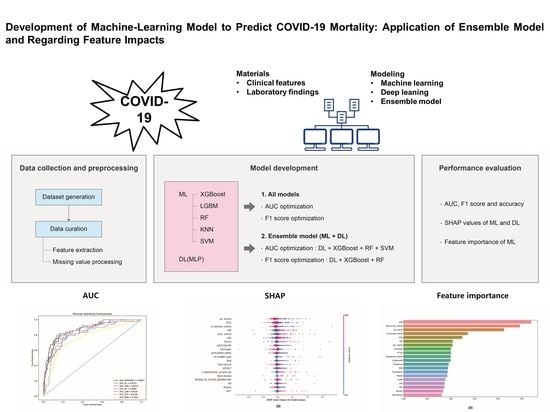
Development of Machine-Learning Model to Predict COVID-19 Mortality: Application of Ensemble Model and Regarding Feature Impacts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Data Collection
2.2. Statistical Data Analyses
2.3. DL Model Selection and Training
2.4. ML Model Selection
2.5. ML Model Development
2.6. Development of an Ensemble Model Using DL and ML
2.7. Performance Measurement
2.8. Shapley Additive exPlanations (SHAP) Method
2.9. Feature Extraction
2.10. Standard Scaling
2.11. Cross-Validation
3. Results
3.1. Demographic and Clinical Characteristics
3.2. Data Features and Parameters
