, S.-J., & Kim, H.-S. (2022). Characterization of Exosomes and Exosomal RNAs Isolated from Post-Mortem Body Fluids for Molecular Forensic Diagnosis. Diagnostics, 12(9), 2153. https://doi.org/10.3390/diagnostics12092153






