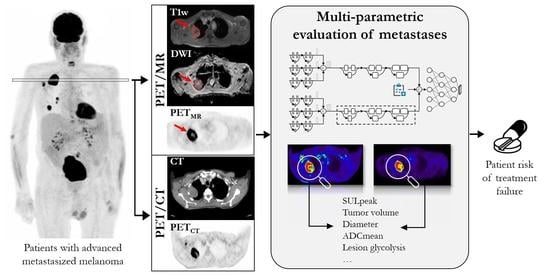
Development of a Hybrid-Imaging-Based Prognostic Index for Metastasized-Melanoma Patients in Whole-Body 18F-FDG PET/CT and PET/MRI Data
Abstract
1. Introduction
2. Material and Methods
2.1. Study Design
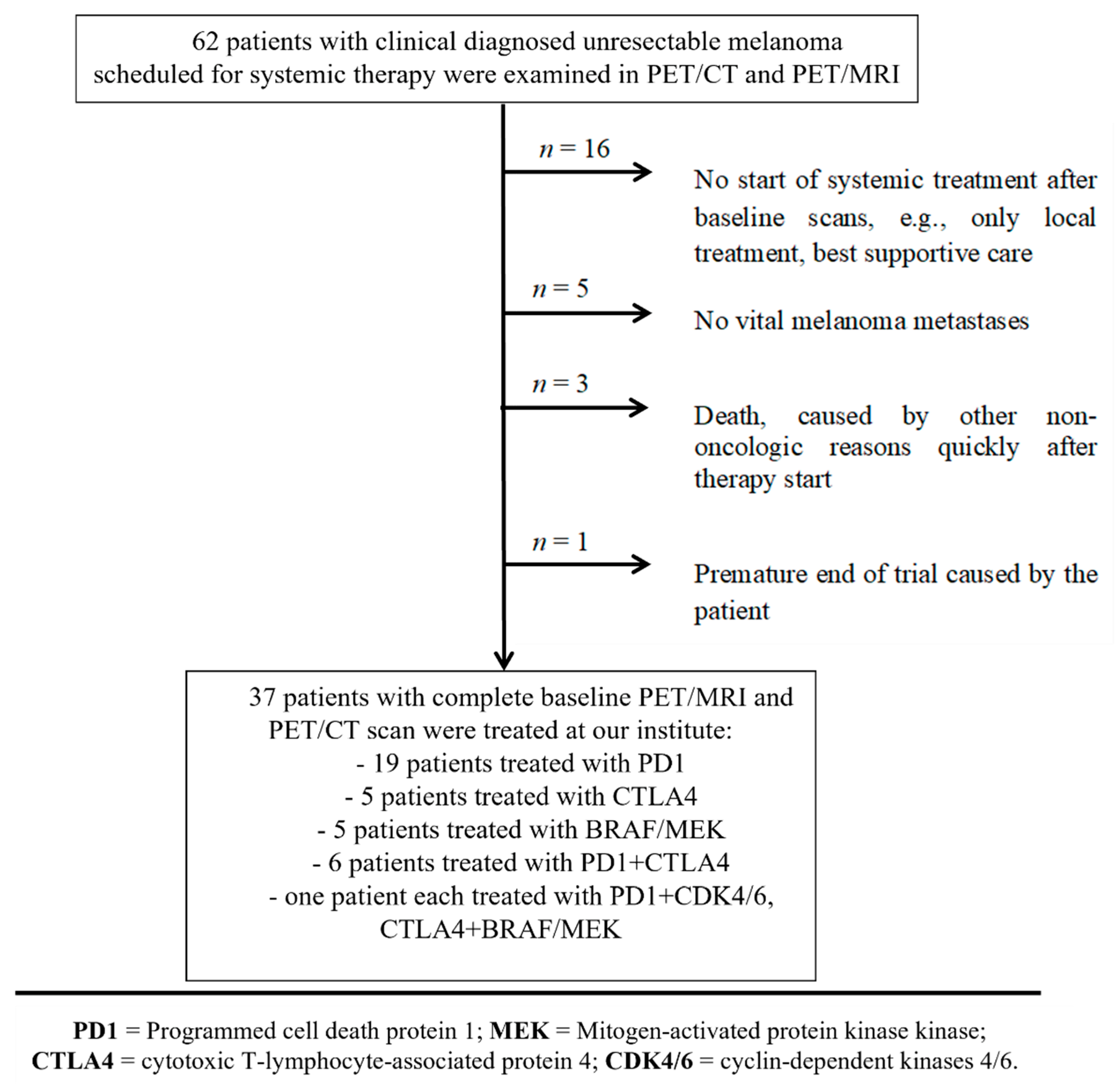
2.2. Patient Cohort and Outcome Evaluation
2.3. PET/CT and PET/MRI Examination Parameters
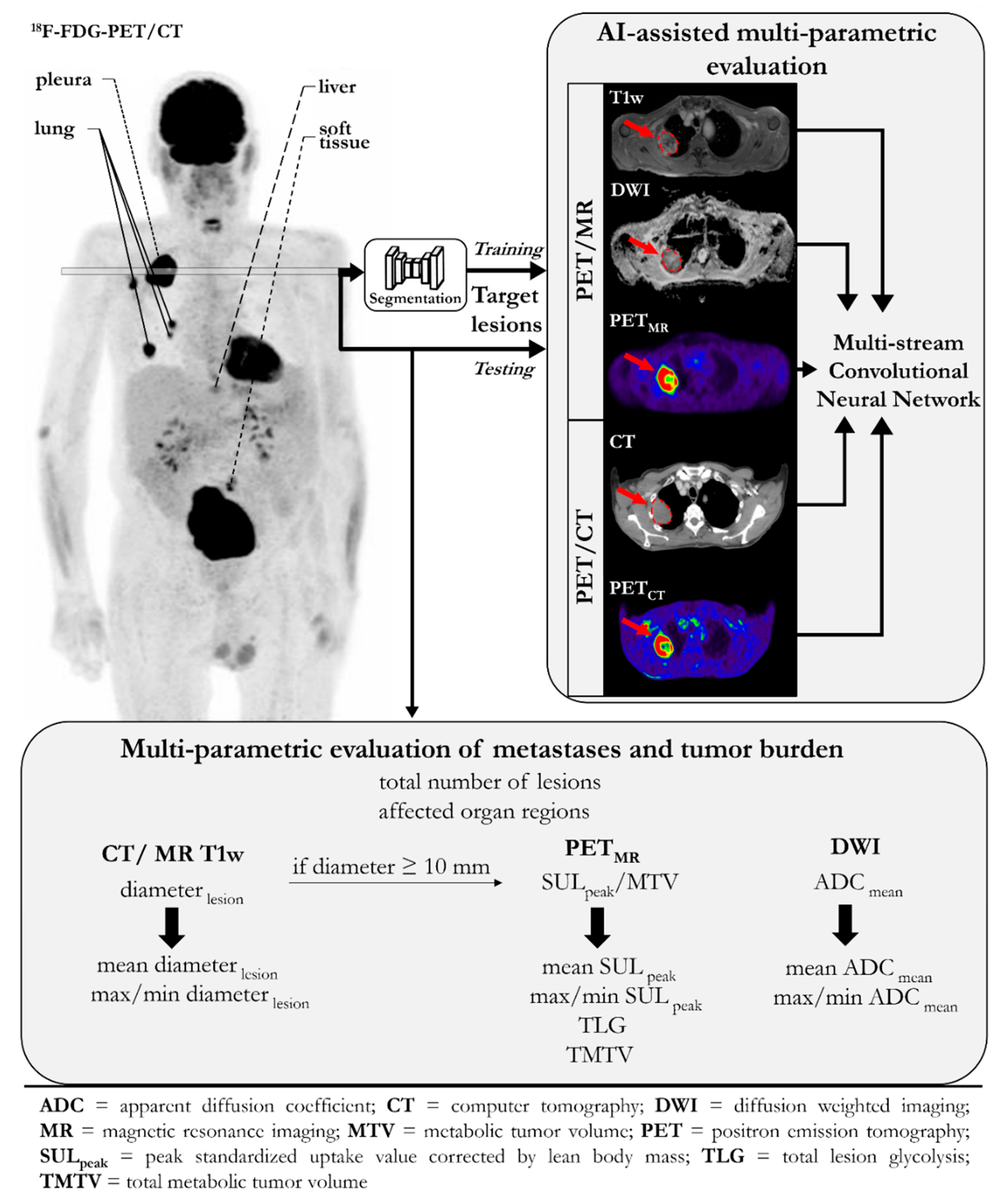
2.4. Multiparametric Evaluation of Metastases and Tumor Burden
2.5. Convolutional Neural Network
2.6. Statistics
3. Results
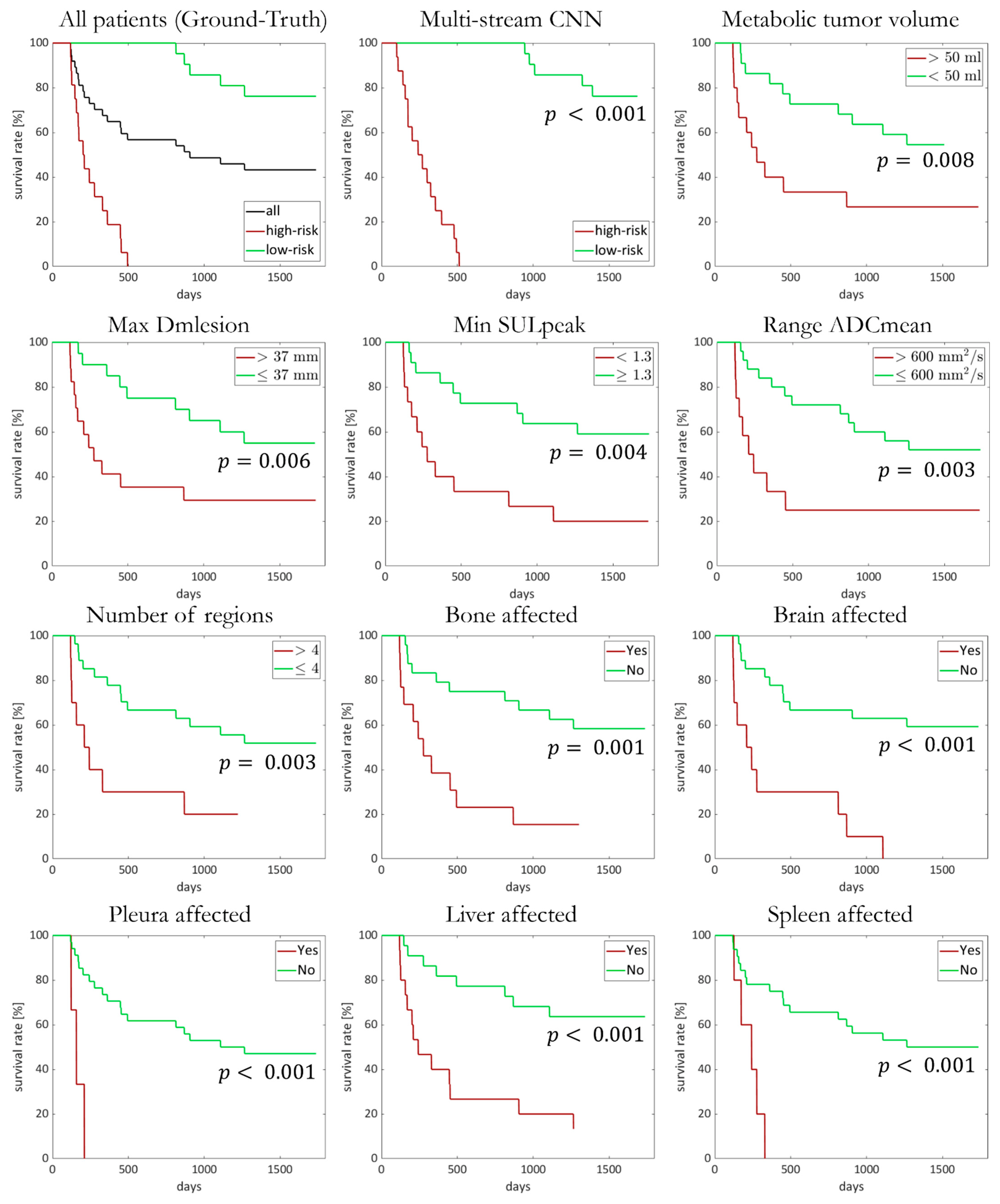
3.1. Treatment Outcome and Overall Survival
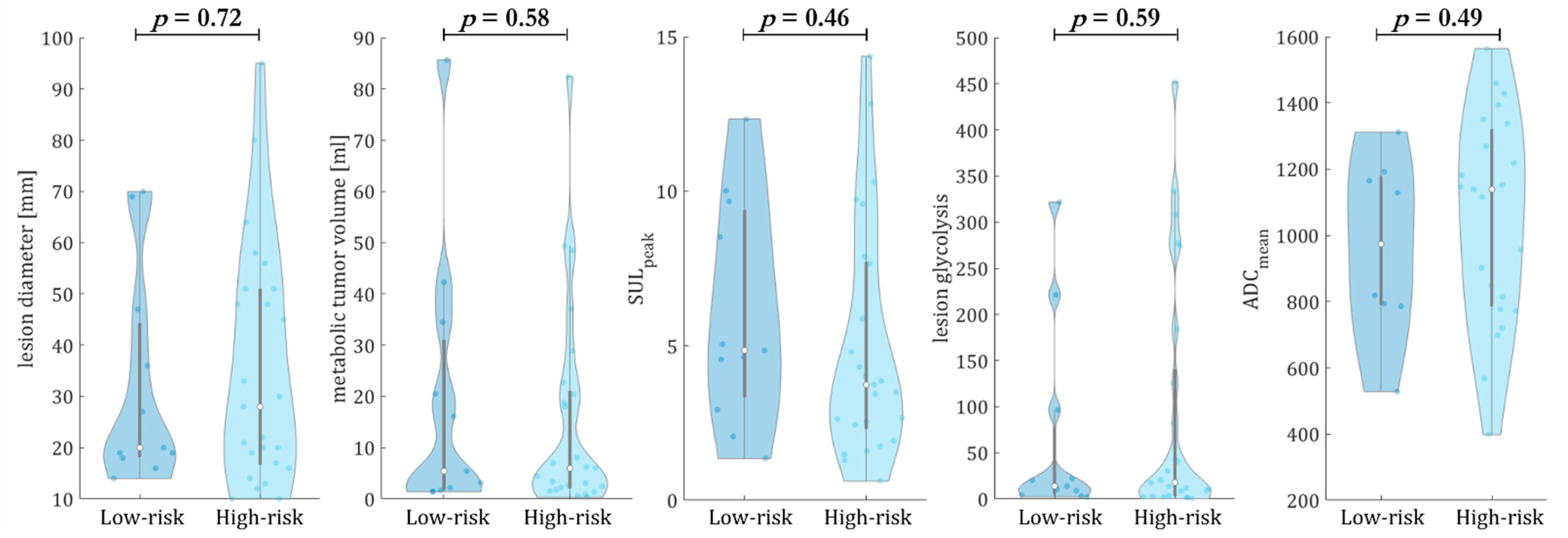
3.2. Multiparametric Evaluation of Metastases and Tumor Burden
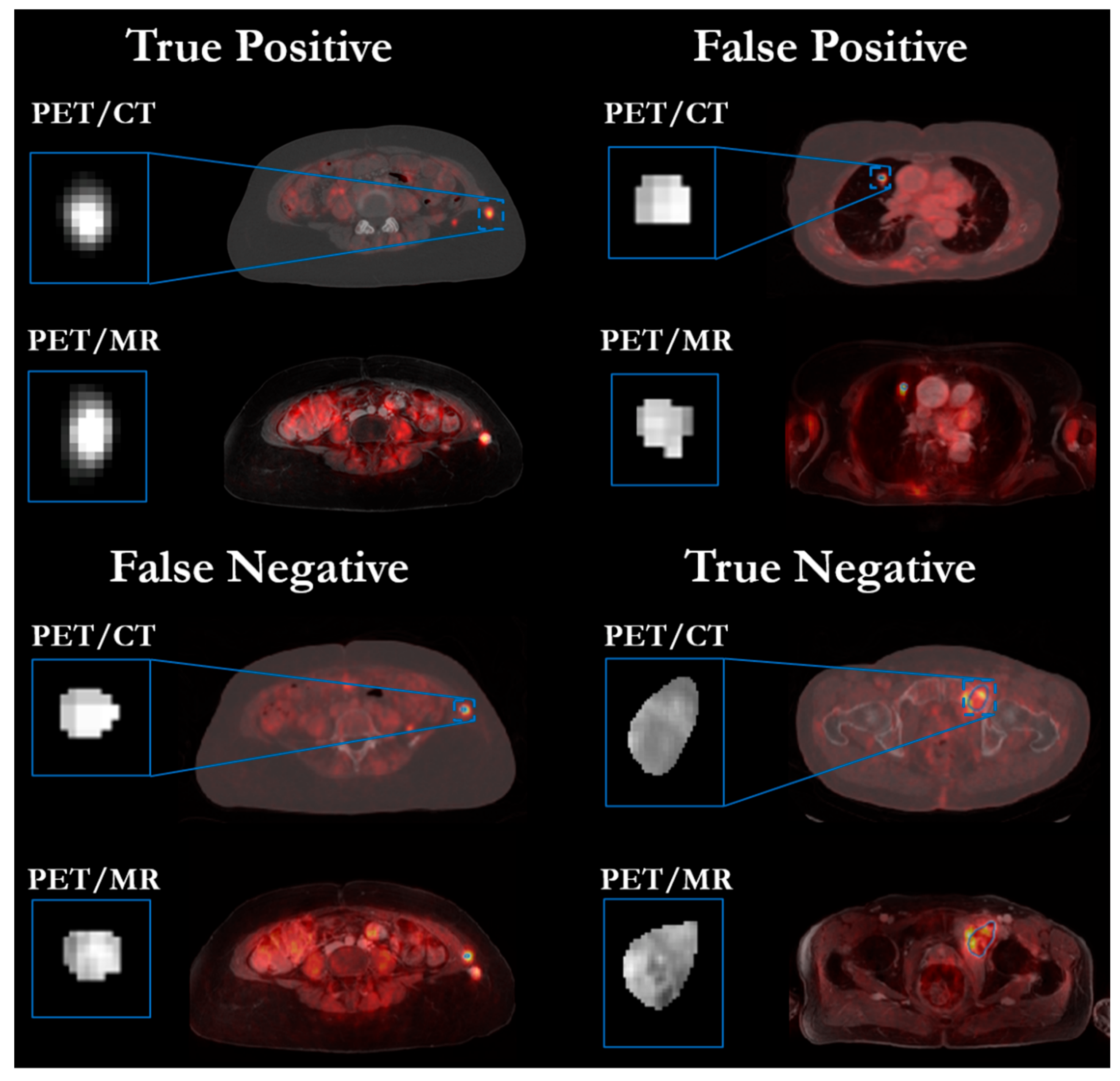
3.3. Convolutional Neural Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Hamid, O.; Robert, C.; Daud, A.; Hodi, F.S.; Hwu, W.-J.; Kefford, R.; Wolchok, J.D.; Hersey, P.; Joseph, R.W.; Weber, J.S.; et al. 5-year survival outcomes in patients (pts) with advanced melanoma treated with pembrolizumab (pembro) in KEYNOTE-001. J. Clin. Oncol. 2018, 36, 9516. [Google Scholar] [CrossRef]
- Long, G.V.; Eroglu, Z.; Infante, J.; Patel, S.; Daud, A.; Johnson, D.B.; Gonzalez, R.; Kefford, R.; Hamid, O.; Schuchter, L.; et al. Long-Term Outcomes in Patients With BRAF V600–Mutant Metastatic Melanoma Who Received Dabrafenib Combined With Trametinib. J. Clin. Oncol. 2018, 36, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2019, 381, 1535–1546. [Google Scholar] [CrossRef]
- Filippi, L.; Bianconi, F.; Schillaci, O.; Spanu, A.; Palumbo, B. The Role and Potential of (18)F-FDG PET/CT in Malignant Melanoma: Prognostication, Monitoring Response to Targeted and Immunotherapy, and Radiomics. Diagnostics 2022, 12, 929. [Google Scholar] [CrossRef]
- Rivas, A.; Delyon, J.; Martineau, A.; Blanc, E.; Allayous, C.; Da Meda, L.; Merlet, P.; Lebbe, C.; Baroudjian, B.; Vercellino, L. 18FDG PET Assessment of Therapeutic Response in Patients with Advanced or Metastatic Melanoma Treated with First-Line Immune Checkpoint Inhibitors. Cancers 2022, 14, 3190. [Google Scholar] [CrossRef]
- Seith, F.; Forschner, A.; Schmidt, H.; Pfannenberg, C.; Guckel, B.; Nikolaou, K.; la Fougere, C.; Garbe, C.; Schwenzer, N. 18F-FDG-PET detects complete response to PD1-therapy in melanoma patients two weeks after therapy start. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 95–101. [Google Scholar] [CrossRef]
- Wong, A.; Callahan, J.; Keyaerts, M.; Neyns, B.; Mangana, J.; Aberle, S.; Herschtal, A.; Fullerton, S.; Milne, D.; Iravani, A.; et al. (18)F-FDG PET/CT based spleen to liver ratio associates with clinical outcome to ipilimumab in patients with metastatic melanoma. Cancer Imaging 2020, 20, 36. [Google Scholar] [CrossRef]