3.3. DL (Neural Network) Performance Optimized by the AUC and F1 Score
3.4. Performances of ML Models Optimized by AUC
3.5. ML Model Results Optimized According to F1 Score
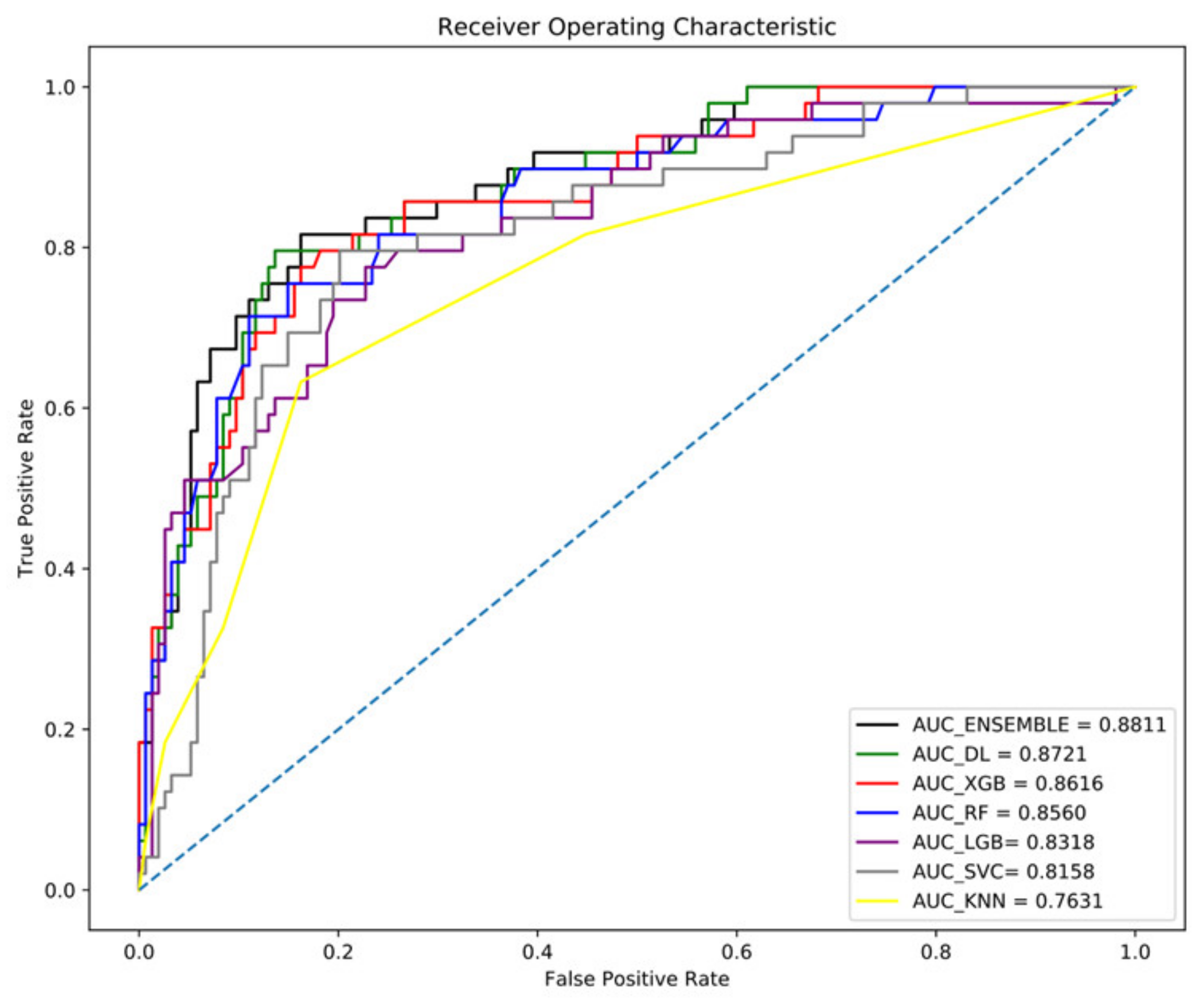
3.6. Ensemble Model of DL and ML Models
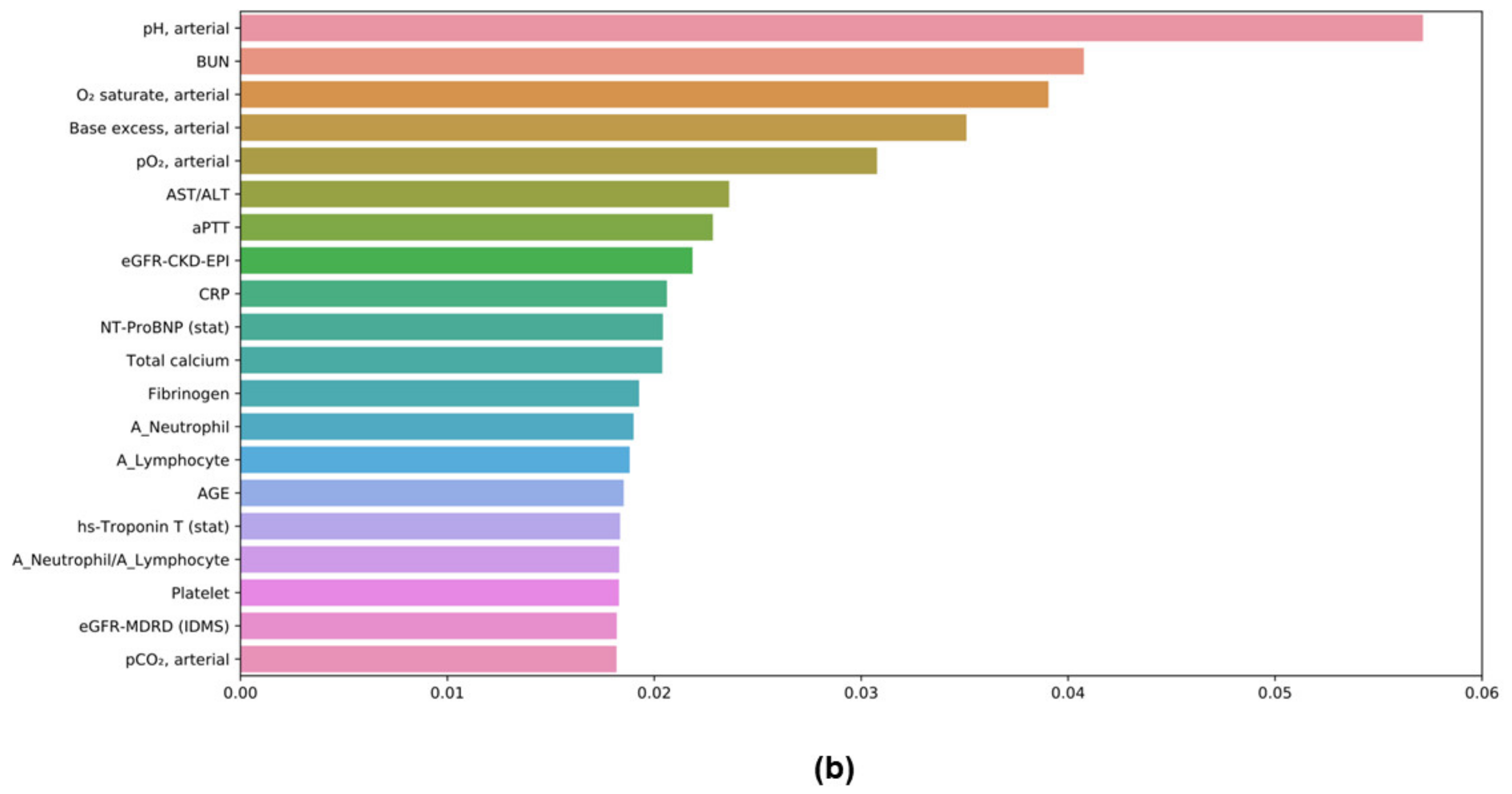
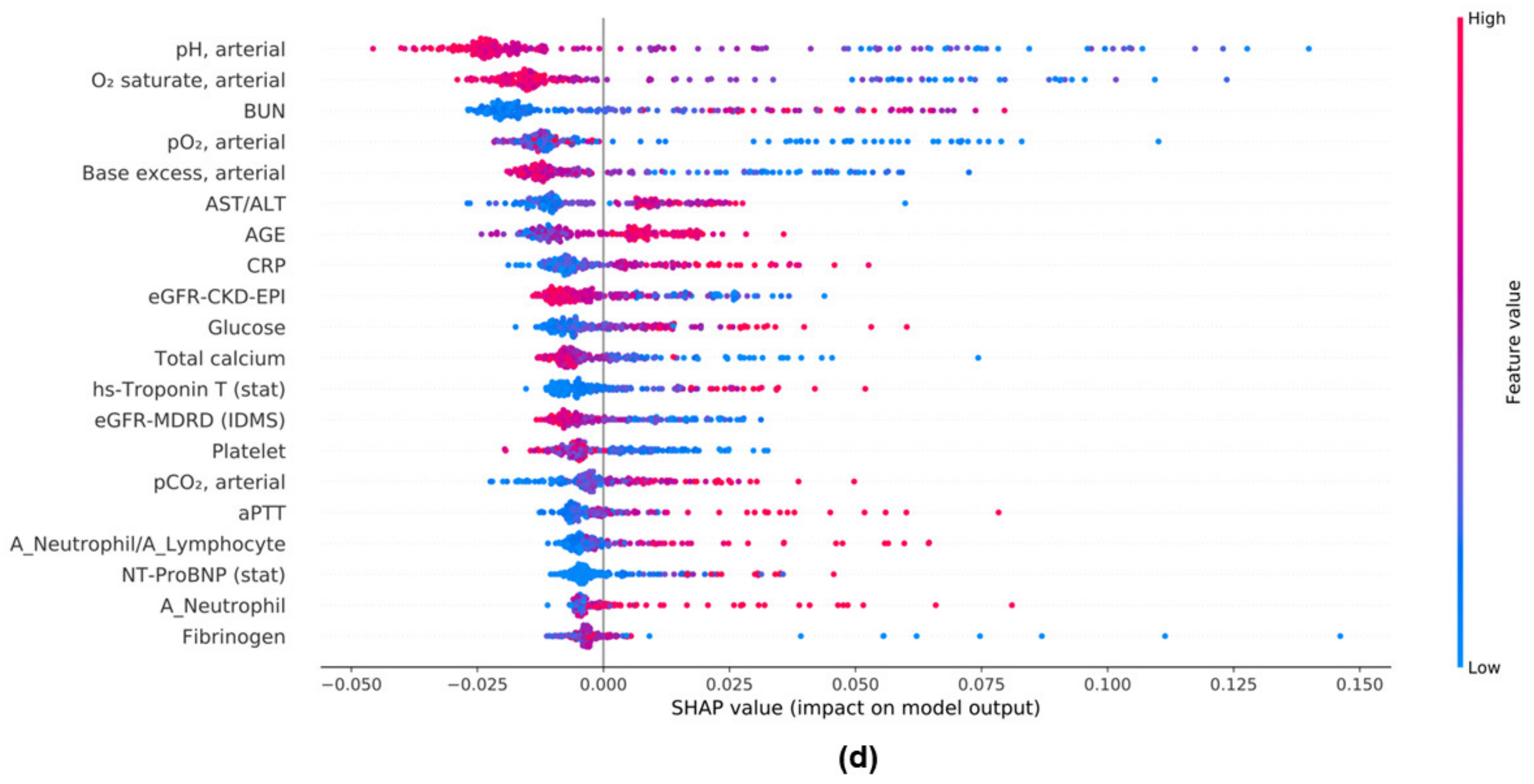
