Abstract
The world is grappling with the coronavirus disease 2019 (COVID-19) pandemic, the causative agent of which is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). COVID-19 symptoms are similar to the common cold, including fever, sore throat, cough, muscle and chest pain, brain fog, dyspnoea, anosmia, ageusia, and headache. The manifestation of the disease can vary from being asymptomatic to severe life-threatening conditions warranting hospitalization and ventilation support. Furthermore, the emergence of mutecated variants of concern (VOCs) is paramount to the devastating effect of the pandemic. This highly contagious virus and its emergent variants challenge the available advanced viral diagnostic methods for high-accuracy testing with faster result yields. This review is to shed light on the natural history, pathology, molecular biology, and efficient diagnostic methods of COVID-19, detecting SARS-CoV-2 in collected samples. We reviewed the gold standard RT-qPCR method for COVID-19 diagnosis to confer a better understanding and application to combat the COVID-19 pandemic. This comprehensive review may further develop awareness about the management of the COVID-19 pandemic.
1. Introduction
Coronavirus disease 2019 (COVID-19) is a highly contagious communicable disease of the present time caused by a novel coronavirus, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) [1]. It is believed that this viral infection was initiated with a zoonotic transfer from a seafood market in Wuhan, China [2]. Initially, the viral outbreak was considered endemic in China but, within a few weeks, the SARS-CoV-2 infection causing COVID-19 was declared a global pandemic by the World Health Organization (WHO) on 11 March 2020 [3]. Till now, the virus has infected 535,863,950 individuals worldwide and is infecting new individuals consistently, developing new clusters of infection (https://covid19.who.int/, accessed on 16 June 2022). We have lost 6,314,972 people and persons aged 65 and older with compromised immunity and with underlying medical conditions, such as chronic lung or liver disease, asthma, diabetes, severe heart problems, etc., are at significant risk of illness, morbidity, and mortality (https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-care/underlyingconditions.html, accessed on 16 June 2022). The detailed chronology and epidemiology of the virus are discussed elsewhere [4]. After the identification of SARS-CoV-2 as the etiological agent of the illness, a race against time was started to develop rapid and efficient diagnostic methods, opening a new avenue for diagnostic innovations [5]. With the availability of the viral genome sequence, quantitative polymerase chain reaction (qPCR) was rapidly adopted as a reliable test for the diagnosis of infection [6]. Although exponential new studies propose novel therapeutic interventions and vaccines, there is a knowledge gap for understanding COVID-19 pathogenesis thoroughly and devising effective strategies to combat the virus in an attempt to alleviate human suffering.
Despite efficient testing and tracing of the infected individuals being central to the countermeasures against the management of the COVID-19, inaccurate testing can undermine these measures against the spread of the infection [7]. Contrarily, a false-positive result can cause avoidable psychological distress, besides wasting resources to manage the nonpatient [8]. This review attempts to encapsulate the current knowledge of the viral pathophysiology with disease diagnosis and to critically analyze the reverse transcriptase-polymerase chain reaction (RT-qPCR) technique, one of the gold standards for the detection of SARS-CoV-2 infection [9,10,11,12,13].
2. History of SARS-CoV-2 or Epidemiology
SARS-CoV-2 belongs to the genus ß-coronavirus of the coronaviridae family of viruses [14,15]. This family comprises enveloped and positive-sense, linear, single-stranded RNA viruses [16]. Among RNA viruses, coronaviruses contain the largest genome [17]. The genome of the early isolate of SARS-CoV-2 from Wuhan is 29,903 nucleotides and the genome size of all other isolates is approximately 30 kb, which is typical of coronaviruses [18,19]. A distinguishing feature of these viruses is the presence of spike-like projections on the surface, which appear like a crown under the electron microscope [11,16]. Hence, these viruses are named coronaviruses as “corona” means “crown” in Latin. These viruses can infect various animals, including humans, are spread through tiny respiratory droplets by direct or indirect contact with infected objects, and can cause respiratory illnesses like the common cold and severe acute respiratory syndrome [16,20]. Out of a total of seven, four human infections caused by coronavirus causing common cold and infection of the upper respiratory tract with mild symptoms are HKU1, NL63, OC43, and 229E [21,22]. These viruses spread via coughing and sneezing and cause mild upper respiratory illness in adults. HKU1, NL63, OC43, and 229E contribute to 15–30% of common cold cases in human adults but can cause severe life-threatening lower respiratory tract infections in immunocompromised individuals, infants, and older person [21]. Coronaviruses responsible for NL63 and 229E are believed to have originated from bat reservoirs, but OC43 and HKU1 are considered rodent-associated [23,24,25]. In addition to human coronaviruses, there are other coronaviruses that exclusively infect animals. Furthermore, the interspecies transmission of these animal viruses to human beings are an emerging threat to human health [26]. Among other animals, wild bats are considered a reservoir of coronaviruses because of their sequence similarity to human coronaviruses [26]. It was reported that SARS-CoV-2 is genetically similar to BatCoV RaTG13 (a BatCoV), indicating that bats might be the natural reservoir of SARS-CoV-2 [4].
In the past two decades, coronaviruses caused two epidemics, SARS-CoV [27,28] and the Middle East respiratory syndrome (MERS) [29]. In February 2003, severe acute respiratory syndrome (SARS) originated in China. The virus spread from China to Hong Kong and to other Asian countries [30]. The spread of SARS proclaimed a new and efficient medium of transmission of viruses by international air travel. The disease symptoms were fever, cough, chest pain, dyspnea, and hypoxemia (low blood oxygen levels), and the global fatality rate was 11% [30,31,32]. There was no available treatment or vaccine against SARS, and the control of the outbreak has relied completely on precaution, detection, and surveillance. Significant steps toward this were disinfection of aircraft and cruise vessels, identification of patients, isolation of patients, contact tracing, and quarantine of symptomatic and asymptomatic persons having any contact with SARS-infected individuals. Additionally, social distancing, wearing masks, frequent handwashing with soap, and use of alcohol-based sanitizer were significant steps in controlling the disease [30,33]. In 2012, MERS was identified in several countries like Saudi Arabia, the United Arab Emirates, and Korea [34]. Symptoms of MERS were similar to SARS-CoV and included fever, cough, and dyspnea, sometimes accompanied by pneumonia and gastrointestinal problems [29]. According to WHO, the fatality rate that was due to MERS was approximately 35% and the lesson learned from SARS proved helpful in controlling MERS. All the precautionary measures of SARS were considered, in addition to prohibiting direct or indirect contact with dromedary camels, as camels were the primary host of MERS [35].
These SARS and MERS infections foreboded a more challenging situation and an upcoming menace. COVID-19 is the third severe disease caused by coronaviruses and poses a severe threat to the world economy and public health. On 11 February 2020, the International Committee on Taxonomy of Viruses (ICTV) named the etiologic agent of COVID-19 as SARS-CoV-2, which was previously known as the 2019 novel coronavirus [15]. This name was proposed because of the higher homology of this new coronavirus with SARS-CoV, the causative agent of SARS [15]. Although SARS-CoV and SARS-CoV-2 are related, these two viruses possess differences. Unlike common-cold-causing human coronaviruses (229E, NL63, OC43, and HKU1) where the infection is confined to the upper respiratory tract, SARS-CoV, MERS-CoV, and SARS-CoV-2 spread from the upper respiratory tract and cause severe infection in the lower respiratory tract, leading to acute lung injury (ALI) and multi-organ failure, eliciting fatal outcomes [36]. Another noteworthy similarity between SARS-CoV and SARS-CoV-2 is the use of ACE2 as a receptor for entry into the host cell [37]. Human coronaviruses (hCoVs) are a constant threat to human health because of their emergence and reemergence, as is evident with SARS-CoV, MERS-CoV, and SARS-CoV-2 infections. Despite similarities between these viruses, there exist obvious differences, as a lesson learned from SARS-CoV is helpful in managing and containing MERS-CoV and SARS-CoV-2. The major problem with the current COVID-19 is the worldwide panic associated with the fast spread of misinformation causing an annoying infodemic [38]. Among these hCoVs, there is a difference in genome size; MERS-CoV possesses the largest genome with approximately 30.11 kb followed by SARS-CoV-2 (~29.9 kb) and SARS-CoV (29.75 kb) [39]. A better understanding of the genomes of hCoVs promotes combat against disease outbreaks by devising strategies for diagnostic systems, drug, and vaccine development [40].