- Kudura, K.; Dimitriou, F.; Basler, L.; Forster, R.; Mihic-Probst, D.; Kutzker, T.; Dummer, R.; Mangana, J.; Burger, I.A.; Kreissl, M.C. Prediction of Early Response to Immune Checkpoint Inhibition Using FDG-PET/CT in Melanoma Patients. Cancers 2021, 13, 3830. [Google Scholar] [CrossRef]
- Grzywa, T.M.; Paskal, W.; Włodarski, P.K. Intratumor and Intertumor Heterogeneity in Melanoma. Transl. Oncol. 2017, 10, 956–975. [Google Scholar] [CrossRef]
- Aparicio, S.; Caldas, C. The implications of clonal genome evolution for cancer medicine. N. Engl. J. Med. 2013, 368, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.Y.; Shin, H.-T.; Yun, J.W.; Kim, K.-T.; Kim, J.; Bae, J.S.; Cho, Y.B.; Lee, W.Y.; Yun, S.H.; Park, Y.A.; et al. Intratumor heterogeneity inferred from targeted deep sequencing as a prognostic indicator. Sci. Rep. 2019, 9, 4542. [Google Scholar] [CrossRef] [PubMed]
- Oren, O.; Gersh, B.J.; Bhatt, D.L. Artificial intelligence in medical imaging: Switching from radiographic pathological data to clinically meaningful endpoints. Lancet Digit. Health 2020, 2, e486–e488. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Dildar, M.; Akram, S.; Irfan, M.; Khan, H.U.; Ramzan, M.; Mahmood, A.R.; Alsaiari, S.A.; Saeed, A.H.M.; Alraddadi, M.O.; Mahnashi, M.H. Skin Cancer Detection: A Review Using Deep Learning Techniques. Int. J. Environ. Res. Public Health 2021, 18, 5479. [Google Scholar] [CrossRef] [PubMed]
- Morin, O.; Vallières, M.; Jochems, A.; Woodruff, H.C.; Valdes, G.; Braunstein, S.E.; Wildberger, J.E.; Villanueva-Meyer, J.E.; Kearney, V.; Yom, S.S.; et al. A Deep Look Into the Future of Quantitative Imaging in Oncology: A Statement of Working Principles and Proposal for Change. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1074–1082. [Google Scholar] [CrossRef]
- Parmar, C.; Barry, J.D.; Hosny, A.; Quackenbush, J.; Aerts, H. Data Analysis Strategies in Medical Imaging. Clin. Cancer Res. 2018, 24, 3492–3499. [Google Scholar] [CrossRef]
- Xue, Y.; Chen, S.; Qin, J.; Liu, Y.; Huang, B.; Chen, H. Application of Deep Learning in Automated Analysis of Molecular Images in Cancer: A Survey. Contrast Media Mol. Imaging 2017, 2017, 9512370. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.J.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef]
- Ghafoorian, M.; Karssemeijer, N.; Heskes, T.; van Uder, I.W.M.; de Leeuw, F.E.; Marchiori, E.; van Ginneken, B.; Platel, B. Non-uniform patch sampling with deep convolutional neural networks for white matter hyperintensity segmentation. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1414–1417. [Google Scholar]
