A Simple and Rapid Low-Cost Procedure for Detection of Vancomycin-Resistance Genes in Enterococci Reveals an Outbreak of Vancomycin-Variable Enterococcus faecium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Standard Routine Antibiotic Resistance Testing
2.2. Sample Preparation for New Procedure
2.3. Real-Time PCR
2.4. Characterization of Assays
2.5. Resistance Genes in Clinical Samples and Isolation of VREfm in Two Test Periods
2.6. Routine Screening of Urine Cultures with E. faecium
2.7. Isolation and Typing of VRE from Patient Samples
2.8. Test of Stored Blood Isolates
2.9. Follow-Up on Positive Test Results in a High-Risk Clinical Department
3. Results
3.1. Characterization of Assays
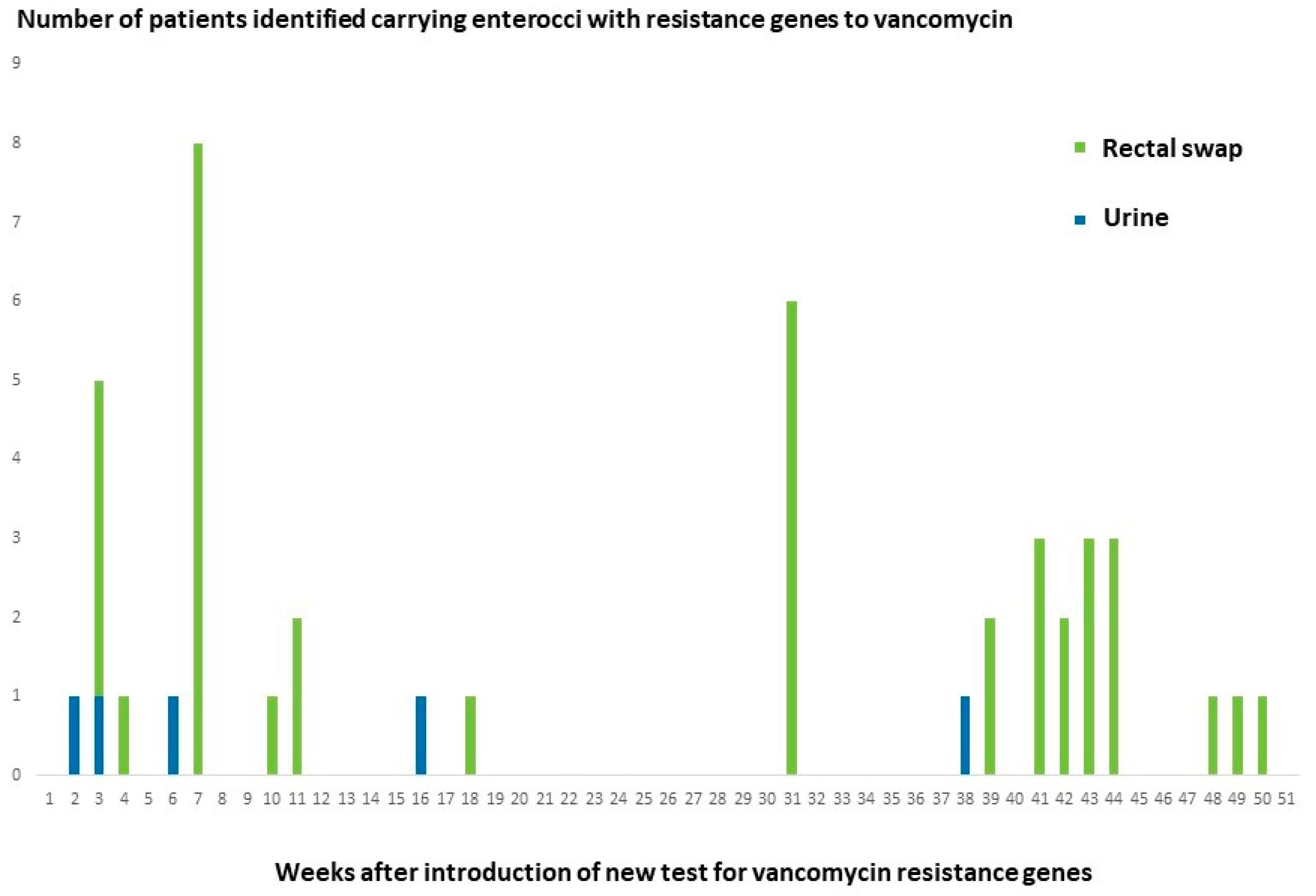
3.2. Results from the Two Test Periods
3.3. Routine Testing of Cultures from Urine Samples
3.4. Vancomycin-Resistance Genes in Isolates from Blood Cultures
3.5. Follow-Up on Positive Test Results in a High-Risk Clinical Department
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crank, C.; O’Driscoll, T. Vancomycin-Resistant Enterococcal Infections: Epidemiology, Clinical Manifestations, and Optimal Management. Infect. Drug Resist. 2015, 8, 217. [Google Scholar] [CrossRef]
- Cetinkaya, Y.; Falk, P.; Mayhall, C.G. Vancomycin-Resistant Enterococci. Clin. Microbiol. Rev. 2000, 13, 686–707. [Google Scholar] [CrossRef]
- Werner, G.; Neumann, B.; Weber, R.E.; Kresken, M.; Wendt, C.; Bender, J.K.; Becker, K.; Borgmann, S.; Diefenbach, A.; Hamprecht, A.; et al. Thirty Years of VRE in Germany—“Expect the Unexpected”: The View from the National Reference Centre for Staphylococci and Enterococci. Drug Resist. Updat. 2020, 53, 100732. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Kattula, D.; Burkert, F. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Available online: http://www.cdc.gov/drugresistance/threat-report-2013/ (accessed on 29 December 2021).