3. Molecular Biology of SARS-CoV-2
SARS-CoV-2 is enveloped in a positive-sense single-stranded RNA virus with a genome size of 29,903 nucleotides (Figure 1) [41,42]. The virion size of this virus varies from 80–120 nm in diameter [43,44]. The nucleotide sequence of SARS-CoV-2 is 79.5% identical to SARS-CoV and 51.8% identical to MERS-CoV [41,45]. This suggests SARS-CoV-2 is closer to SARS-CoV. SARS-CoV and SARS-CoV-2 have similar lengths for most of the proteins. SARS-CoV-2 encodes four structural genes: spike glycoprotein (S), membrane glycoprotein (M), envelope glycoprotein (E), and nucleocapsid (N). The amino acid sequences of these structural genes are ~90% identical with SARS-CoV except the S gene [4,41]. The S protein of SARS-CoV-2 plays a crucial role in the viral entry into the host cell by binding to the host cell-surface receptor angiotensin-converting enzyme-2 (ACE2), and modifications in this protein may lead to different mechanisms and differential intensity of entry into the host cells [46,47,48]. Most of the SARS-CoV-2 non-structural proteins have greater than 85% amino acid sequence identity with SARS-CoV [49]. SARS-CoV-2 possesses four structural proteins: spike glycoprotein (S, 1273 amino acids), envelope glycoprotein (E, 75 amino acids), membrane protein (M, 222 amino acids), and nucleocapsid (N, 419 amino acids) (Figure 1) [41,50]. The N protein is involved in the RNA binding and packaging [50,51]. The most abundant protein in the outer membrane is M-glycoprotein. M and E proteins play a role in viral packaging and the S proteins play a crucial role in host cell binding and infection.
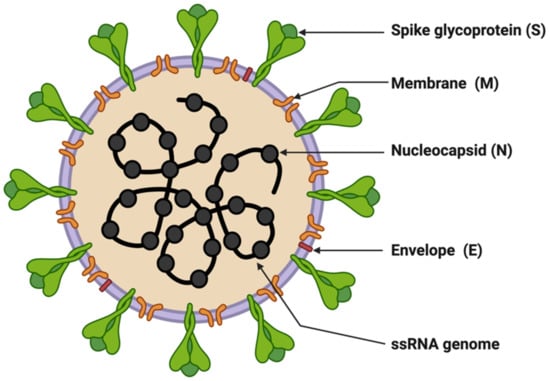
Figure 1.
Structure of SARS-CoV-2. The figure was created with Biorender.com on 8 June 2022.
Entry of SARS-CoV-2 into host cells is mediated by binding the receptor-binding domain (RBD) of the S protein to host cell receptors ACE2 and TMPRSS2, a serine protease that helps in the primming of the S protein [52,53]. ACE2 is present in the lung on pneumocytes II, indicating the lung as the primary target organ of SARS-CoV-2. In addition to this, ACE2 also catalyzes the conversion of regulatory peptides in the cardiovascular system, responding to maintain the homeostatic state, and this activity may account for the rationale behind fatal symptoms including pulmonary embolism or deep venous thrombosis in severe COVID-19 patients [54]. However, the factors contributing to enhanced SARS-CoV-2 transmission are the efficient use of TMPRSS2 compared to SARS-CoV and the higher affinity for ACE2 owing to the modifications in the RBD leading to stabilizing virus-binding hotspots [44,52]. In addition, SARS-CoV-2 entry requires sequential cleavage of the spike glycoprotein at the S1/S2 and the S2’ cleavage sites to mediate membrane fusion. SARS-CoV-2 has a polybasic insertion (PRRAR) at the S1/S2 cleavage site that can be cleaved by furin (furin is a host-cell enzyme in human organs, such as the liver, the lungs, and the small intestines). These factors provide a mechanism called a spring-loaded manner of entry into the host cell, which prohibits endosomal trapping and is accountable for the higher transmissibility of SARS-CoV-2 compared to SARS-CoV [55]. The coronavirus spike (S) glycoprotein is a crucial target for vaccines, therapeutic antibodies, and diagnostics. The SARS-CoV-2 variants of concern (VOCs), Alpha, Delta, and Omicron, have mutations in the S1 subunit of the spike protein, which hosts the RBDs, hence altering the interaction of RBD with host-cell receptor ACE2, resulting in viral entry efficiency into the host cell (Table 1). The Alpha variant has ten modifications in the spike-protein sequence, which results in RBDs being more likely to stay in the ‘up’ position [56].

Table 1.
List of SARS-CoV-2 variants.
The Delta variant hosts multiple mutations in the S1 subunit, including three in the RBD that seem to improve the RBD’s ability to bind to ACE2 and evade the immune system [89]. These multiple mutations in spike proteins enable increased transmission and possible antibody resistance. These variants of SARS-CoV-2 tend to have alterations in furin cleavage sites. In both variants, proline at the 681 position is replaced with other amino acids: in the Alpha, variant proline has been replaced by histidine (P681H), while in the Delta variant, an arginine (P681R) has replaced the proline. These mutations help the virus to transmit into host cells more efficiently. The new Omicron variant has many modifications in the spike protein [90]. Preliminary data indicate that the patients with Omicron infection have mild symptoms, but there is an increased risk of reinfection [91].
4. Diagnostics for COVID-19
Depending on an individual’s age, immune responses, and associated co-morbidities, infection by SARS-CoV-2 leads to highly amassed responses in different individuals ranging from asymptomatic to individuals exhibiting enormously diversified symptoms. Young and healthy people show no or mild symptoms, but they may act as silent carriers and can cause covert infections [92]. Severe COVID-19 cases can end in hospitalization, some necessitating assisted mechanical ventilation, and some cases may be fatal [93].
Identifying infected individuals and asymptomatic viral carriers with rapid and accurate testing has played a pivotal role in containing and mitigating the COVID-19 pandemic. Identification of individuals infected with SARS-CoV-2, either symptomatic or asymptomatic, has prevented further person-to-person disease transmission (https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html, accessed on 16 June 2022). A coalition of multiple methods is in use to diagnose the presence of viral infection in individuals [94]. The primary steps for COVID-19 diagnosis are examining the presence of classical signs and symptoms such as fever or chills, cough, shortness of breath, muscle or body aches, headache, fatigue, sore throat, the new loss of taste or smell, dyspnoea, congestion, or runny nose, nausea or vomiting, conjunctivitis, and gastrointestinal issues (Figure 2) [6]. Furthermore, physical examination of signs including bronchial breath sounds, bronchophony, egophony, wheezing, crackles, rhonchi, and tests such as the anion gap blood test for respiratory acidosis or alkalosis and a complete blood count (CBC) to monitor thrombocytopenia and lymphopenia [95].
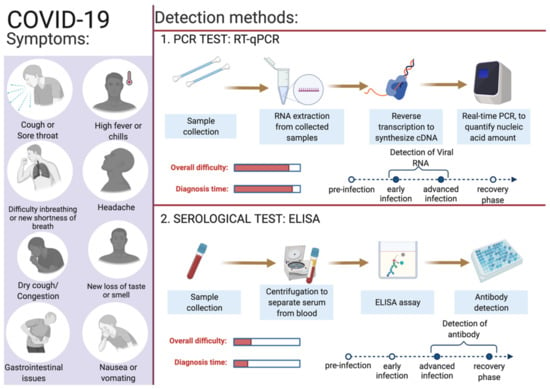
Figure 2.
Overview of COVID-19 symptoms and SARS-CoV-2 detection methods for COVID-19 diagnosis. This figure was created with Biorender.com on 6 June 2022.
