Predicting Systemic Health Features from Retinal Fundus Images Using Transfer-Learning-Based Artificial Intelligence Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset and Design
2.2. Materials
2.3. Procedures
Ground Truth Labeling
2.4. Dataset Subdivision
2.5. Dataset Preprocessing
2.6. Model Training and Testing
2.7. Statistical Analysis
3. Results
3.1. Dataset Characteristics
3.2. AI Models Can Predict Systemic Health Features from Fundus Imaging Alone
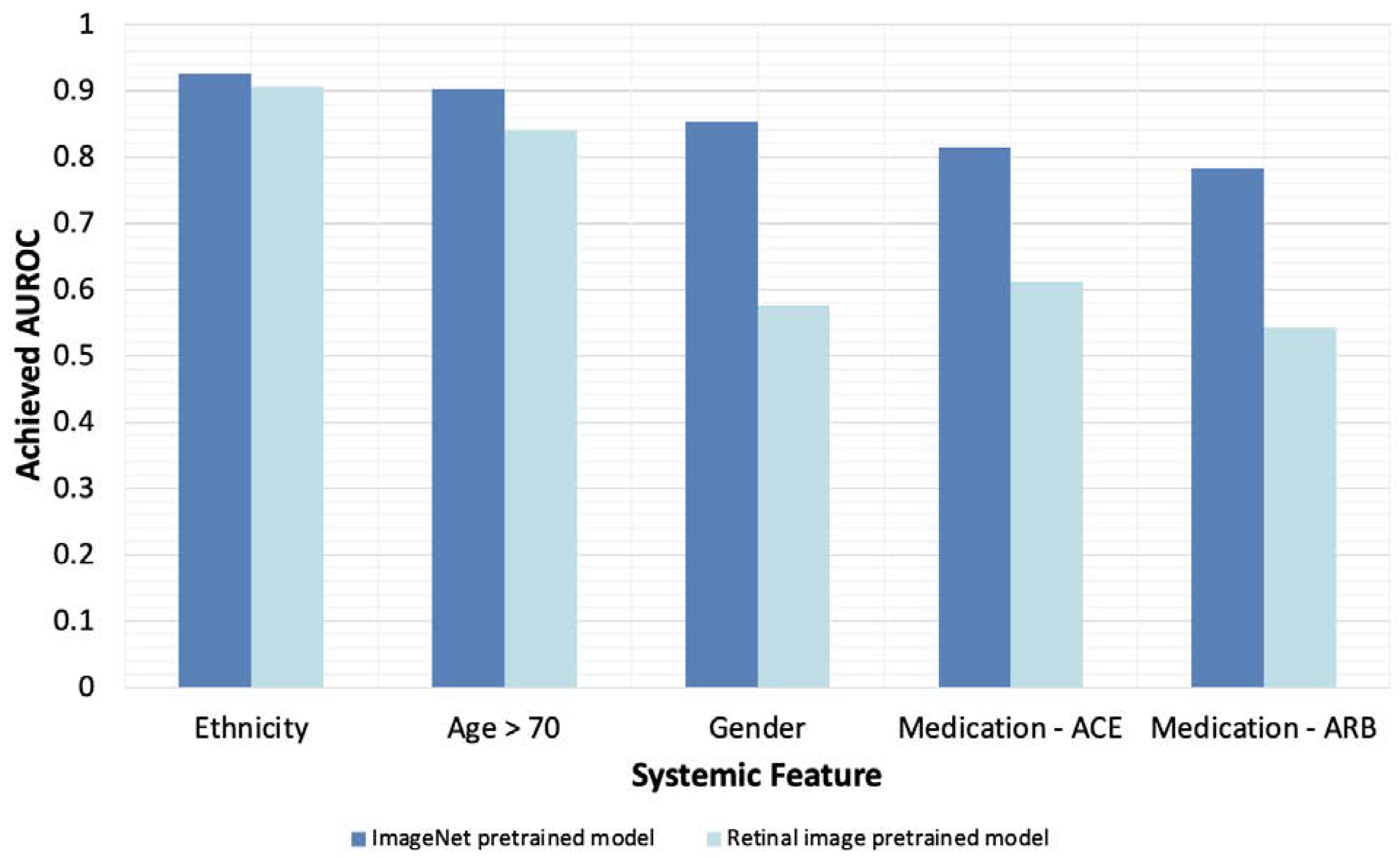
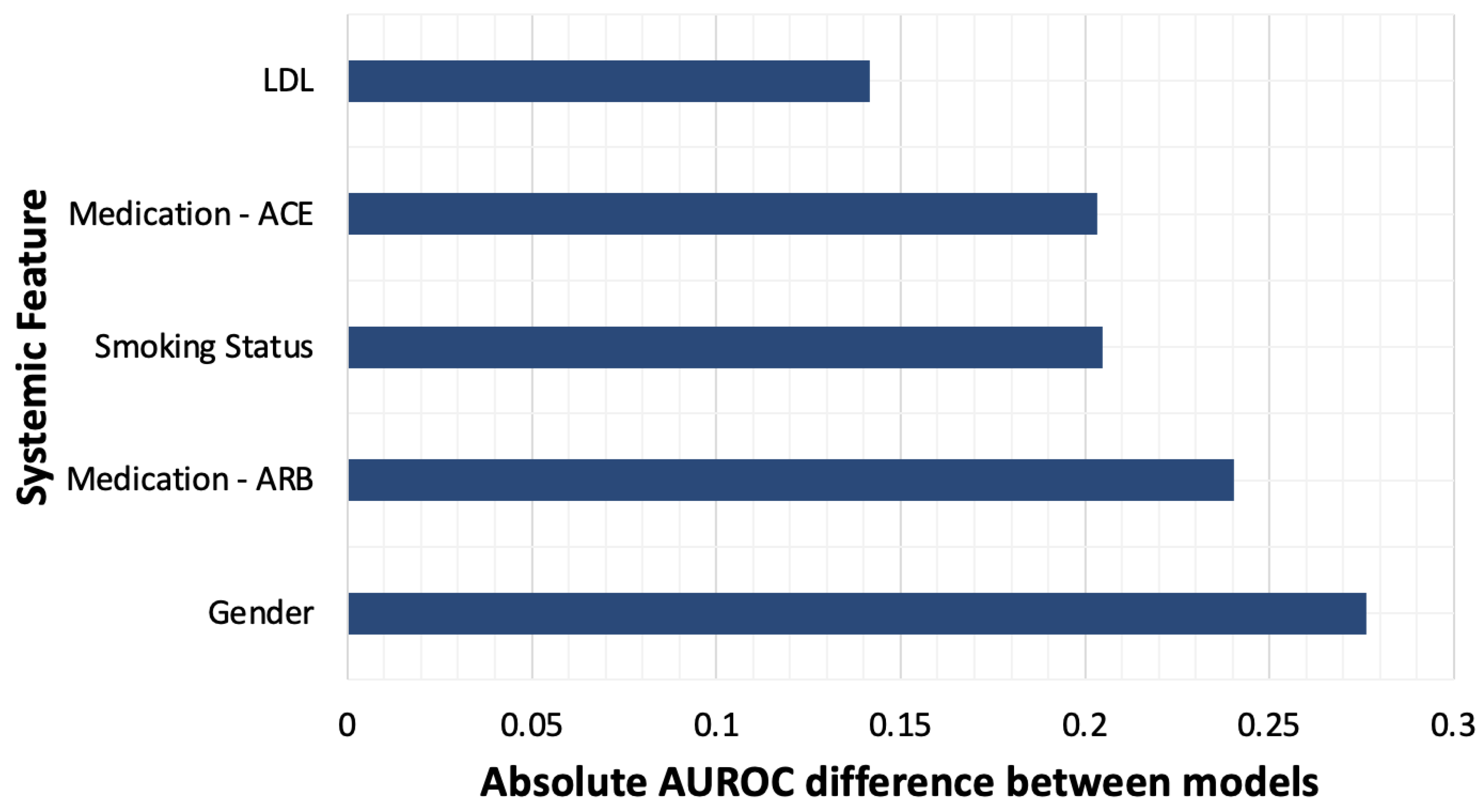
3.3. Pretraining with General Images Optimizes Model Performance
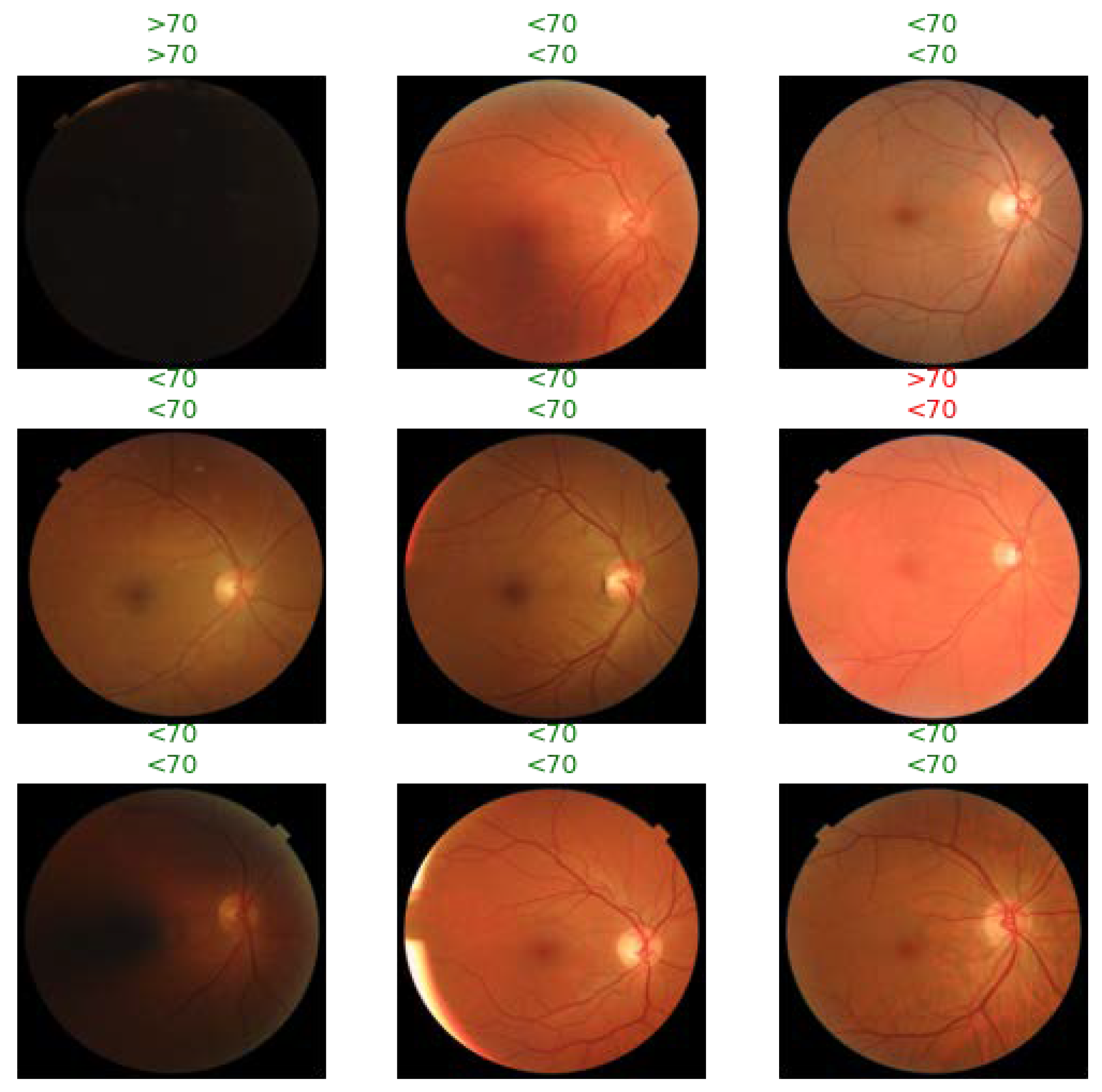
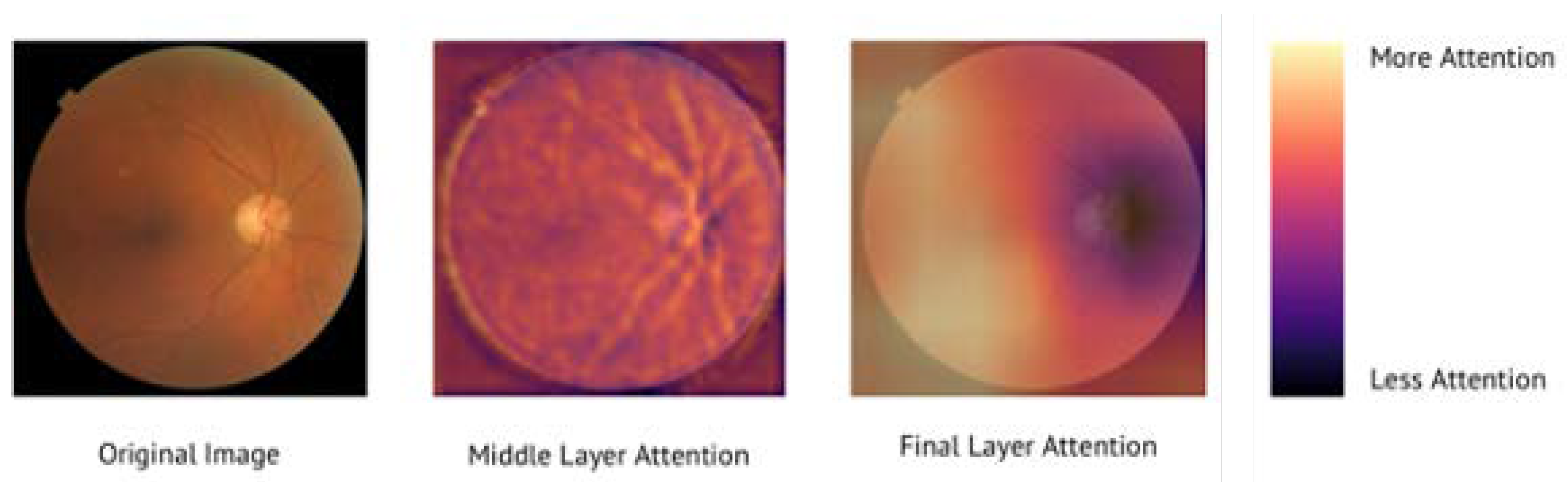
3.4. AI Models Attend to Fundus Images in a Physiologically Valid Manner
3.5. Feature Categories with Missing Data
4. Discussion
4.1. Clinical Significance
4.2. Advantages of Transfer-Learning Techniques
4.3. Addressing Bias in Artificial Intelligence Models
4.4. Limitations and Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abràmoff, M.D.; Garvin, M.K.; Sonka, M. Retinal Imaging and Image Analysis. IEEE Rev. Biomed. Eng. 2010, 3, 169–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadler, Z.; Wollstein, G.; Ishikawa, H.; Schuman, J.S. Clinical Application of Ocular Imaging. Optom. Vis. Sci. 2012, 89, E543–E553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, T.Y.Y. Ocular imaging at the cutting-edge. Eye 2020, 35, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.A.; Bruce, B.B.; Newman, N.J.; Biousse, V. The Use of Retinal Photography in Nonophthalmic Settings and Its Potential for Neurology. Neurologist 2012, 18, 350–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moraru, A.D.; Costin, D.; Moraru, R.L.; Branisteanu, D.C. Artificial intelligence and deep learning in ophthalmology—Present and future (Review). Exp. Ther. Med. 2020, 20, 3469–3473. [Google Scholar] [CrossRef]
- Ludwig, C.A.; Perera, C.; Myung, D.; Greven, M.A.; Smith, S.J.; Chang, R.T.; Leng, T. Automatic Identification of Referral-Warranted Diabetic Retinopathy Using Deep Learning on Mobile Phone Images. Transl. Vis. Sci. Technol. 2020, 9, 60. [Google Scholar] [CrossRef]
- Wintergerst, M.W.; Bejan, V.; Hartmann, V.; Schnorrenberg, M.; Bleckwenn, M.; Weckbecker, K.; Finger, R.P. Telemedical Diabetic Retinopathy Screening in a Primary Care Setting: Quality of Retinal Photographs and Accuracy of Automated Image Analysis. Ophthalmic Epidemiol. 2021, 29, 286–295. [Google Scholar] [CrossRef]
- De Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Askham, H.; Glorot, X.; O’Donoghue, B.; Visentin, D.; et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018, 24, 1342–1350. [Google Scholar] [CrossRef]
- Ting, D.S.W.; Cheung, C.Y.-L.; Lim, G.; Tan, G.S.W.; Quang, N.D.; Gan, A.; Hamzah, H.; Garcia-Franco, R.; Yeo, I.Y.S.; Lee, S.Y.; et al. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. JAMA 2017, 318, 2211–2223. [Google Scholar] [CrossRef]