- Hegstad, K.; Giske, C.G.; Haldorsen, B.; Matuschek, E.; Schønning, K.; Leegaard, T.M.; Kahlmeter, G.; Sundsfjord, A. Performance of the EUCAST Disk Diffusion Method, the CLSI Agar Screen Method, and the Vitek 2 Automated Antimicrobial Susceptibility Testing System for Detection of Clinical Isolates of Enterococci with Low- and Medium-Level VanB-Type Vancomycin Resistance: A Multicenter Study. J. Clin. Microbiol. 2014, 52, 1582–1589. [Google Scholar] [CrossRef]
- Viswanath, L.S.; Sugumar, M.; Chandra Murthy Peela, S.; Walia, K.; Sistla, S. Detection of Vancomycin Variable Enterococci (VVE) among Clinical Isolates of Enterococcus Faecium Collected across India-First Report from the Subcontinent. Indian J. Med. Microbiol. 2022, 40, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.A.; Pedersen, M.S.; Nielsen, L.G.; Ma, C.M.G.; Søes, L.M.; Worning, P.; Østergaard, C.; Westh, H.; Pinholt, M.; Schønning, K. Emergence of a Vancomycin-Variable Enterococcus Faecium ST1421 Strain Containing a Deletion in VanX. J. Antimicrob. Chemother. 2018, 73, 2936–2940. [Google Scholar] [CrossRef]
- Downing, M.A.; Xiong, J.; Eshaghi, A.; McGeer, A.; Patel, S.N.; Johnstone, J. Vancomycin-Variable Enterococcal Bacteremia. J. Clin. Microbiol. 2015, 53, 3951–3953. [Google Scholar] [CrossRef]
- Kohler, P.; Eshaghi, A.; Kim, H.C.; Plevneshi, A.; Green, K.; Willey, B.M.; McGeer, A.; Patel, S.N. Prevalence of Vancomycin-Variable Enterococcus Faecium (VVE) among VanA-Positive Sterile Site Isolates and Patient Factors Associated with VVE Bacteremia. PLoS ONE 2018, 13, e0193926. [Google Scholar] [CrossRef]
- Gagnon, S.; Lévesque, S.; Lefebvre, B.; Bourgault, A.M.; Labbé, A.C.; Roger, M. VanA-Containing Enterococcus Faecium Susceptible to Vancomycin and Teicoplanin Because of Major Nucleotide Deletions in Tn1546. J. Antimicrob. Chemother. 2011, 66, 2758–2762. [Google Scholar] [CrossRef] [PubMed]
- Merlino, J.; Gray, T. Vancomycin Variable Enterococcus (VVE), E. Faecium, Harbouring the VanA Gene Complex. Pathology 2021, 53, 680–682. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M.; Justesen, U.S.; Pinholt, M.; Roer, L.; Kaya, H.; Worning, P.; Nygaard, S.; Kemp, M.; Clausen, M.E.; Nielsen, K.L.; et al. Surveillance of Vancomycin-Resistant Enterococci Reveals Shift in Dominating Clones and National Spread of a Vancomycin-Variable VanA Enterococcus Faecium ST1421-CT1134 Clone, Denmark, 2015 to March 2019. Eurosurveillance 2019, 24, 1900503. [Google Scholar] [CrossRef]
- Szakacs, T.A.; Kalan, L.; McConnell, M.J.; Eshaghi, A.; Shahinas, D.; McGeer, A.; Wright, G.D.; Low, D.E.; Patel, S.N. Outbreak of Vancomycin-Susceptible Enterococcus Faecium Containing the Wild-Type VanA Gene. J. Clin. Microbiol. 2014, 52, 1682–1686. [Google Scholar] [CrossRef] [PubMed]
- Sivertsen, A.; Pedersen, T.; Larssen, K.W.; Bergh, K.; Rønning, T.G.; Radtke, A.; Hegstad, K. A Silenced VanA Gene Cluster on a Transferable Plasmid Caused an Outbreak of Vancomycin-Variable Enterococci. Antimicrob. Agents Chemother. 2016, 60, 4119–4127. [Google Scholar] [CrossRef] [PubMed]
- Thaker, M.N.; Kalan, L.; Waglechner, N.; Eshaghi, A.; Patel, S.N.; Poutanen, S.; Willey, B.; Coburn, B.; McGeer, A.; Low, D.E.; et al. Vancomycin-Variable Enterococci Can Give Rise to Constitutive Resistance during Antibiotic Therapy. Antimicrob. Agents Chemother. 2015, 59, 1405–1410. [Google Scholar] [CrossRef] [PubMed]