SARS-CoV-2 enters the human body as respiratory aerosols; samples from the oropharyngeal or nasopharyngeal are primarily used for viral detection. This virus travels from the upper respiratory tract to the lower respiratory tract, where viral replication occurs. Primarily, the upper respiratory system samples such as oropharyngeal swabs (OPS) and nasopharyngeal swabs (NPSs) are in use for COVID-19 diagnosis [96,97]. Other samples such as saliva, bronchoalveolar lavage (BAL), pleural fluid, tracheal aspirates, blood, urine, and fecal material can also be used for the detection of SARS-CoV-2 infection. For the monitoring and prognosis of the disease at every stage, effective diagnostic tests play a pivotal role. Since the initial report of the SARS-CoV-2 infection, numerous assay kits and tests have been developed for the purpose of COVID-19 diagnosis. Predominantly, there are two types of diagnostic methods in use: the first category is molecular genetics-based (viral test) and the second is serological-based (antibody test) (Figure 2). Among these reverse-transcriptase PCR, isothermal nucleic acid amplification, hybridization microarray assay, serological/immunological SARS-CoV-2 antibody ELISA, and chest CT are promising. In Table 2, a list of different diagnosis methods in use for COVID-19 diagnosis is provided. Advanced molecular biology techniques using polymerase chain reaction (PCR) in real time is a rapid testing method for SARS-CoV-2 infection. This technique is convenient and in use owing to the availability of the genome sequence of SARS-CoV-2. Adapting the PCR technique for COVID-19 diagnosis was straightforward as this technique is in use for the diagnosis of several other diseases, including previous coronavirus infections [11]. The following section describes in detail the use of gold standard RT-qPCR methods to detect the presence of SARS-CoV-2 in collected samples.

Table 2.
List of different diagnostic methods in use.
4.1. Reverse-Transcriptase PCR (RT-qPCR)
The nucleic acid amplification test (NAAT) by RT-qPCR is a sensitive, accurate, and globally accepted gold standard diagnostic method for the SARS-CoV-2 detection [9,10,108]. PCR is being used as a diagnostic test to detect pathogens, novel infections, and antimicrobial resistance profiling [11,109]. PCR is a precise and sensitive method to detect nucleic acids and possesses the potential to generate billions of copies of target DNA from a single copy [109]. This technique relied on an enzyme-driven process for amplifying short regions of DNA in vitro. The requirement of this method is information on at least partial sequences of the target DNA for designing oligonucleotide primers that hybridize specifically to the target sequences [109]. In clinical settings, real-time RT-qPCR is a revolutionary advancement where detection and expression analysis of gene(s) can be carried out in real time, as PCR reaction progresses, and amplification and analysis are done simultaneously in a closed system. This closed system further helps to minimize false-positive results associated with the amplification product contamination [110]. In addition to this, RT-qPCR is fast, sensitive, and reproducible; with the use of automated instrumentation, these features are further enhanced. Recently, NAAT have included other techniques such as isothermal amplification platforms with nicking endonuclease amplification reaction (NEAR), loop-mediated isothermal amplification (LAMP), and transcription-mediated amplification (TMA) [111]. A detailed overview of the RT-qPCR method for SARS-CoV-2 detection is depicted in Figure 3.
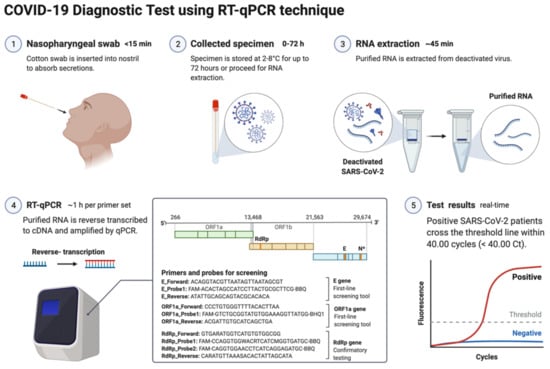
Figure 3.
Schematic representation of COVID-19 diagnostic test using RT-PCR. This figure was created with Biorender.com on 18 May 2022.
4.2. Specimens for Detection of SARS-CoV-2
The genetic material of SARS-CoV-2 (RNA) is first converted into complementary DNA (cDNA) by the action of RNA-dependent DNA polymerase (reverse transcriptase) prior to the actual amplification. For this, viral RNA can be collected from diverse specimens such as ocular secretions, saliva, sputum, bronchoalveolar lavage (BAL), blood, and fecal material, but upper respiratory system samples such as oropharyngeal swabs (OPS) and nasopharyngeal swabs (NPSs) are widely in use [96,97]. In detecting SARS-CoV-2 in various samples, limit of detection (LoD) plays a crucial role [112]. Presently, the best-of-class assay has LoD of ~100 copies of viral RNA per milliliters of transport media; assays with higher LoDs may result in a false negative [112]. Though OPS and NPSs are primarily in use because of lower LoDs, there is a recommendation for the use of combined swabs for COVID-19 diagnosis to avoid false-negative results [113]. Saliva has also been used as a reliable, noninvasive approach for SARS-CoV-2 detection and disease progression [114]. The advantages of using saliva for diagnosis are self-collection, reduced transmission risk during the sample collection, and also a lesser requirement of PPE, trained healthcare professionals, transportation, and storage costs [115]. Importantly, viral load over the course of the infection is detrimental to the analytical sensitivity of assays. It was reported in several studies that the viral load of SARS-CoV-2 peaks during or even shortly before the onset of symptoms and decreases rapidly within the first seven days [115,116]. Furthermore, the virus can be detected in samples for longer periods from the onset of symptoms, usually for 20 days or longer in some patients [117]. There are specific guidelines for sample collection for different specimens by the CDC (https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html, accessed on 16 June 2022). For NPSs and OPS, collecting using only synthetic fiber swabs with thin plastic or wire shafts specifically designed for sampling nasopharyngeal mucosa is recommended. For this patient, the head needs to be tilted back 70 degrees and the swab needs to be inserted slowly into the nostril to contact the nasopharynx. Thereafter, gently rub and roll the swab and leave it for a few seconds to absorb secretions; remove it slowly and place it in the transport tube. These samples can be stored at 2–8 °C for up to 72 h; for longer duration, samples must be stored at −70 °C. Extracted nucleic acid samples must be stored at −70 °C or lower. The collected specimen must be transported to the laboratory while maintaining a cold chain of 2–4 °C throughout [118].
4.3. Biomarkers/Genes Used for RT-qPCR
According to Centers for Disease Control and Prevention (CDC) and WHO guidelines, the RNA samples are reverse-transcribed into cDNA using different primers specific for the open reading frame 1ab (ORF1ab), ORF8, RNA-dependent RNA polymerase (RdRp), hemagglutinin-esterase (HE), and the nucleocapsid genes N1, N2, envelope genes (E), spike genes (S), and transmembrane gene (M), while human RNase P is used as control (Table S1). Some other controls in use for each reaction are no template control, 2019-nCoV positive control, and human specimen control (CDC 2020) [119,120,121]. Additionally, ORF1ab and RdRp are included in RT-qPCR reactions to rule out any potential cross-reactivity, which may occur with other coronaviruses, and to avoid chances of genetic drift in the SARS-CoV-2 genome [122]. As per the CDC recommendation, screening must be done targeting nucleocapsid genes (N1 and N2), but the WHO recommendations require targeting E genes, which must be followed by confirmation using the RdRp gene [122]. Though there is less impact on the detection of SARS-CoV-2 because of emergent variants as most mutations accumulated in the S gene and not in other genes, which are a common target for detection assays. Some VOCs of SARS-CoV-2 (Alfa and Omicron) provides negative results or weaker signals with S-gene RT-qPCR assays, while positive ones with other genes (https://www.ecdc.europa.eu/sites/default/files/documents/Methods-for-the-detection-and-characterisation-of-SARS-CoV-2-variants-first-update.pdf, accessed on 16 June 2022). This effect of no detection of the S gene or weaker signals is referred as S-gene target failure (SGTF) and is due to deletion at nt207–212 (Δ69–70) [123]. Alfa and the majority of Omicron variants of SARS-CoV-2 give negative RT-qPCR results using the S gene, but positive ones with ORF1 and the N gene [124].