- Brosch, T.; Tang, L.Y.W.; Yoo, Y.; Li, D.K.B.; Traboulsee, A.; Tam, R. Deep 3D Convolutional Encoder Networks With Shortcuts for Multiscale Feature Integration Applied to Multiple Sclerosis Lesion Segmentation. IEEE Trans. Med. Imaging 2016, 35, 1229–1239. [Google Scholar] [CrossRef]
- Setio, A.A.; Ciompi, F.; Litjens, G.; Gerke, P.; Jacobs, C.; van Riel, S.J.; Wille, M.M.; Naqibullah, M.; Sanchez, C.I.; van Ginneken, B. Pulmonary Nodule Detection in CT Images: False Positive Reduction Using Multi-View Convolutional Networks. IEEE Trans. Med. Imaging 2016, 35, 1160–1169. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, J.; Hamarneh, G. Multi-Resolution-Tract CNN with Hybrid Pretrained and Skin-Lesion Trained Layers; Spring: Cham, Switzerland; pp. 164–171.
- Aljohani, K.; Turki, T. Automatic Classification of Melanoma Skin Cancer with Deep Convolutional Neural Networks. Ai 2022, 3, 512–525. [Google Scholar] [CrossRef]
- Li, R.; Zhang, W.; Suk, H.I.; Wang, L.; Li, J.; Shen, D.; Ji, S. Deep learning based imaging data completion for improved brain disease diagnosis. Med. Image Comput. Comput. Assist. Interv. 2014, 17, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Anthimopoulos, M.; Christodoulidis, S.; Ebner, L.; Christe, A.; Mougiakakou, S. Lung Pattern Classification for Interstitial Lung Diseases Using a Deep Convolutional Neural Network. IEEE Trans. Med. Imaging 2016, 35, 1207–1216. [Google Scholar] [CrossRef]
- Fakoor, R.; Ladhak, F.; Nazi, A.; Huber, M. Using deep learning to enhance cancer diagnosis and classification. In Proceedings of the International Conference on Machine Learning, Atlanta, GA, USA, 16–21 June 2013. [Google Scholar]
- Codella, N.C.; Nguyen, Q.-B.; Pankanti, S.; Gutman, D.A.; Helba, B.; Halpern, A.C.; Smith, J.R. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J. Res. Dev. 2017, 61, 5:1–5:15. [Google Scholar]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P. Automated Melanoma Recognition in Dermoscopy Images via Very Deep Residual Networks. IEEE Trans. Med. Imaging 2017, 36, 994–1004. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Jafari, M.H.; Nasr-Esfahani, E.; Karimi, N.; Soroushmehr, S.M.R.; Samavi, S.; Najarian, K. Extraction of skin lesions from non-dermoscopic images for surgical excision of melanoma. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 1021–1030. [Google Scholar] [CrossRef]
- Nasr-Esfahani, E.; Samavi, S.; Karimi, N.; Soroushmehr, S.M.R.; Jafari, M.H.; Ward, K.; Najarian, K. Melanoma detection by analysis of clinical images using convolutional neural network. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 1373–1376. [Google Scholar]