3.7. Feature Importances in ML Models
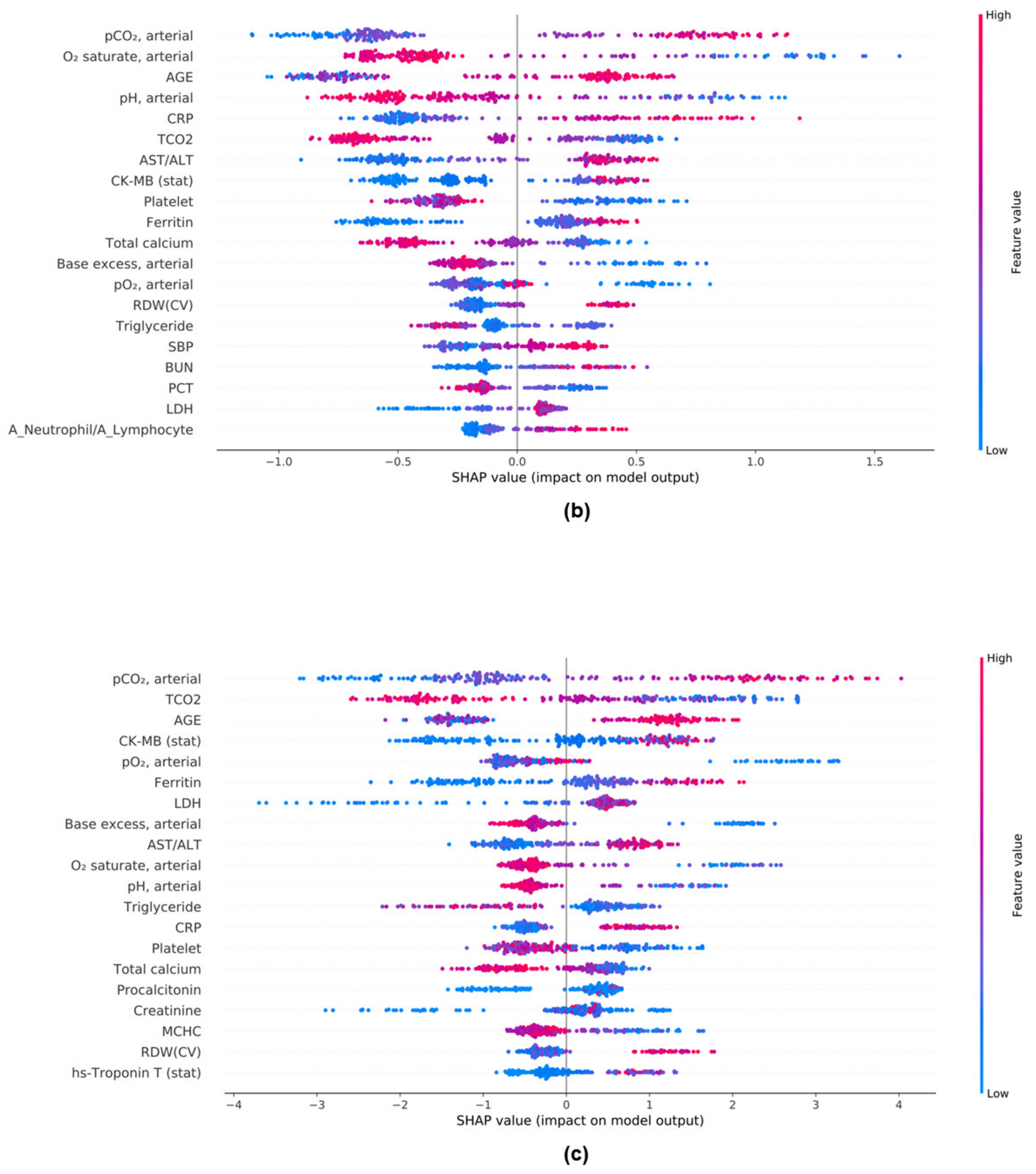
3.8. Evaluation of the Model Performance Impact When Using the Mean SHAP Value in DL and ML Models
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Coronavirus Disease (COVID-19). Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 31 December 2019).