The RT-qPCR reaction can be performed in either one or two steps [125,126]. In the conventional two-step RT-qPCR, the reactions for cDNA synthesis and amplification of DNA are conducted separately in two sequential steps, while in one-step RT-qPCR, both the above-mentioned cDNA synthesis and DNA amplification reactions are performed in a single step within one tube containing the requirements to accomplish the entire assay [125]. In detecting SARS-CoV-2 for COVID-19 diagnosis, this one-step RT-qPCR is preferred over the two-step method owing to it being fast and efficient and involving limited sample handling, minimal experimental errors, and a reduced bench time [97,125]. This is followed by cDNA being amplified using fluorescent-based quantitative PCR assays to allow sensitive detection and quantification of the viral RNA [97]. Figure 4 shows the mechanistic steps of DNA amplification and its detection. The qPCR reaction steps are similar to the PCR steps, with initial denaturation of the template at 95 °C for 5–10 min followed by cyclic steps including denaturation (95 °C, 15–20 s), primer/probe annealing (60 °C, 15–20 s), and primer extension (72 °C, 1 min) for gene amplification. Annealing temperature plays a critical role in efficient amplification of the gene of interest and requires optimization and varies from template to template. The annealing temperature determines the qPCR efficiency and depends on the melting temperature (Tm) and is well-established for SARS-CoV-2 detection using different regions of the RNA genome. The qPCR is thereafter continued for 35–45 cycles; during each cycle, the template DNA amount is doubled, resulting in an increase in fluorescent signals. In Figure 4, the sigmoidal curve represents a typical result of the qPCR results, and this helps us interpret the assay outcomes. This curve has three distinct phases: up to cycle 15 or so the curve is near the baseline, in the second phase there is a strong upswing of the cure, usually between 15–30 cycles, and in this phase the amplification signal crosses the threshold. In the third phase, generally after 30 cycles there is a plateau where amplification tapers off and ceases to grow. This curve helps in determining the cycle threshold (Ct) value; this is the point where the curve first clearly rises off the baseline to a statistically significant degree. Crossing this noise threshold is the basis for calling a sample positive in the qualitative assay and the Ct value is the basis for the generation of the standard curve used in the quantifying template in quantitative PCR.
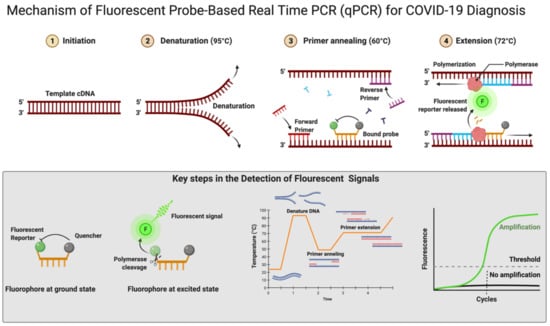
Figure 4.
Mechanism of fluorescent probe-based real-time PCR (qPCR) for COVID-19 diagnosis. Figure was created with Biorender.com on 20 May 2022.
4.4. Reagents (Dyes)
In real-time RT-qPCR, the monitoring of amplification can be done in real time using fluorescent DNA-intercalating dyes such as SYBER green. This dye can bind nonspecifically to the double-stranded DNA generated during the amplification process [127]. There is a more popular alternative approach that uses a fluorescent-labeled internal DNA probe that specifically anneals within the target amplification region and a quencher molecule; this is the case with TaqMan assays [97]. In the TaqMan assay, a fluorescent-labeled oligonucleotide (short DNA molecule) probe is added that is labeled at both the 5′ and 3′ ends. In this, a fluorescent reporter is placed at the 5′ end of the probe and a quencher at the 3′ end, which is also fluorescently labeled. Until there is no amplification, both the 5′ reporter and 3′ quencher are in close proximity and no signal is detected. A fluorescent signal is detected only after the 5′ end reporter and the 3′ end quencher are separated (Figure 4). This separation of reporter and quencher usually takes place because of the enzymatic reaction during RT-qPCR, where the probe is incorporated into the PCR product. The TaqMan assay is more specific and sensitive as it depends upon two processes: first, the primer binding to its specific target sequences and, second, the probe binding to a specific complementary sequence in the downstream region of the primer [128]. An automated system further repeats the amplification process for up to approximately 40 cycles until the viral cDNA can be detected, usually by a fluorescent or electrical signal [129]. There is an effort for the rapid development of fully automated RT-qPCR methods and machines that can be used for quick, accurate results. There are high-throughput machines available that can be used to test 35,000 samples per day and this is further scalable up to 150,000 assays per day. The TaqMan RT-qPCR assay is considered highly sensitive and reproducible; hence, this method can produce reliable results [130]. Using two or more probes, real-time multiplex PCR can be performed to simultaneously detect multiple targets in a single reaction [131,132]. Figure 4 shows the mechanistic details of fluorescent probe-based real-time PCR.
4.5. Ct Value/Threshold Value
In the process of real-time PCR, the target genes are amplified and doubled with each cycle; thus, amplification occurs exponentially. As amplification proceeds, an increasing number of targets become available and the fluorescent signal increases exponentially, producing an exponential curve. The cycle threshold (Ct) value refers to the number of cycles of amplification required for the fluorescence signal of the PCR product or nucleic acid target to be detected or measurable and crossing a threshold or cut-off value is an indication of a positive RT-qPCR test result of a subjected sample [133,134]. This fluorescent signal intensity reflects the amounts of DNA amplicons present at the particular time; generally, after 30–35 cycles the viral cDNA can be quantified, even starting with a very small amount of viral RNA [126].
On the basis of internal controls, RT-qPCR tests can be either qualitative or quantitative, and this affects how a Ct value can be interpreted. In a qualitative RT-qPCR test, known amounts of virus are used to determine whether the Ct values are associated to determine positive and negative test results. In testing a specimen, a Ct value helps interpret a test result as positive or negative, but it cannot be used to determine the exact amount of virus present in an individual patient specimen. In a quantitative RT-qPCR test, a range of known numbers of genome copies (reference samples) are tested as a control in each RT-qPCR reaction; comparing the Ct value of a specimen to the Ct values from the reference samples, the test can calculate the copy number of target nucleic acid. The US Food and Drug Administration (FDA) emergency use authorization (EUA) has approved all SARS-CoV-2 RT-qPCR diagnostic kits only for qualitative test purposes (www.cdc.gov/coronavirus/2019, accessed on 16 June 2022). A list of available RT-qPCR kits for the detection of SARS-CoV-2 approved by the FDA under EUA is provided in the Supplementary Materials (Table S1).
The correlation between Ct value and viral load may be a useful tool for comparison purposes of certain populations including symptomatic and asymptomatic populations. Despite an association between the Ct value and the amount of genetic material in the tested samples, attempting to correlate Ct values and the amount of virus in the original specimen may be faulty. The Ct values of tested samples can be affected by various factors other than viral load, including but not limited to improper collection or storage, processing, or the sensitivity level of the test performed. Thus, a high Ct value can result from factors unrelated to the amount of virus present in the specimen. Hence, Ct values should not be used to infer a relationship with the viral load from a specimen, nor should they be used to determine the level of infection risk (https://www.cdc.gov/coronavirus/2019-ncov/lab/faqs.html, accessed on 16 June 2022). Some countries including India provide Ct values in RT-qPCR results. The significance of this Ct value is that it determines infectivity; a Ct value of 35 or lower is considered COVID-19 positive, while a patient with a Ct value higher than 35 is considered negative for COVID-19.
Additionally, concerns with the experimental result are false-positive or false-negative detections. A false-positive test refers to a false indication for infection present without any infection or presence of virus, while a false-negative test leads to patients declared to be “uninfected”, despite being infected [135]. The main reason for false-positive results is laboratory error, sample contamination, and cross-reactivity or off-target reactions (the test cross-reacting with something that is not SARS-CoV-2), while false-negative RT-PCR results can be due to a low level of viral RNA, improper sample collection, loss or damage during transportation, inefficient extraction, and improper storage conditions. Furthermore, positive PCR results indicate the presence of viral RNA, but this may not necessarily confirm the presence of the infectious virus. Finally, PCR positivity depends on specimen types; it declines more rapidly while using NP swabs compared to the sputum [136].
5. Limitations of RT-qPCR Detection Technique for SARS-CoV-2
Despite wide acceptance and use of the real-time PCR (qPCR) method as a gold standard molecular test of choice with high specificity and accuracy, it has limitations. This method demands professional skilled personnel and is associated with a high cost of instruments and a laboratory setup with a biosafety level 2 cabinet. This method requires absolute cleanliness and a sterile environment because of the high sensitivity of the assay, which can be contaminated easily and may sequalae in false-positive results. A false-positive result may occur because of contamination; furthermore, this can occur because of the presence of shedding of viral residual RNA in recovered patients. Furthermore, a false negative is the prime concern of many available commercial RT-qPCR kits because of lower diagnostic efficiency than optimal [137,138]. Considering the high incidence of false negative RT-qPCR results, the US Food and Drug Administration (FDA) has concluded that a negative RT-qPCR does not completely rule out the SARS-CoV-2 infection. To overcome the challenges of conventional RT-qPCR, many biomedical companies have developed diagnostic platforms that are fully automated and take less time to declare results [139].