- Dirks, I.; Keyaerts, M.; Neyns, B.; Vandemeulebroucke, J. Computer-aided detection and segmentation of malignant melanoma lesions on whole-body (18)F-FDG PET/CT using an interpretable deep learning approach. Comput. Methods Programs Biomed. 2022, 221, 106902. [Google Scholar] [CrossRef]
- Cireşan, D.C.; Giusti, A.; Gambardella, L.M.; Schmidhuber, J. Mitosis Detection in Breast Cancer Histology Images with Deep Neural Networks; Springer: Berlin/Heidelberg, Germany, 2013; pp. 411–418. [Google Scholar]
- Wang, H.; Cruz-Roa, A.; Basavanhally, A.; Gilmore, H.; Shih, N.; Feldman, M.; Tomaszewski, J.; Gonzalez, F.; Madabhushi, A. Mitosis detection in breast cancer pathology images by combining handcrafted and convolutional neural network features. J. Med. Imaging 2014, 1, 034003. [Google Scholar] [CrossRef]
- Cruz-Roa, A.; Basavanhally, A.; González, F.; Gilmore, H.; Feldman, M.; Ganesan, S.; Shih, N.; Tomaszewski, J.; Madabhushi, A. Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. In Proceedings of the Medical Imaging 2014: Digital Pathology, San Diego, CA, USA, 15–20 February 2014; p. 904103. [Google Scholar]
- Roth, H.R.; Lu, L.; Seff, A.; Cherry, K.M.; Hoffman, J.; Wang, S.; Liu, J.; Turkbey, E.; Summers, R.M. A new 2.5D representation for lymph node detection using random sets of deep convolutional neural network observations. MICCAI Int. Conf. Med. Image Comput. Comput. Assist. Interv. 2014, 17, 520–527. [Google Scholar] [CrossRef]
- Shin, H.-C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Barbu, A.; Lu, L.; Roth, H.; Seff, A.; Summers, R.M. An analysis of robust cost functions for CNN in computer-aided diagnosis. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2018, 6, 253–258. [Google Scholar] [CrossRef]
- Roth, H.R.; Lu, L.; Liu, J.; Yao, J.; Seff, A.; Cherry, K.; Kim, L.; Summers, R.M. Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans. Med. Imaging 2015, 35, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.; Hansen, M.B.; Tietze, A.; Mouridsen, K. Prediction of Tissue Outcome and Assessment of Treatment Effect in Acute Ischemic Stroke Using Deep Learning. Stroke 2018, 49, 1394–1401. [Google Scholar] [CrossRef]
- Deist, T.M.; Dankers, F.; Valdes, G.; Wijsman, R.; Hsu, I.C.; Oberije, C.; Lustberg, T.; van Soest, J.; Hoebers, F.; Jochems, A.; et al. Machine learning algorithms for outcome prediction in (chemo)radiotherapy: An empirical comparison of classifiers. Med. Phys. 2018, 45, 3449–3459. [Google Scholar] [CrossRef] [PubMed]