- Abràmoff, M.D.; Lavin, P.T.; Birch, M.; Shah, N.; Folk, J.C. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digit. Med. 2018, 1, 39. [Google Scholar] [CrossRef]
- Gargeya, R.; Leng, T. Automated Identification of Diabetic Retinopathy Using Deep Learning. Ophthalmology 2017, 124, 962–969. [Google Scholar] [CrossRef]
- Brown, J.; Campbell, J.P.; Beers, A.; Chang, K.; Ostmo, S.; Chan, R.V.P.; Dy, J.; Erdogmus, D.; Ioannidis, S.; Kalpathy-Cramer, J.; et al. Automated Diagnosis of Plus Disease in Retinopathy of Prematurity Using Deep Convolutional Neural Networks. JAMA Ophthalmol. 2018, 136, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Ataer-Cansizoglu, E.; Bolon-Canedo, V.; Campbell, J.P.; Bozkurt, A.; Erdogmus, D.; Kalpathy-Cramer, J.; Patel, S.; Jonas, K.; Chan, R.V.P.; Ostmo, S.; et al. Computer-Based Image Analysis for Plus Disease Diagnosis in Retinopathy of Prematurity: Performance of the “i-ROP” System and Image Features Associated With Expert Diagnosis. Transl. Vis. Sci. Technol. 2015, 4, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrara, D.; Newton, E.M.; Lee, A.Y. Artificial intelligence-based predictions in neovascular age-related macular degeneration. Curr. Opin. Ophthalmol. 2021, 32, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Fu, D.J.; Liefers, B.; Faes, L.; Glinton, S.; Wagner, S.; Struyven, R.; Pontikos, N.; Keane, P.A.; Balaskas, K. Clinically relevant deep learning for detection and quantification of geographic atrophy from optical coherence tomography: A model development and external validation study. Lancet Digit. Heal. 2021, 3, e665–e675. [Google Scholar] [CrossRef]