6. Future Perspectives and Conclusions
COVID-19 is one of the deadliest pandemics in world history and requires the unmet attention of each and every citizen of the world to control the pandemic. The first step in containing SARS-CoV-2 is the detection of infected persons and appropriate isolation and treatment. Toward this aim, the need for efficient detection methods is imperative, with fast, accurate, and reliable result outputs. For this, RT-qPCR-based detection is the gold standard method of the present time and constant improvements for better and faster screening are in progress. Furthermore, asymptomatic individuals act as carriers and transmit the virus unknowingly. To stop this carrier-mediated viral transmission, all individuals must be tested for infection on a regular basis. Hence, the development of herd immunity using a vaccine regimen, discovery, and availability of suitable therapeutics and regular testing may help us fight against the COVID-19 pandemic.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/diagnostics12061503/s1, Table S1: Overview of RT-PCR kits for the diagnosis of COVID-19, approved by the FDA under an emergency use authorization (EUA).
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.S.; Smith, D.W. COVID-19: A novel zoonotic disease caused by a coronavirus from China: What we know and What we don’t. Microbiol. Aust. 2020, 41, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020, 91, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Sethuraman, N.; Jeremiah, S.S.; Ryo, A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA 2020, 323, 2249–2251. [Google Scholar] [CrossRef]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef]
- Escandon, K.; Rasmussen, A.L.; Bogoch, I.I.; Murray, E.J.; Escandon, K.; Popescu, S.V.; Kindrachuk, J. COVID-19 false dichotomies and a comprehensive review of the evidence regarding public health, COVID-19 symptomatology, SARS-CoV-2 transmission, mask wearing, and reinfection. BMC Infect. Dis. 2021, 21, 710. [Google Scholar] [CrossRef]
- Woloshin, S.; Patel, N.; Kesselheim, A.S. False Negative Tests for SARS-CoV-2 Infection—Challenges and Implications. N. Engl. J. Med. 2020, 383, e38. [Google Scholar] [CrossRef]
- Liu, R.; Han, H.; Liu, F.; Lv, Z.; Wu, K.; Liu, Y.; Feng, Y.; Zhu, C. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin. Chim. Acta 2020, 505, 172–175. [Google Scholar] [CrossRef]
- Drame, M.; Tabue Teguo, M.; Proye, E.; Hequet, F.; Hentzien, M.; Kanagaratnam, L.; Godaert, L. Should RT-PCR be considered a gold standard in the diagnosis of COVID-19? J. Med. Virol. 2020, 92, 2312–2313. [Google Scholar] [CrossRef]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef] [PubMed]
- Chau, C.H.; Strope, J.D.; Figg, W.D. COVID-19 Clinical Diagnostics and Testing Technology. Pharmacotherapy 2020, 40, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Waller, J.V.; Kaur, P.; Tucker, A.; Lin, K.K.; Diaz, M.J.; Henry, T.S.; Hope, M. Diagnostic Tools for Coronavirus Disease (COVID-19): Comparing CT and RT-PCR Viral Nucleic Acid Testing. AJR Am. J. Roentgenol. 2020, 215, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Machhi, J.; Herskovitz, J.; Senan, A.M.; Dutta, D.; Nath, B.; Oleynikov, M.D.; Blomberg, W.R.; Meigs, D.D.; Hasan, M.; Patel, M.; et al. The Natural History, Pathobiology, and Clinical Manifestations of SARS-CoV-2 Infections. J. Neuroimmune Pharm. 2020, 15, 359–386. [Google Scholar] [CrossRef]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Pal, M.; Berhanu, G.; Desalegn, C.; Kandi, V. Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An Update. Cureus 2020, 12, e7423. [Google Scholar] [CrossRef]
- Mousavizadeh, L.; Ghasemi, S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2021, 54, 159–163. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Flamholz, A.; Phillips, R.; Milo, R. SARS-CoV-2 (COVID-19) by the numbers. eLife 2020, 9, e57309. [Google Scholar] [CrossRef]
- Smith, E.C.; Denison, M.R. Implications of altered replication fidelity on the evolution and pathogenesis of coronaviruses. Curr. Opin. Virol. 2012, 2, 519–524. [Google Scholar] [CrossRef]
- Ye, Z.W.; Yuan, S.; Yuen, K.S.; Fung, S.Y.; Chan, C.P.; Jin, D.Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020, 16, 1686–1697. [Google Scholar] [CrossRef]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). Encycl. Virol. 2021, 2, 428–440. [Google Scholar] [CrossRef]
- Van der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.; Wolthers, K.C.; Wertheim-van Dillen, P.M.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a new human coronavirus. Nat. Med. 2004, 10, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Huang, Y.; Lau, S.K.; Yuen, K.Y. Coronavirus genomics and bioinformatics analysis. Viruses 2010, 2, 1804–1820. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.X.; Ng, Y.L.; Tam, J.P.; Liu, D.X. Human Coronaviruses: A Review of Virus-Host Interactions. Diseases 2016, 4, 26. [Google Scholar] [CrossRef]
- Irwin, R.J.; McEwen, S.A.; Clarke, R.C.; Meek, A.H. The prevalence of verocytotoxin-producing Escherichia coli and antimicrobial resistance patterns of nonverocytotoxin-producing Escherichia coli and Salmonella in Ontario broiler chickens. Can. J. Vet. Res. 1989, 53, 411–418. [Google Scholar]
- Poon, L.L.; Chu, D.K.; Chan, K.H.; Wong, O.K.; Ellis, T.M.; Leung, Y.H.; Lau, S.K.; Woo, P.C.; Suen, K.Y.; Yuen, K.Y.; et al. Identification of a novel coronavirus in bats. J. Virol. 2005, 79, 2001–2009. [Google Scholar] [CrossRef]
- Peiris, J.S.; Lai, S.T.; Poon, L.L.; Guan, Y.; Yam, L.Y.; Lim, W.; Nicholls, J.; Yee, W.K.; Yan, W.W.; Cheung, M.T.; et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef]
- Satija, N.; Lal, S.K. The molecular biology of SARS coronavirus. Ann. N. Y. Acad. Sci. 2007, 1102, 26–38. [Google Scholar] [CrossRef]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Hung, L.S. The SARS epidemic in Hong Kong: What lessons have we learned? J. R. Soc. Med. 2003, 96, 374–378. [Google Scholar] [CrossRef]
- Hung, E.C.; Chim, S.S.; Chan, P.K.; Tong, Y.K.; Ng, E.K.; Chiu, R.W.; Leung, C.B.; Sung, J.J.; Tam, J.S.; Lo, Y.M. Detection of SARS coronavirus RNA in the cerebrospinal fluid of a patient with severe acute respiratory syndrome. Clin. Chem. 2003, 49, 2108–2109. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Qi, T.; Liu, L.; Ling, Y.; Qian, Z.; Li, T.; Li, F.; Xu, Q.; Zhang, Y.; Xu, S.; et al. Clinical progression of patients with COVID-19 in Shanghai, China. J. Infect. 2020, 80, e1–e6. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, A.S.; Tomlinson, B.; Cockram, C.S.; Thomas, G.N. Lessons from the severe acute respiratory syndrome outbreak in Hong Kong. Emerg. Infect. Dis. 2003, 9, 1042–1045. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.Y.; Kang, J.M.; Ha, Y.E.; Park, G.E.; Lee, J.Y.; Ko, J.H.; Lee, J.Y.; Kim, J.M.; Kang, C.I.; Jo, I.J.; et al. MERS-CoV outbreak following a single patient exposure in an emergency room in South Korea: An epidemiological outbreak study. Lancet 2016, 388, 994–1001. [Google Scholar] [CrossRef]
- Mackay, I.M.; Arden, K.E. MERS coronavirus: Diagnostics, epidemiology and transmission. Virol. J. 2015, 12, 222. [Google Scholar] [CrossRef]