- Jojoa Acosta, M.F.; Caballero Tovar, L.Y.; Garcia-Zapirain, M.B.; Percybrooks, W.S. Melanoma diagnosis using deep learning techniques on dermatoscopic images. BMC Med. Imaging 2021, 21, 6. [Google Scholar] [CrossRef]
- Nie, D.; Zhang, H.; Adeli, E.; Liu, L.; Shen, D. 3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients. Med. Image Comput. Comput. Assist. Interv. 2016, 9901, 212–220. [Google Scholar] [CrossRef]
- Bychkov, D.; Linder, N.; Turkki, R.; Nordling, S.; Kovanen, P.E.; Verrill, C.; Walliander, M.; Lundin, M.; Haglund, C.; Lundin, J. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 2018, 8, 3395. [Google Scholar] [CrossRef]
- Lucia, F.; Visvikis, D.; Desseroit, M.C.; Miranda, O.; Malhaire, J.P.; Robin, P.; Pradier, O.; Hatt, M.; Schick, U. Prediction of outcome using pretreatment (18)F-FDG PET/CT and MRI radiomics in locally advanced cervical cancer treated with chemoradiotherapy. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 768–786. [Google Scholar] [CrossRef]
- Seith, F.; Forschner, A.; Weide, B.; Guckel, B.; Schwartz, M.; Schwenck, J.; Othman, A.E.; Fenchel, M.; Garbe, C.; Nikolaou, K.; et al. Is there a link between very early changes of primary and secondary lymphoid organs in (18)F-FDG-PET/MRI and treatment response to checkpoint inhibitor therapy? J. Immunother. Cancer 2020, 8, e000656. [Google Scholar] [CrossRef] [PubMed]
- Padhani, A.R. Imaging in the evaluation of cancer. In Recommendations for Cross-Sectional Imaging in Cancer Management, 2nd ed.; Nicholson, T., Ed.; The Royal College of Radiologists: London, UK, 2014. [Google Scholar]
- Meignan, M.; Sasanelli, M.; Casasnovas, R.O.; Luminari, S.; Fioroni, F.; Coriani, C.; Masset, H.; Itti, E.; Gobbi, P.G.; Merli, F.; et al. Metabolic tumour volumes measured at staging in lymphoma: Methodological evaluation on phantom experiments and patients. Eur. J. Nucl. Med. Mol. Imaging 2014, 41, 1113–1122. [Google Scholar] [CrossRef]
- Seban, R.D.; Moya-Plana, A.; Antonios, L.; Yeh, R.; Marabelle, A.; Deutsch, E.; Schwartz, L.H.; Gomez, R.G.H.; Saenger, Y.; Robert, C.; et al. Prognostic 18F-FDG PET biomarkers in metastatic mucosal and cutaneous melanoma treated with immune checkpoint inhibitors targeting PD-1 and CTLA-4. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 2301–2312. [Google Scholar] [CrossRef] [PubMed]
- Zagoruyko, S.; Komodakis, N. Wide residual networks. arXiv 2016, arXiv:1605.07146. [Google Scholar]
- Hepp, T.; Fischer, M.; Winkelmann, M.T.; Baldenhofer, S.; Kuestner, T.; Nikolaou, K.; Yang, B.; Gatidis, S. Fully Automated Segmentation and Shape Analysis of the Thoracic Aorta in Non-contrast-enhanced Magnetic Resonance Images of the German National Cohort Study. J. Thorac. Imaging 2020, 35, 389–398. [Google Scholar] [CrossRef]