- Bogunović, H.; Montuoro, A.; Baratsits, M.; Karantonis, M.G.; Waldstein, S.M.; Schlanitz, F.; Schmidt-Erfurth, U. Machine Learning of the Progression of Intermediate Age-Related Macular Degeneration Based on OCT Imaging. Investig. Opthalmology Vis. Sci. 2017, 58, BIO141–BIO150. [Google Scholar] [CrossRef] [Green Version]
- Treder, M.; Lauermann, J.L.; Eter, N. Automated detection of exudative age-related macular degeneration in spectral domain optical coherence tomography using deep learning. Graefe’s Arch. Clin. Exp. Ophthalmol. 2017, 256, 259–265. [Google Scholar] [CrossRef]
- Lee, C.S.; Baughman, D.M.; Lee, A.Y. Deep Learning Is Effective for Classifying Normal versus Age-Related Macular Degeneration OCT Images. Ophthalmol. Retin. 2017, 1, 322–327. [Google Scholar] [CrossRef]
- Liu, H.; Li, L.; Wormstone, I.M.; Qiao, C.; Zhang, C.; Liu, P.; Li, S.; Wang, H.; Mou, D.; Pang, R.; et al. Development and Validation of a Deep Learning System to Detect Glaucomatous Optic Neuropathy Using Fundus Photographs. JAMA Ophthalmol. 2019, 137, 1353–1360. [Google Scholar] [CrossRef]