- Vos, L.M.; Bruyndonckx, R.; Zuithoff, N.P.A.; Little, P.; Oosterheert, J.J.; Broekhuizen, B.D.L.; Lammens, C.; Loens, K.; Viveen, M.; Butler, C.C.; et al. Lower respiratory tract infection in the community: Associations between viral aetiology and illness course. Clin. Microbiol. Infect. 2021, 27, 96–104. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Zhu, Z.; Lian, X.; Su, X.; Wu, W.; Marraro, G.A.; Zeng, Y. From SARS and MERS to COVID-19: A brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir. Res. 2020, 21, 224. [Google Scholar] [CrossRef]
- Al-Qahtani, A.A. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): Emergence, history, basic and clinical aspects. Saudi J. Biol. Sci. 2020, 27, 2531–2538. [Google Scholar] [CrossRef]
- Llanes, A.; Restrepo, C.M.; Caballero, Z.; Rajeev, S.; Kennedy, M.A.; Lleonart, R. Betacoronavirus Genomes: How Genomic Information has been Used to Deal with Past Outbreaks and the COVID-19 Pandemic. Int. J. Mol. Sci. 2020, 21, 4546. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses Executive Committee. The new scope of virus taxonomy: Partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 2020, 5, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Ren, L.L.; Wang, Y.M.; Wu, Z.Q.; Xiang, Z.C.; Guo, L.; Xu, T.; Jiang, Y.Z.; Xiong, Y.; Li, Y.J.; Li, X.W.; et al. Identification of a novel coronavirus causing severe pneumonia in human: A descriptive study. Chin. Med. J. 2020, 133, 1015–1024. [Google Scholar] [CrossRef]
- Lau, Y.L.; Peiris, J.S. Pathogenesis of severe acute respiratory syndrome. Curr. Opin. Immunol. 2005, 17, 404–410. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef]
- Mariano, G.; Farthing, R.J.; Lale-Farjat, S.L.M.; Bergeron, J.R.C. Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be. Front. Mol. Biosci. 2020, 7, 605236. [Google Scholar] [CrossRef]
- Das, A.; Ahmed, R.; Akhtar, S.; Begum, K.; Banu, S. An overview of basic molecular biology of SARS-CoV-2 and current COVID-19 prevention strategies. Gene Rep. 2021, 23, 101122. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Sun, P.D. High affinity binding of SARS-CoV-2 spike protein enhances ACE2 carboxypeptidase activity. J. Biol. Chem. 2020, 295, 18579–18588. [Google Scholar] [CrossRef]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef]
- Gobeil, S.M.; Janowska, K.; McDowell, S.; Mansouri, K.; Parks, R.; Stalls, V.; Kopp, M.F.; Manne, K.; Li, D.; Wiehe, K.; et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science 2021, 373. [Google Scholar] [CrossRef]
- McCallum, M.; Bassi, J.; De Marco, A.; Chen, A.; Walls, A.C.; Di Iulio, J.; Tortorici, M.A.; Navarro, M.J.; Silacci-Fregni, C.; Saliba, C.; et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 2021, 373, 648–654. [Google Scholar] [CrossRef]
- Carroll, T.; Fox, D.; van Doremalen, N.; Ball, E.; Morris, M.K.; Sotomayor-Gonzalez, A.; Servellita, V.; Rustagi, A.; Yinda, C.K.; Fritts, L.; et al. The B.1.427/1.429 (epsilon) SARS-CoV-2 variants are more virulent than ancestral B.1 (614G) in Syrian hamsters. PLoS Pathog. 2022, 18, e1009914. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Simulundu, E.; Mupeta, F.; Chanda-Kapata, P.; Saasa, N.; Changula, K.; Muleya, W.; Chitanga, S.; Mwanza, M.; Simusika, P.; Chambaro, H.; et al. First COVID-19 case in Zambia—Comparative phylogenomic analyses of SARS-CoV-2 detected in African countries. Int. J. Infect. Dis. 2021, 102, 455–459. [Google Scholar] [CrossRef]
- Wink, P.L.; Volpato, F.C.Z.; Monteiro, F.L.; Willig, J.B.; Zavascki, A.P.; Barth, A.L.; Martins, A.F. First identification of SARS-CoV-2 lambda (C.37) variant in Southern Brazil. Infect. Control. Hosp. Epidemiol. 2021, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Darvishi, M.; Rahimi, F.; Talebi Bezmin Abadi, A. SARS-CoV-2 Lambda (C.37): An emerging variant of concern? Gene Rep. 2021, 25, 101378. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.E.; Davila-Barclay, A.; Salvatierra, G.; Gonzalez, L.; Cuicapuza, D.; Solis, L.; Marcos-Carbajal, P.; Huancachoque, J.; Maturrano, L.; Tsukayama, P. The Emergence of Sars-CoV-2 Variant Lambda (C.37) in South America. Microbiol. Spectr. 2021, 9, e0078921. [Google Scholar] [CrossRef] [PubMed]
- Lee, R. B.1.1.7: What We Know about the Novel SARS-CoV-2 Variant. Available online: https://asm.org/Articles/2021/January/B-1-1-7-What-We-Know-About-the-Novel-SARS-CoV-2-Va (accessed on 12 June 2022).
- Tang, J.W.; Tambyah, P.A.; Hui, D.S. Emergence of a new SARS-CoV-2 variant in the UK. J. Infect. 2021, 82, e27–e28. [Google Scholar] [CrossRef]
- Wall, E.C.; Wu, M.; Harvey, R.; Kelly, G.; Warchal, S.; Sawyer, C.; Daniels, R.; Hobson, P.; Hatipoglu, E.; Ngai, Y.; et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet 2021, 397, 2331–2333. [Google Scholar] [CrossRef]
- Lovelace, B., Jr. WHO Says Delta Is Becoming the Dominant COVID Variant Globally. Available online: https://www.cnbc.com/2021/06/18/who-says-delta-is-becoming-the-dominant-covid-variant-globally.html (accessed on 12 June 2022).
- Voloch, C.M.; da Silva Francisco, R., Jr.; de Almeida, L.G.P.; Cardoso, C.C.; Brustolini, O.J.; Gerber, A.L.; Guimaraes, A.P.C.; Mariani, D.; da Costa, R.M.; Ferreira, O.C., Jr.; et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021, 95, e00119-21. [Google Scholar] [CrossRef]
- Nonaka, C.K.V.; Graf, T.; Barcia, C.A.L.; Costa, V.F.; de Oliveira, J.L.; Passos, R.D.H.; Bastos, I.N.; de Santana, M.C.B.; Santos, I.M.; de Sousa, K.A.F.; et al. SARS-CoV-2 variant of concern P.1 (Gamma) infection in young and middle-aged patients admitted to the intensive care units of a single hospital in Salvador, Northeast Brazil, February 2021. Int. J. Infect. Dis. 2021, 111, 47–54. [Google Scholar] [CrossRef]
- Annavajhala, M.K.; Mohri, H.; Wang, P.; Nair, M.; Zucker, J.E.; Sheng, Z.; Gomez-Simmonds, A.; Kelley, A.L.; Tagliavia, M.; Huang, Y.; et al. Emergence and expansion of SARS-CoV-2 B.1.526 after identification in New York. Nature 2021, 597, 703–708. [Google Scholar] [CrossRef]
- Yang, W.; Greene, S.K.; Peterson, E.R.; Li, W.; Mathes, R.; Graf, L.; Lall, R.; Hughes, S.; Wang, J.; Fine, A. Epidemiological characteristics of the B.1.526 SARS-CoV-2 variant. Sci. Adv. 2022, 8, eabm0300. [Google Scholar] [CrossRef]
- Delli Compagni, E.; Mangone, I.; Bonfini, B.; Di Gennaro, A.; Teodori, L.; Leone, A.; Casaccia, C.; Portanti, O.; Averaimo, D.; Zilli, K.; et al. Whole-Genome Sequences of SARS-CoV-2 Lineage B.1.525 Strains (Variant eta) Detected from Patients in the Abruzzo Region (Central Italy) during Spring 2021. Microbiol. Resour. Announc. 2021, 10, e0061821. [Google Scholar] [CrossRef]
- Ozer, E.A.; Simons, L.M.; Adewumi, O.M.; Fowotade, A.A.; Omoruyi, E.C.; Adeniji, J.A.; Olayinka, O.A.; Dean, T.J.; Zayas, J.; Bhimalli, P.P.; et al. Multiple expansions of globally uncommon SARS-CoV-2 lineages in Nigeria. Nat. Commun. 2022, 13, 688. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.; Kemp, S.A.; Datir, R.; Saito, A.; Meng, B.; Rakshit, P.; Takaori-Kondo, A.; Kosugi, Y.; Uriu, K.; Kimura, I.; et al. SARS-CoV-2 B.1.617 Mutations L452R and E484Q Are Not Synergistic for Antibody Evasion. J. Infect. Dis. 2021, 224, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Haw, N.J.L.; Canal, E.M.R.; Zuasula, J., Jr.; Loreche, M.J.; Bernadas, J. Epidemiological characteristics of the SARS-CoV-2 Theta variant (P.3) in the Central Visayas region, Philippines, 30 October 2020-16 February 2021. West. Pac. Surveill. Response J. 2022, 13, 1–3. [Google Scholar] [CrossRef]