- Peduzzi, P.; Concato, J.; Feinstein, A.R.; Holford, T.R. Importance of events per independent variable in proportional hazards regression analysis II. Accuracy and precision of regression estimates. J. Clin. Epidemiol. 1995, 48, 1503–1510. [Google Scholar] [CrossRef]
- Mariotto, A.B.; Enewold, L.; Zhao, J.; Zeruto, C.A.; Yabroff, K.R. Medical Care Costs Associated with Cancer Survivorship in the United States. Cancer Epidemiol. Biomark. Prev. 2020, 29, 1304–1312. [Google Scholar] [CrossRef]
- Weber, J.S.; Kähler, K.C.; Hauschild, A. Management of Immune-Related Adverse Events and Kinetics of Response with Ipilimumab. J. Clin. Oncol. 2012, 30, 2691–2697. [Google Scholar] [CrossRef] [PubMed]
- Pak, K.; Cheon, G.J.; Nam, H.Y.; Kim, S.J.; Kang, K.W.; Chung, J.K.; Kim, E.E.; Lee, D.S. Prognostic value of metabolic tumor volume and total lesion glycolysis in head and neck cancer: A systematic review and meta-analysis. J. Nucl. Med. 2014, 55, 884–890. [Google Scholar] [CrossRef]
- Son, S.H.; Kang, S.M.; Jeong, S.Y.; Lee, S.W.; Lee, S.J.; Lee, J.; Ahn, B.C. Prognostic Value of Volumetric Parameters Measured by Pretreatment 18F FDG PET/CT in Patients With Cutaneous Malignant Melanoma. Clin. Nucl. Med. 2016, 41, e266–e273. [Google Scholar] [CrossRef]
- Pires da Silva, I.; Lo, S.; Quek, C.; Gonzalez, M.; Carlino, M.S.; Long, G.V.; Menzies, A.M. Site-specific response patterns, pseudoprogression, and acquired resistance in patients with melanoma treated with ipilimumab combined with anti-PD-1 therapy. Cancer 2020, 126, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Schoder, H.; Teng, R.; Humm, J.L.; Ni, A.; Wolchok, J.D.; Weber, W.A. Prognostic value of baseline metabolic tumor volume measured on (18)F-fluorodeoxyglucose positron emission tomography/computed tomography in melanoma patients treated with ipilimumab therapy. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Hlongwa, K.N.; Mokoala, K.M.G.; Matsena-Zingoni, Z.; Vorster, M.; Sathekge, M.M. The Use of (18)F-FDG PET/CT Metabolic Parameters in Predicting Overall Survival in Patients Undergoing Restaging for Malignant Melanoma. Diagnostics 2022, 12, 590. [Google Scholar] [CrossRef] [PubMed]
- Acciardo, S.; Mignion, L.; Joudiou, N.; Bouzin, C.; Baurain, J.F.; Gallez, B.; Jordan, B.F. Imaging markers of response to combined BRAF and MEK inhibition in BRAF mutated vemurafenib-sensitive and resistant melanomas. Oncotarget 2018, 9, 16832–16846. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bilen, M.A.; Shabto, J.M.; Martini, D.J.; Liu, Y.; Lewis, C.; Collins, H.; Akce, M.; Kissick, H.; Carthon, B.C.; Shaib, W.L.; et al. Sites of metastasis and association with clinical outcome in advanced stage cancer patients treated with immunotherapy. BMC Cancer 2019, 19, 857. [Google Scholar] [CrossRef]