- Kihara, Y.; Heeren, T.F.C.; Lee, C.S.; Wu, Y.; Xiao, S.; Tzaridis, S.; Holz, F.G.; Issa, P.C.; Egan, C.A.; Lee, A.Y. Estimating Retinal Sensitivity Using Optical Coherence Tomography With Deep-Learning Algorithms in Macular Telangiectasia Type 2. JAMA Netw. Open 2019, 2, e188029. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Heo, T.-Y.; Kim, A.; Kim, J.; Han, K.; Yun, J.; Min, J. Development of a Fundus Image-Based Deep Learning Diagnostic Tool for Various Retinal Diseases. J. Pers. Med. 2021, 11, 321. [Google Scholar] [CrossRef] [PubMed]
- Antaki, F.; Coussa, R.G.; Kahwati, G.; Hammamji, K.; Sebag, M.; Duval, R. Accuracy of automated machine learning in classifying retinal pathologies from ultra-widefield pseudocolour fundus images. Br. J. Ophthalmol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Savoy, M. IDx-DR for Diabetic Retinopathy Screening. Am. Fam. Physician 2020, 101, 307–308. [Google Scholar] [PubMed]
- Bhaskaranand, M.; Ramachandra, C.; Bhat, S.; Cuadros, J.; Nittala, M.G.; Sadda, S.R.; Solanki, K. The Value of Automated Diabetic Retinopathy Screening with the EyeArt System: A Study of More Than 100,000 Consecutive Encounters from People with Diabetes. Diabetes Technol. Ther. 2019, 21, 635–643. [Google Scholar] [CrossRef] [Green Version]
- Commissioner O of the. FDA Permits Marketing of Artificial Intelligence-Based Device to Detect Certain Diabetes-Related Eye Problems. FDA. Available online: https://www.fda.gov/news-events/press-announcements/fda-permits-marketing-artificial-intelligence-based-device-detect-certain-diabetes-related-eye (accessed on 23 January 2022).