- Halfmann, P.J.; Kuroda, M.; Armbrust, T.; Theiler, J.; Balaram, A.; Moreno, G.K.; Accola, M.A.; Iwatsuki-Horimoto, K.; Valdez, R.; Stoneman, E.; et al. Characterization of the SARS-CoV-2 B.1.621 (Mu) variant. Sci. Transl. Med. 2022, eabm4908. [Google Scholar] [CrossRef]
- Barrera-Avalos, C.; Luraschi, R.; Acuna-Castillo, C.; Vidal, M.; Mella-Torres, A.; Inostroza-Molina, A.; Vera, R.; Vargas, S.; Hernandez, I.; Perez, C.; et al. Description of Symptoms Caused by the Infection of the SARS-CoV-2 B.1.621 (Mu) Variant in Patients With Complete CoronaVac Vaccination Scheme: First Case Report From Santiago of Chile. Front. Public Health 2022, 10, 797569. [Google Scholar] [CrossRef]
- Manouana, G.P.; Nzamba Maloum, M.; Bikangui, R.; Oye Bingono, S.O.; Ondo Nguema, G.; Honkpehedji, J.Y.; Rossatanga, E.G.; Zoa-Assoumou, S.; Pallerla, S.R.; Rachakonda, S.; et al. Emergence of B.1.1.318 SARS-CoV-2 viral lineage and high incidence of alpha B.1.1.7 variant of concern in the Republic of Gabon. Int. J. Infect. Dis. 2022, 114, 151–154. [Google Scholar] [CrossRef]
- Scheepers, C.; Everatt, J.; Amoako, D.G.; Tegally, H.; Wibmer, C.K.; Mnguni, A.; Ismail, A.; Mahlangu, B.; Lambson, B.E.; Richardson, S.I.; et al. Emergence and phenotypic characterization of C.1.2, a globally detected lineage that rapidly accumulated mutations of concern. medRxiv 2021. [Google Scholar] [CrossRef]
- Colson, P.; Delerce, J.; Burel, E.; Dahan, J.; Jouffret, A.; Fenollar, F.; Yahi, N.; Fantini, J.; La Scola, B.; Raoult, D. Emergence in Southern France of a new SARS-CoV-2 variant of probably Cameroonian origin harbouring both substitutions N501Y and E484K in the spike protein. medRxiv 2021. [Google Scholar] [CrossRef]
- Ren, S.Y.; Wang, W.B.; Gao, R.D.; Zhou, A.M. Omicron variant (B.1.1.529) of SARS-CoV-2: Mutation, infectivity, transmission, and vaccine resistance. World J. Clin. Cases 2022, 10, 1–11. [Google Scholar] [CrossRef]
- World Health Organization. Enhancing Response to Omicron SARS-CoV-2 Variant. Available online: https://www.who.int/publications/m/item/enhancing-readiness-for-omicron-(b.1.1.529)-technical-brief-and-priority-actions-for-member-states (accessed on 12 June 2022).
- Chen, J.; Wei, G.W. Omicron BA.2 (B.1.1.529.2): High Potential for Becoming the Next Dominant Variant. J. Phys. Chem. Lett. 2022, 13, 3840–3849. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, S.; Sarkar, R. Mutational and phylogenetic analyses of the two lineages of the Omicron variant. J. Med. Virol. 2022, 94, 1777–1779. [Google Scholar] [CrossRef] [PubMed]
- Desingu, P.A.; Nagarajan, K.; Dhama, K. Emergence of Omicron third lineage BA.3 and its importance. J. Med. Virol. 2022, 94, 1808–1810. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Epidemiological Update: SARS-CoV-2 Omicron Sub-Lineages BA.4 and BA.5. Available online: https://www.ecdc.europa.eu/en/news-events/epidemiological-update-sars-cov-2-omicron-sub-lineages-ba4-and-ba5 (accessed on 12 June 2022).
- Worl Health Organization. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 12 June 2022).
- Khateeb, J.; Li, Y.; Zhang, H. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Crit. Care 2021, 25, 244. [Google Scholar] [CrossRef]
- Callaway, E.; Ledford, H. How bad is Omicron? What scientists know so far. Nature 2021, 600, 197–199. [Google Scholar] [CrossRef]
- Pulliam, J.R.C.; van Schalkwyk, C.; Govender, N.; von Gottberg, A.; Cohen, C.; Groome, M.J.; Dushoff, J.; Mlisana, K.; Moultrie, H. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv 2021. [Google Scholar] [CrossRef]
- Oran, D.P.; Topol, E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection: A Narrative Review. Ann. Intern. Med. 2020, 173, 362–367. [Google Scholar] [CrossRef]
- Richardson, S.; Hirsch, J.S.; Narasimhan, M.; Crawford, J.M.; McGinn, T.; Davidson, K.W.; The Northwell COVID-19 Research Consortium; Barnaby, D.P.; Becker, L.B.; Chelico, J.D.; et al. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA 2020, 323, 2052–2059. [Google Scholar] [CrossRef]
- Wu, S.Y.; Yau, H.S.; Yu, M.Y.; Tsang, H.F.; Chan, L.W.C.; Cho, W.C.S.; Shing Yu, A.C.; Yuen Yim, A.K.; Li, M.J.W.; Wong, Y.K.E.; et al. The diagnostic methods in the COVID-19 pandemic, today and in the future. Expert Rev. Mol. Diagn. 2020, 20, 985–993. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Xia, J.; Tong, J.; Liu, M.; Shen, Y.; Guo, D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J. Med. Virol. 2020, 92, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent. Sci. 2020, 6, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. COVID-19 Testing: What You Need to Know. Available online: https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/testing.html (accessed on 12 June 2022).
- Kashir, J.; Yaqinuddin, A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med. Hypotheses 2020, 141, 109786. [Google Scholar] [CrossRef] [PubMed]
- Khan, P.; Aufdembrink, L.M.; Engelhart, A.E. Isothermal SARS-CoV-2 Diagnostics: Tools for Enabling Distributed Pandemic Testing as a Means of Supporting Safe Reopenings. ACS Synth. Biol. 2020, 9, 2861–2880. [Google Scholar] [CrossRef]
- James, A.S.; Alawneh, J.I. COVID-19 Infection Diagnosis: Potential Impact of Isothermal Amplification Technology to Reduce Community Transmission of SARS-CoV-2. Diagnostics 2020, 10, 399. [Google Scholar] [CrossRef]
- Gorzalski, A.J.; Tian, H.; Laverdure, C.; Morzunov, S.; Verma, S.C.; VanHooser, S.; Pandori, M.W. High-Throughput Transcription-mediated amplification on the Hologic Panther is a highly sensitive method of detection for SARS-CoV-2. J. Clin. Virol. 2020, 129, 104501. [Google Scholar] [CrossRef]
- Rahman, M.R.; Hossain, M.A.; Mozibullah, M.; Mujib, F.A.; Afrose, A.; Shahed-Al-Mahmud, M.; Apu, M.A.I. CRISPR is a useful biological tool for detecting nucleic acid of SARS-CoV-2 in human clinical samples. Biomed. Pharm. 2021, 140, 111772. [Google Scholar] [CrossRef]
- Vindeirinho, J.M.; Pinho, E.; Azevedo, N.F.; Almeida, C. SARS-CoV-2 Diagnostics Based on Nucleic Acids Amplification: From Fundamental Concepts to Applications and Beyond. Front. Cell Infect. Microbiol. 2022, 12, 799678. [Google Scholar] [CrossRef]
- Lee, E.Y.P.; Ng, M.Y.; Khong, P.L. COVID-19 pneumonia: What has CT taught us? Lancet Infect. Dis. 2020, 20, 384–385. [Google Scholar] [CrossRef]
- Sidiq, Z.; Hanif, M.; Dwivedi, K.K.; Chopra, K.K. Benefits and limitations of serological assays in COVID-19 infection. Indian J. Tuberc 2020, 67, S163–S166. [Google Scholar] [CrossRef]
- Karp, D.G.; Danh, K.; Espinoza, N.F.; Seftel, D.; Robinson, P.V.; Tsai, C.T. A serological assay to detect SARS-CoV-2 antibodies in at-home collected finger-prick dried blood spots. Sci. Rep. 2020, 10, 20188. [Google Scholar] [CrossRef] [PubMed]
- Van Kasteren, P.B.; van der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Rothman, R.E. PCR-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004, 4, 337–348. [Google Scholar] [CrossRef]
- Deepak, S.; Kottapalli, K.; Rakwal, R.; Oros, G.; Rangappa, K.; Iwahashi, H.; Masuo, Y.; Agrawal, G. Real-Time PCR: Revolutionizing Detection and Expression Analysis of Genes. Curr. Genom. 2007, 8, 234–251. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Nucleic Acid Amplification Tests (NAATs). Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/naats.html (accessed on 15 June 2022).