- Sefidgar, M.; Soltani, M.; Raahemifar, K.; Bazmara, H.; Nayinian, S.M.; Bazargan, M. Effect of tumor shape, size, and tissue transport properties on drug delivery to solid tumors. J. Biol. Eng. 2014, 8, 12. [Google Scholar] [CrossRef]
- Poklepovic, A.S.; Carvajal, R.D. Prognostic Value of Low Tumor Burden in Patients With Melanoma. Oncology 2018, 32, e90–e96. [Google Scholar]
- Galon, J.; Mlecnik, B.; Bindea, G.; Angell, H.K.; Berger, A.; Lagorce, C.; Lugli, A.; Zlobec, I.; Hartmann, A.; Bifulco, C.; et al. Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J. Pathol. 2014, 232, 199–209. [Google Scholar] [CrossRef]

| Sex | Age | Therapeutic Agent | Response † | OS | Risk | |
|---|---|---|---|---|---|---|
| 1 | m | 49 | PD1-Ab | progress | 330 * | High |
| 2 | m | 52 | CTLA4-Ab + BRAF/MEK inhibitor | progress | 127 * | High |
| 3 | f | 59 | CTLA4-Ab | progress | 495 * | High |
| 4 | m | 75 | PD1-Ab | partial response | 1266 * | Low |
| 5 | f | 74 | PD1-Ab | progress | 814 * | High |
| 6 | f | 74 | CTLA4-Ab | progress | 170 * | High |
| 7 | f | 53 | PD1-Ab | progress | 148 * | High |
| 8 | m | 51 | CTLA4-Ab | complete response | 1737 | Low |
| 9 | f | 49 | PD1-Ab | complete response | 1732 | Low |
| 10 | m | 59 | CTLA4-Ab | progress | 453 * | High |
| 11 | f | 62 | BRAF inhibitor | progress | 277 * | High |
| 12 | f | 52 | CTLA4-Ab + V-TEC | progress | 1456 | High |
| 13 | m | 84 | PD1-Ab | partial response | 1512 | Low |
| 14 | m | 35 | BRAF/MEK inhibitor | progress | 210 * | High |
| 15 | f | 64 | PD1-Ab | progress | 202 * | High |
| 16 | m | 81 | BRAF/MEK inhibitor | partial response | 869 * | Low |
| 17 | f | 58 | PD1-Ab | progress | 1108 * | High |
| 18 | f | 83 | PD1-Ab | progress | 448 * | High |
| 19 | m | 75 | PD1-Ab | progress | 158 * | High |
| 20 | m | 66 | PD1-Ab | stable disease | 906 * | Low |
| 21 | m | 76 | BRAF/MEK inhibitor | partial response | 243 * | High |
| 22 | m | 73 | PD1-Ab | partial response | 1302 | Low |
| 23 | m | 64 | PD1-Ab | progress | 1240 | High |
| 24 | m | 60 | PD1-Ab | complete response | 1172 | Low |
| 25 | f | 67 | PD1-Ab + CTLA4-Ab | partial response | 1222 | Low |
| 26 | f | 68 | PD1-Ab + CTLA4-Ab | partial response | 1157 | Low |
| 27 | f | 50 | BRAF/MEK inhibitor | complete response | 362 * | High |
| 28 | m | 56 | PD1-Ab | progress (iRECIST, stable disease) | 1101 | High |
| 29 | f | 40 | PD1-Ab | stable disease | 1020 | Low |
| 30 | f | 73 | PD1-Ab | progress | 1021 | High |
| 31 | f | 82 | PD1-Ab + CTLA4-Ab | progress | 976 | High |