- Coburn, B.; Low, D.E.; Patel, S.N.; Poutanen, S.M.; Shahinas, D.; Eshaghi, A.; Willey, B.M.; McGeera, A. Vancomycin-Variable Enterococcus Faecium: In Vivo Emergence of Vancomycin Resistance in a Vancomycin-Susceptible Isolate. J. Clin. Microbiol. 2014, 52, 1766–1767. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.M.; Janice, J.; Sivertsen, A.; Sjögren, I.; Sundsfjord, A.; Hegstad, K. Alternative VanHAX Promoters and Increased VanA-Plasmid Copy Number Resurrect Silenced Glycopeptide Resistance in Enterococcus Faecium. J. Antimicrob. Chemother. 2021, 76, 876–882. [Google Scholar] [CrossRef]
- Jung, Y.H.; Lee, Y.S.; Lee, S.Y.; Yoo, J.S.; Yoo, J.I.; Kim, H.S.; Kim, O.; Yu, J.-y. Structure and Transfer of the VanA Cluster in VanA-Positive, Vancomycin-Susceptible Enterococcus Faecium, and Its Revertant Mutant. Diagn. Microbiol. Infect. Dis. 2014, 80, 148–150. [Google Scholar] [CrossRef]
- Choi, H.J.; Nam, D.; Peck, K.R.; Song, J.H.; Shin, D.; Ko, K.S. Loss of Vancomycin Resistance Not Completely Dependent on the Tn1546 Element in Enterococcus Faecium Isolates. Diagn. Microbiol. Infect. Dis. 2011, 69, 105–110. [Google Scholar] [CrossRef]
- Fang, H.; Ohlsson, A.K.; Jiang, G.X.; Ullberg, M. Screening for Vancomycin-Resistant Enterococci: An Efficient and Economical Laboratory-Developed Test. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 261–265. [Google Scholar] [CrossRef]
- Ludwig, W.; Schleifer, K.H. How Quantitative Is Quantitative PCR with Respect to Cell Counts? Syst. Appl. Microbiol. 2000, 23, 556–562. [Google Scholar] [CrossRef]
- De Been, M.; Pinholt, M.; Top, J.; Bletz, S.; Mellmann, A.; Van Schaik, W.; Brouwer, E.; Rogers, M.; Kraat, Y.; Bonten, M.; et al. Core Genome Multilocus Sequence Typing Scheme for High- Resolution Typing of Enterococcus Faecium. J. Clin. Microbiol. 2015, 53, 3788–3797. [Google Scholar] [CrossRef] [PubMed]
- Holzknecht, B.J.; Hansen, D.S.; Nielsen, L.; Kailow, A.; Jarløv, J.O. Screening for Vancomycin-Resistant Enterococci with Xpert® VanA/VanB: Diagnostic Accuracy and Impact on Infection Control Decision Making. New Microbes New Infect. 2017, 16, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Ayobami, O.; Willrich, N.; Reuss, A.; Eckmanns, T.; Markwart, R. The Ongoing Challenge of Vancomycin-Resistant Enterococcus Faecium and Enterococcus Faecalis in Europe: An Epidemiological Analysis of Bloodstream Infections. Emerg. Microbes Infect. 2020, 9, 1180–1193. [Google Scholar] [CrossRef] [PubMed]
- Mak, A.; Miller, M.A.; Chong, G.; Monczak, Y. Comparison of PCR and Culture for Screening of Vancomycin-Resistant Enterococci: Highly Disparate Results for VanA and VanB. J. Clin. Microbiol. 2009, 47, 4136–4137. [Google Scholar] [CrossRef]
- Graham, M.; Ballard, S.A.; Grabsch, E.A.; Johnson, P.D.R.; Grayson, M.L. High Rates of Fecal Carriage of Nonenterococcal VanB in Both Children and Adults. Antimicrob. Agents Chemother. 2008, 52, 1195–1197. [Google Scholar] [CrossRef] [PubMed]
- Ballard, S.A.; Pertile, K.K.; Lim, M.; Johnson, P.D.R.; Grayson, M.L. Molecular Characterization of VanB Elements in Naturally Occurring Gut Anaerobes. Antimicrob. Agents Chemother. 2005, 49, 1688–1694. [Google Scholar] [CrossRef]