- Arnaout, R.; Lee, R.A.; Lee, G.R.; Callahan, C.; Yen, C.F.; Smith, K.P.; Arora, R.; Kirby, J.E. SARS-CoV2 Testing: The Limit of Detection Matters. bioRxiv 2020. [Google Scholar] [CrossRef]
- Sharma, K.; Aggarwala, P.; Gandhi, D.; Mathias, A.; Singh, P.; Sharma, S.; Negi, S.S.; Bhargava, A.; Das, P.; Gaikwad, U.; et al. Comparative analysis of various clinical specimens in detection of SARS-CoV-2 using rRT-PCR in new and follow up cases of COVID-19 infection: Quest for the best choice. PLoS ONE 2021, 16, e0249408. [Google Scholar] [CrossRef]
- Medeiros da Silva, R.C.; Nogueira Marinho, L.C.; de Araujo Silva, D.N.; Costa de Lima, K.; Pirih, F.Q.; Luz de Aquino Martins, A.R. Saliva as a possible tool for the SARS-CoV-2 detection: A review. Travel Med. Infect. Dis. 2020, 38, 101920. [Google Scholar] [CrossRef]
- To, K.K.; Tsang, O.T.; Yip, C.C.; Chan, K.H.; Wu, T.C.; Chan, J.M.; Leung, W.S.; Chik, T.S.; Choi, C.Y.; Kandamby, D.H.; et al. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin. Infect. Dis. 2020, 71, 841–843. [Google Scholar] [CrossRef]
- Becherer, L.; Borst, N.; Bakheit, M.; Frischmann, S.; Zengerle, R.; von Stetten, F. Loop-mediated isothermal amplification (LAMP)—Review and classification of methods for sequence-specific detection. Anal. Methods 2020, 12, 717–746. [Google Scholar] [CrossRef]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- Shrestha, L.B.; Pokharel, K. Standard Operating Procedure for Specimen Collection, Packaging and Transport for Diagnosis of SARS-COV-2. JNMA J. Nepal Med. Assoc. 2020, 58, 627–629. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Yip, C.C.; To, K.K.; Tang, T.H.; Wong, S.C.; Leung, K.H.; Fung, A.Y.; Ng, A.C.; Zou, Z.; Tsoi, H.W.; et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J. Clin. Microbiol. 2020, 58, e00310-20. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.W.; Pan, Y.; Cheng, S.M.S.; Hui, K.P.Y.; Krishnan, P.; Liu, Y.; Ng, D.Y.M.; Wan, C.K.C.; Yang, P.; Wang, Q.; et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brunink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58, e00512-20. [Google Scholar] [CrossRef] [PubMed]
- Dudas, G.; Hong, S.L.; Potter, B.I.; Calvignac-Spencer, S.; Niatou-Singa, F.S.; Tombolomako, T.B.; Fuh-Neba, T.; Vickos, U.; Ulrich, M.; Leendertz, F.H.; et al. Emergence and spread of SARS-CoV-2 lineage B.1.620 with variant of concern-like mutations and deletions. Nat. Commun. 2021, 12, 5769. [Google Scholar] [CrossRef]
- Erster, O.; Beth-Din, A.; Asraf, H.; Levy, V.; Kabat, A.; Mannasse, B.; Azar, R.; Shifman, O.; Lazar, S.; Mandelboim, M.; et al. SPECIFIC DETECTION OF SARS-COV-2 B.1.1.529 (OMICRON) VARIANT BY FOUR RT-qPCR DIFFERENTIAL ASSAYS. medRxiv 2021. [Google Scholar] [CrossRef]
- Bustin, S.A.; Nolan, T. RT-qPCR Testing of SARS-CoV-2: A Primer. Int. J. Mol. Sci. 2020, 21, 3004. [Google Scholar] [CrossRef]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef]
- Higuchi, R.; Dollinger, G.; Walsh, P.S.; Griffith, R. Simultaneous amplification and detection of specific DNA sequences. Nat. Biotechnol. 1992, 10, 413–417. [Google Scholar] [CrossRef]
- Tajadini, M.; Panjehpour, M.; Javanmard, S.H. Comparison of SYBR Green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Adv. Biomed. Res. 2014, 3, 85. [Google Scholar] [CrossRef] [PubMed]
- VanGuilder, H.D.; Vrana, K.E.; Freeman, W.M. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 2008, 44, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Leutenegger, C.M.; Boretti, F.S.; Mislin, C.N.; Flynn, J.N.; Schroff, M.; Habel, A.; Junghans, C.; Koenig-Merediz, S.A.; Sigrist, B.; Aubert, A.; et al. Immunization of cats against feline immunodeficiency virus (FIV) infection by using minimalistic immunogenic defined gene expression vector vaccines expressing FIV gp140 alone or with feline interleukin-12 (IL-12), IL-16, or a CpG motif. J. Virol. 2000, 74, 10447–10457. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tsuji, S.; Iguchi, Y.; Shibata, N.; Teramura, I.; Kitagawa, T.; Yamanaka, H. Real-time multiplex PCR for simultaneous detection of multiple species from environmental DNA: An application on two Japanese medaka species. Sci. Rep. 2018, 8, 9138. [Google Scholar] [CrossRef]
- Elnifro, E.M.; Ashshi, A.M.; Cooper, R.J.; Klapper, P.E. Multiplex PCR: Optimization and application in diagnostic virology. Clin. Microbiol. Rev. 2000, 13, 559–570. [Google Scholar] [CrossRef]
- Engelmann, I.; Alidjinou, E.K.; Ogiez, J.; Pagneux, Q.; Miloudi, S.; Benhalima, I.; Ouafi, M.; Sane, F.; Hober, D.; Roussel, A.; et al. Preanalytical Issues and Cycle Threshold Values in SARS-CoV-2 Real-Time RT-PCR Testing: Should Test Results Include These? ACS Omega 2021, 6, 6528–6536. [Google Scholar] [CrossRef]
- Caraguel, C.G.; Stryhn, H.; Gagne, N.; Dohoo, I.R.; Hammell, K.L. Selection of a cutoff value for real-time polymerase chain reaction results to fit a diagnostic purpose: Analytical and epidemiologic approaches. J. Vet. Diagn. Invest. 2011, 23, 2–15. [Google Scholar] [CrossRef]
- Mouliou, D.S.; Gourgoulianis, K.I. False-positive and false-negative COVID-19 cases: Respiratory prevention and management strategies, vaccination, and further perspectives. Expert Rev. Respir. Med. 2021, 15, 993–1002. [Google Scholar] [CrossRef]
- Wolfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Muller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef]
- Younes, N.; Al-Sadeq, D.W.; Al-Jighefee, H.; Younes, S.; Al-Jamal, O.; Daas, H.I.; Yassine, H.M.; Nasrallah, G.K. Challenges in Laboratory Diagnosis of the Novel Coronavirus SARS-CoV-2. Viruses 2020, 12, 582. [Google Scholar] [CrossRef]
- Xu, J.; Wu, R.; Huang, H.; Zheng, W.; Ren, X.; Wu, N.; Ji, B.; Lv, Y.; Liu, Y.; Mi, R. Computed Tomographic Imaging of 3 Patients With Coronavirus Disease 2019 Pneumonia With Negative Virus Real-time Reverse-Transcription Polymerase Chain Reaction Test. Clin. Infect. Dis. 2020, 71, 850–852. [Google Scholar] [CrossRef] [PubMed]
- Habli, Z.; Saleh, S.; Zaraket, H.; Khraiche, M.L. COVID-19 in-vitro Diagnostics: State-of-the-Art and Challenges for Rapid, Scalable, and High-Accuracy Screening. Front. Bioeng. Biotechnol. 2020, 8, 605702. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).