- Wagner, S.K.; Fu, D.J.; Faes, L.; Liu, X.; Huemer, J.; Khalid, H.; Ferraz, D.; Korot, E.; Kelly, C.; Balaskas, K.; et al. Insights into Systemic Disease through Retinal Imaging-Based Oculomics. Transl. Vis. Sci. Technol. 2020, 9, 6. [Google Scholar] [CrossRef] [Green Version]
- MacGillivray, T.J.; Trucco, E.; Cameron, J.R.; Dhillon, B.; Houston, J.G.; Van Beek, E.J.R. Retinal imaging as a source of biomarkers for diagnosis, characterization and prognosis of chronic illness or long-term conditions. Br. J. Radiol. 2014, 87, 20130832. [Google Scholar] [CrossRef] [Green Version]
- Günthner, R.; Hanssen, H.; Hauser, C.; Angermann, S.; Lorenz, G.; Kemmner, S.; Matschkal, J.; Braunisch, M.C.; Kuechle, C.; Renders, L.; et al. Impaired Retinal Vessel Dilation Predicts Mortality in End-Stage Renal Disease. Circ. Res. 2019, 124, 1796–1807. [Google Scholar] [CrossRef]
- Dong, L.; He, W.; Zhang, R.; Ge, Z.; Wang, Y.X.; Zhou, J.; Xu, J.; Shao, L.; Wang, Q.; Yan, Y.; et al. Artificial Intelligence for Screening of Multiple Retinal and Optic Nerve Diseases. JAMA Netw. Open 2022, 5, e229960. [Google Scholar] [CrossRef]
- Yip, W.; Ong, P.G.; Teo, B.W.; Cheung, C.; Tai, E.S.; Cheng, C.-Y.; Lamoureux, E.; Wong, T.Y.; Sabanayagam, C. Retinal Vascular Imaging Markers and Incident Chronic Kidney Disease: A Prospective Cohort Study. Sci. Rep. 2017, 7, 9374. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Liu, X.; Xu, J.; Yuan, J.; Cai, W.; Chen, T.; Wang, K.; Gao, Y.; Nie, S.; Xu, X.; et al. Deep-learning models for the detection and incidence prediction of chronic kidney disease and type 2 diabetes from retinal fundus images. Nat. Biomed. Eng. 2021, 5, 533–545. [Google Scholar] [CrossRef]
- Coronado, I.; Abdelkhaleq, R.; Yan, J.; Marioni, S.S.; Jagolino-Cole, A.; Channa, R.; Pachade, S.; Sheth, S.A.; Giancardo, L. Towards Stroke Biomarkers on Fundus Retinal Imaging: A Comparison Between Vasculature Embeddings and General Purpose Convolutional Neural Networks. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Jalisco, Mexico, 9 December 2021; pp. 3873–3876. [Google Scholar] [CrossRef]
- Chang, J.; Ko, A.; Park, S.M.; Choi, S.; Kim, K.; Kim, S.M.; Yun, J.M.; Kang, U.; Shin, I.H.; Shin, J.Y.; et al. Association of Cardiovascular Mortality and Deep Learning-Funduscopic Atherosclerosis Score derived from Retinal Fundus Images. Am. J. Ophthalmol. 2020, 217, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.Y.; Xu, D.; Cheng, C.-Y.; Sabanayagam, C.; Tham, Y.-C.; Yu, M.; Rim, T.H.; Chai, C.Y.; Gopinath, B.; Mitchell, P.; et al. A deep-learning system for the assessment of cardiovascular disease risk via the measurement of retinal-vessel calibre. Nat. Biomed. Eng. 2020, 5, 498–508. [Google Scholar] [CrossRef]
- Coppola, G.; di Renzo, A.; Ziccardi, L.; Martelli, F.; Fadda, A.; Manni, G.; Barboni, P.; Pierelli, F.; Sadun, A.A.; Parisi, V. Optical Coherence Tomography in Alzheimer’s Disease: A Meta-Analysis. PLoS ONE 2015, 10, e0134750. [Google Scholar] [CrossRef]
- Ko, F.; Muthy, Z.A.; Gallacher, J.; Sudlow, C.; Rees, G.; Yang, Q.; Keane, P.A.; Petzold, A.; Khaw, P.T.; Reisman, C.; et al. Association of Retinal Nerve Fiber Layer Thinning With Current and Future Cognitive Decline. JAMA Neurol. 2018, 75, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Mutlu, U.; Colijn, J.M.; Ikram, M.A.; Bonnemaijer, P.W.M.; Licher, S.; Wolters, F.J.; Tiemeier, H.; Koudstaal, P.J.; Klaver, C.C.W. Association of Retinal Neurodegeneration on Optical Coherence Tomography With Dementia. JAMA Neurol. 2018, 75, 1256–1263. [Google Scholar] [CrossRef] [PubMed]
- Moons, L.; De Groef, L. Multimodal retinal imaging to detect and understand Alzheimer’s and Parkinson’s disease. Curr. Opin. Neurobiol. 2021, 72, 1–7. [Google Scholar] [CrossRef]