| 32 | f | 53 | PD1-Ab + CTLA4-Ab | partial response | 847 | Low |
| 33 | m | 44 | PD1-Ab | progress | 834 | High |
| 34 | m | 40 | PD1-AK + CDK4/6 | progress | 123 * | High |
| 35 | f | 57 | PD1-Ab + CTLA4-Ab | progress | 174 * | High |
| 36 | f | 73 | PD1-Ab | progress | 675 | High |
| 37 | m | 61 | PD1-Ab + CTLA4-Ab | progress | 120 * | High |
| Mean | Range | |
|---|---|---|
| Number of lesions per patient | 26 | 2–200+ |
| Total metabolic tumor volume (mL) | 106 | 1–1982 |
| Total lesion glycolysis | 560 | 1–13,341 |
| Average lesion size per patient (mm) | 16 | 5–48 |
| Intraindividual range in lesion size (mm) | 41 | 4–295 |
| Average ADCmean per patient (mm2/s) | 1039 | 459–1782 |
| Intraindividual range in ADCmean (mm2/s) | 599 | 4–1513 |
| Average SULpeak per patient | 3.4 | 0.6–11.2 |
| Intraindividual range in SULpeak | 4.0 | 0.0–13.9 |
| Number of affected organ regions | 3 | 1–8 |
| Number of Patients with Metastases in Certain Organ Regions | ||
| Lymph nodes | 17 | |
| Soft tissue | 18 | |
| Bone | 13 | |
| Liver | 15 | |
| Spleen | 5 | |
| Lung | 17 | |
| Pleura | 3 | |
| Brain | 10 | |
| Other viscera (excluding liver and spleen) | 9 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Küstner, T.; Vogel, J.; Hepp, T.; Forschner, A.; Pfannenberg, C.; Schmidt, H.; Schwenzer, N.F.; Nikolaou, K.; la Fougère, C.; Seith, F. Development of a Hybrid-Imaging-Based Prognostic Index for Metastasized-Melanoma Patients in Whole-Body 18F-FDG PET/CT and PET/MRI Data. Diagnostics 2022, 12, 2102. https://doi.org/10.3390/diagnostics12092102
Küstner T, Vogel J, Hepp T, Forschner A, Pfannenberg C, Schmidt H, Schwenzer NF, Nikolaou K, la Fougère C, Seith F. Development of a Hybrid-Imaging-Based Prognostic Index for Metastasized-Melanoma Patients in Whole-Body 18F-FDG PET/CT and PET/MRI Data. Diagnostics. 2022; 12(9):2102. https://doi.org/10.3390/diagnostics12092102
Chicago/Turabian StyleKüstner, Thomas, Jonas Vogel, Tobias Hepp, Andrea Forschner, Christina Pfannenberg, Holger Schmidt, Nina F. Schwenzer, Konstantin Nikolaou, Christian la Fougère, and Ferdinand Seith. 2022. "Development of a Hybrid-Imaging-Based Prognostic Index for Metastasized-Melanoma Patients in Whole-Body 18F-FDG PET/CT and PET/MRI Data" Diagnostics 12, no. 9: 2102. https://doi.org/10.3390/diagnostics12092102
APA StyleKüstner, T., Vogel, J., Hepp, T., Forschner, A., Pfannenberg, C., Schmidt, H., Schwenzer, N. F., Nikolaou, K., la Fougère, C., & Seith, F. (2022). Development of a Hybrid-Imaging-Based Prognostic Index for Metastasized-Melanoma Patients in Whole-Body 18F-FDG PET/CT and PET/MRI Data. Diagnostics, 12(9), 2102. https://doi.org/10.3390/diagnostics12092102