- Marbjerg, L.; Stougaard, C.L.; Sørensen, S.-A.G.; Thomsen, A.V.; Wang, L.; Andersen, L.; Andersen, T.E.; Kallipolitis, B.; Kemp, M. A New Tool for Analyses of Whole Genome Sequences Reveals Dissemination of Specific Strains of Vancomycin-Resistant Enterococcus Faecium in a Hospital. Front. Med. 2021, 8, 733676. [Google Scholar] [CrossRef]
| Gene | Oligo Sequence | Reference |
|---|---|---|
| vanA-F | 5′-AGT CAA TAC TCT GCC CGG TTT C-3′ | Fang et al. [20] |
| vanA-R | 5′-GCA GCG GCC ATC ATA CG-3′ | |
| vanA-P | 5′-FAM-CGT CAT ACA GTC GTT ATC-MGB-3′ | |
| vanB-F | 5′-GGR AAC GAG GAT TTG ATT G-3′ | This study |
| vanB-R | 5′-CGT GGC TCA RCC GGA TT-3′ | |
| vanB-P | 5′-VIC-CGG CGA AGT GGA TC-MGB-3′ | |
| 23S-F | 5′-AGA AAT TCC AAA CGA ACT TG-3′ | Ludwig and Schleifer [21] |
| 23S-R | 5′-CAG TGC TCT ACC TCC ATC ATT-3′ | |
| 23S-P | 5′-FAM-TGG TTC TCT CCG AAA TAG CTT TAGGGC TA-BHQ1-3′ |
| Test Period | Number of Urine Samples Tested | Number of Samples Reported with Culture of Vancomycin Resistant Enterococci Based on Phenotypic Resistance Testing with Subsequent Identification of Resistance Genes by PCR | Number of Primary Cultures Positive for vanA or vanB Gene |
|---|---|---|---|
| One month from 2017 | 340 | 2 vanA gene, 4 vanB gene | 3 vanA gene, 4 vanB gene |
| One month from 2018 | 357 | 0 | 16 vanA gene |
| Patient # | Number of Urine Samples Positive for vanA or vanB Gene | Species | MLST Sequence Type (ST) and Core Genome MLST Cluster Type (CT) |
|---|---|---|---|
| 1 | 1 sample positive for vanA gene | E. faecium | ST1421-CT1134 |
| 2 | 1 sample positive for vanA gene | E. faecium | ST1421-CT1134 |
| 3 | 7 samples positive for vanA gene | E. faecium | ST1421-CT1134 |
| 4 | 2 samples positive for vanA gene | E. faecium | ST1421-CT1134 |
| 5 | 1 sample positive for vanA gene | - | No isolate obtained |
| 6 | 1 sample positive for vanA gene | - | No isolate obtained |
| 7 | 1 sample positive for vanA gene | E. faecium | ST80-CT993 |
| 8 | 1 sample positive for vanA gene | - | No isolate obtained |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdullah, H.M.; Marbjerg, L.H.; Andersen, L.; Hoegh, S.V.; Kemp, M. A Simple and Rapid Low-Cost Procedure for Detection of Vancomycin-Resistance Genes in Enterococci Reveals an Outbreak of Vancomycin-Variable Enterococcus faecium. Diagnostics 2022, 12, 2120. https://doi.org/10.3390/diagnostics12092120
Abdullah HM, Marbjerg LH, Andersen L, Hoegh SV, Kemp M. A Simple and Rapid Low-Cost Procedure for Detection of Vancomycin-Resistance Genes in Enterococci Reveals an Outbreak of Vancomycin-Variable Enterococcus faecium. Diagnostics. 2022; 12(9):2120. https://doi.org/10.3390/diagnostics12092120
Chicago/Turabian StyleAbdullah, Hozan Muhammed, Lis Høy Marbjerg, Lise Andersen, Silje Vermedal Hoegh, and Michael Kemp. 2022. "A Simple and Rapid Low-Cost Procedure for Detection of Vancomycin-Resistance Genes in Enterococci Reveals an Outbreak of Vancomycin-Variable Enterococcus faecium" Diagnostics 12, no. 9: 2120. https://doi.org/10.3390/diagnostics12092120
APA StyleAbdullah, H. M., Marbjerg, L. H., Andersen, L., Hoegh, S. V., & Kemp, M. (2022). A Simple and Rapid Low-Cost Procedure for Detection of Vancomycin-Resistance Genes in Enterococci Reveals an Outbreak of Vancomycin-Variable Enterococcus faecium. Diagnostics, 12(9), 2120. https://doi.org/10.3390/diagnostics12092120