- Normando, E.M.; Davis, B.; De Groef, L.; Nizari, S.; Turner, L.A.; Ravindran, N.; Pahlitzsch, M.; Brenton, J.; Malaguarnera, G.; Guo, L.; et al. The retina as an early biomarker of neurodegeneration in a rotenone-induced model of Parkinson’s disease: Evidence for a neuroprotective effect of rosiglitazone in the eye and brain. Acta Neuropathol. Commun. 2016, 4, 86. [Google Scholar] [CrossRef] [Green Version]
- Betzler, B.K.; Rim, T.H.; Sabanayagam, C.; Cheng, C.-Y. Artificial Intelligence in Predicting Systemic Parameters and Diseases From Ophthalmic Imaging. Front. Digit. Heal. 2022, 4, 889445. [Google Scholar] [CrossRef]
- Lau, A.Y.; Mok, V.; Lee, J.; Fan, Y.; Zeng, J.; Lam, B.Y.K.; Wong, A.; Kwok, C.; Lai, M.; Zee, B. Retinal image analytics detects white matter hyperintensities in healthy adults. Ann. Clin. Transl. Neurol. 2018, 6, 98–105. [Google Scholar] [CrossRef]
- Poplin, R.; Varadarajan, A.V.; Blumer, K.; Liu, Y.; McConnell, M.V.; Corrado, G.S.; Peng, L.; Webster, D.R. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat. Biomed. Eng. 2018, 2, 158–164. [Google Scholar] [CrossRef]
- Mitani, A.; Huang, A.; Venugopalan, S.; Corrado, G.S.; Peng, L.; Webster, D.R.; Hammel, N.; Liu, Y.; Varadarajan, A.V. Detection of anaemia from retinal fundus images via deep learning. Nat. Biomed. Eng. 2019, 4, 18–27. [Google Scholar] [CrossRef]
- Wisely, C.E.; Wang, D.; Henao, R.; Grewal, D.S.; Thompson, A.C.; Robbins, C.B.; Yoon, S.P.; Soundararajan, S.; Polascik, B.W.; Burke, J.R.; et al. Convolutional neural network to identify symptomatic Alzheimer’s disease using multimodal retinal imaging. Br. J. Ophthalmol. 2020, 106, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Smith, G.; Guo, H.; Liu, B.; Pan, Z.; Wang, Z.; Xiong, S.; Fang, R. Modular machine learning for Alzheimer’s disease classification from retinal vasculature. Sci. Rep. 2021, 11, 238. [Google Scholar] [CrossRef] [PubMed]
- Korot, E.; Pontikos, N.; Liu, X.; Wagner, S.K.; Faes, L.; Huemer, J.; Balaskas, K.; Denniston, A.K.; Khawaja, A.; Keane, P.A. Predicting sex from retinal fundus photographs using automated deep learning. Sci. Rep. 2021, 11, 10286. [Google Scholar] [CrossRef] [PubMed]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef] [Green Version]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Han, D.; Liu, Q.; Fan, W. A new image classification method using CNN transfer learning and web data augmentation. Expert Syst. Appl. 2018, 95, 43–56. [Google Scholar] [CrossRef]
- Morid, M.A.; Borjali, A.; Del Fiol, G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput. Biol. Med. 2020, 128, 104115. [Google Scholar] [CrossRef]
- Kim, H.E.; Cosa-Linan, A.; Santhanam, N.; Jannesari, M.; Maros, M.E.; Ganslandt, T. Transfer learning for medical image classification: A literature review. BMC Med. Imaging 2022, 22, 104115. [Google Scholar] [CrossRef]
- Diabetic Retinopathy Screening. EyePACS. Available online: https://www.eyepacs.com/ (accessed on 9 May 2022).
- Ozbulak, G.; Ekenel, H.K. Initialization of convolutional neural networks by Gabor filters. In Proceedings of the 2018 26th Signal Processing and Communications Applications Conference (SIU), Izmir, Turkey, 2–5 May 2018; pp. 1–4. [Google Scholar] [CrossRef]
- Char, D.S.; Shah, N.H.; Magnus, D. Implementing Machine Learning in Health Care—Addressing Ethical Challenges. N. Engl. J. Med. 2018, 378, 981–983. [Google Scholar] [CrossRef] [Green Version]
- Challen, R.; Denny, J.; Pitt, M.; Gompels, L.; Edwards, T.; Tsaneva-Atanasova, K. Artificial intelligence, bias and clinical safety. BMJ Qual. Saf. 2019, 28, 231–237. [Google Scholar] [CrossRef] [PubMed]

| Demographic Feature | N | Proportion of Dataset (%) |
|---|---|---|
| Unique participants | 760 | – |
| Total fundus images | 1277 | – |
| Right eyes | 650 | 50.9 |
| Left eyes | 627 | 49.1 |
| Sex | ||
| Male | 432 | 54.7 |
| Female | 358 | 45.3 |
| Age (years) | ||
| 20–29 | 23 | 2.9 |
| 30–39 | 59 | 7.5 |
| 40–49 | 130 | 16.5 |
| 50–59 | 196 | 24.8 |
| 60–69 | 203 | 25.7 |
| 70–79 | 126 | 15.9 |
| 80–89 | 46 | 5.8 |
| 90–99 | 7 | 0.9 |
| Race | ||
| Asian | 202 | 25.6 |
| African American/Black | 253 | 32 |
| White | 68 | 8.6 |
| Native American/Pacific Islander | 18 | 2.3 |
| Other/Unknown | 249 | 31.5 |
| Ethnicity | ||
| Hispanic/Latino | 173 | 21.9 |
| Non-Hispanic/Latino | 547 | 69.2 |
| Other/Unknown | 70 | 8.9 |
| Comorbidities | ||
| Cardiac Disease | 669 | 88 |
| Stroke | 584 | 76.8 |
| Hypertension | 696 | 91.6 |
| Diabetic Retinopathy | 90 | 11.8 |
| Systemic Feature | AUROC | Optimized F1 Score | Sensitivity | Specificity |
|---|---|---|---|---|
| Ethnicity | 0.926 | 0.871 | 0.86 | 0.886 |
| Age > 70 | 0.902 | 0.873 | 0.862 | 0.869 |
| Gender | 0.852 | 0.758 | 0.742 | 0.774 |
| Medication—ACEi | 0.815 | 0.804 | 0.811 | 0.791 |
| Medication—ARB | 0.783 | 0.707 | 0.7 | 0.708 |
| LDL | 0.766 | 0.718 | 0.694 | 0.714 |
| HDL | 0.756 | 0.711 | 0.692 | 0.722 |
| Smoking status | 0.732 | 0.697 | 0.632 | 0.713 |
| HbA1c | 0.708 | 0.669 | 0.638 | 0.634 |
| Cardiac disease | 0.7 | 0.669 | 0.625 | 0.598 |
| Medication—Aspirin | 0.696 | 0.681 | 0.673 | 0.685 |
| Hypertension | 0.687 | 0.695 | 0.643 | 0.623 |
| Systemic Feature | AUROC of ImageNet Pre-Trained Model | AUROC of Retinal Image Pre-Trained Model |
|---|---|---|
| Gender | 0.852 | 0.576 |
| Medication—ARB | 0.783 | 0.542 |
| Smoking Status | 0.732 | 0.528 |
| Medication—ACEi | 0.815 | 0.612 |
| LDL | 0.766 | 0.624 |
| Hypertension | 0.687 | 0.585 |
| HDL | 0.756 | 0.667 |
| Cardiac Disease | 0.7 | 0.623 |
| HbA1c | 0.708 | 0.64 |
| Age > 70 | 0.902 | 0.84 |
| Medication—Aspirin | 0.696 | 0.638 |
| Ethnicity | 0.926 | 0.907 |
| Mean AUROC | 0.777 | 0.648 |
| Systemic Feature | Images with Corresponding Patient Data | Images without Corresponding Patient Data |
|---|---|---|
| Ethnicity | 1182 | 95 |
| Gender | 1277 | 0 |
| LDL | 1129 | 148 |
| HDL | 291 | 986 |
| Smoking status | 1247 | 30 |
| Age > 70 | 1277 | 0 |
| Cardiac disease | 1277 | 0 |
| HbA1c | 1183 | 60 |
| Hypertension | 1277 | 0 |
| Medication—ARB | 1277 | 0 |
| Medication—ACEi | 1277 | 0 |
| Medication—Aspirin | 1277 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, N.C.; Perera, C.; Dow, E.R.; Chen, K.M.; Mahajan, V.B.; Mruthyunjaya, P.; Do, D.V.; Leng, T.; Myung, D. Predicting Systemic Health Features from Retinal Fundus Images Using Transfer-Learning-Based Artificial Intelligence Models. Diagnostics 2022, 12, 1714. https://doi.org/10.3390/diagnostics12071714
Khan NC, Perera C, Dow ER, Chen KM, Mahajan VB, Mruthyunjaya P, Do DV, Leng T, Myung D. Predicting Systemic Health Features from Retinal Fundus Images Using Transfer-Learning-Based Artificial Intelligence Models. Diagnostics. 2022; 12(7):1714. https://doi.org/10.3390/diagnostics12071714
Chicago/Turabian StyleKhan, Nergis C., Chandrashan Perera, Eliot R. Dow, Karen M. Chen, Vinit B. Mahajan, Prithvi Mruthyunjaya, Diana V. Do, Theodore Leng, and David Myung. 2022. "Predicting Systemic Health Features from Retinal Fundus Images Using Transfer-Learning-Based Artificial Intelligence Models" Diagnostics 12, no. 7: 1714. https://doi.org/10.3390/diagnostics12071714
APA StyleKhan, N. C., Perera, C., Dow, E. R., Chen, K. M., Mahajan, V. B., Mruthyunjaya, P., Do, D. V., Leng, T., & Myung, D. (2022). Predicting Systemic Health Features from Retinal Fundus Images Using Transfer-Learning-Based Artificial Intelligence Models. Diagnostics, 12(7), 1714. https://doi.org/10.3390/diagnostics12071714