- Korea Disease Control and Prevention Agency. Available online: https://www.kdca.go.kr/ (accessed on 15 February 2022).
- Nedyalkova, M.; Vasighi, M.; Sappati, S.; Kumar, A.; Madurga, S.; Simeonov, V. Inhibition Ability of Natural Compounds on Receptor-Binding Domain of SARS-CoV2: An In Silico Approach. Pharmaceuticals 2021, 14, 1328. [Google Scholar] [CrossRef] [PubMed]
- Nedyalkova, M.; Simeonov, V. Partitioning Pattern of Natural Products Based on Molecular Properties Descriptors Representing Drug-Likeness. Symmetry 2021, 13, 546. [Google Scholar] [CrossRef]
- Sutton, D.; Fuchs, K.; D’Alton, M.; Goffman, D. Universal Screening for SARS-CoV-2 in Women Admitted for Delivery. N. Engl. J. Med. 2020, 382, 2163–2164. [Google Scholar] [CrossRef] [PubMed]
- Baggett, T.P.; Keyes, H.; Sporn, N.; Gaeta, J.M. Prevalence of SARS-CoV-2 Infection in Residents of a Large Homeless Shelter in Boston. JAMA 2020, 323, 2191–2192. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.H.; Tornatore, J.M.; Lawrence, K.E.; Illuzzi, J.L.; Sussman, L.S.; Lipkind, H.S.; Pettker, C.M. Prevalence of SARS-CoV-2 Among Patients Admitted for Childbirth in Southern Connecticut. JAMA 2020, 323, 2520–2522. [Google Scholar] [CrossRef] [PubMed]
- Louie, J.K.; Scott, H.M.; DuBois, A.; Sturtz, N.; Lu, W.; Stoltey, J.; Masinde, G.; Cohen, S.; Sachdev, D.; Philip, S.; et al. Lessons from Mass-Testing for Coronavirus Disease 2019 in Long-Term Care Facilities for the Elderly in San Francisco. Clin. Infect. Dis. 2021, 72, 2018–2020. [Google Scholar] [CrossRef]
- Kasper, M.R.; Geibe, J.R.; Sears, C.L.; Riegodedios, A.J.; Luse, T.; Von Thun, A.M.; McGinnis, M.B.; Olson, N.; Houskamp, D.; Fenequito, R.; et al. An Outbreak of Covid-19 on an Aircraft Carrier. N. Engl. J. Med. 2020, 383, 2417–2426. [Google Scholar] [CrossRef]
- Letizia, A.G.; Ramos, I.; Obla, A.; Goforth, C.; Weir, D.L.; Ge, Y.; Bamman, M.M.; Dutta, J.; Ellis, E.; Estrella, L.; et al. SARS-CoV-2 Transmission among Marine Recruits during Quarantine. N. Engl. J. Med. 2020, 383, 2407–2416. [Google Scholar] [CrossRef]
- World Health Organization. Dashboard. Available online: https://covid19.who.int/ (accessed on 20 January 2022).
- Stokes, E.K.; Zambrano, L.D.; Anderson, K.N.; Marder, E.P.; Raz, K.M.; El Burai Felix, S.; Tie, Y.; Fullerton, K.E. Coronavirus Disease 2019 Case Surveillance—United States, January 22–May 30, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 759–765. [Google Scholar] [CrossRef]
- Zhang, Z.; Yao, W.; Wang, Y.; Long, C.; Fu, X. Wuhan and Hubei COVID-19 mortality analysis reveals the critical role of timely supply of medical resources. J. Infect. 2020, 81, 147–178. [Google Scholar] [CrossRef]
- Shim, E.; Mizumoto, K.; Choi, W.; Chowell, G. Estimating the Risk of COVID-19 Death During the Course of the Outbreak in Korea, February–May 2020. J. Clin. Med. 2020, 9, 1641. [Google Scholar] [CrossRef] [PubMed]
- Knaus, W.A.; Draper, E.A.; Wagner, D.P.; Zimmerman, J.E. APACHE II: A severity of disease classification system. Crit. Care Med. 1985, 13, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, J.R.; Lemeshow, S.; Saulnier, F. A new Simplified Acute Physiology Score (SAPS II) based on a European/North American multicenter study. JAMA 1993, 270, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.L.; Moreno, R.; Takala, J.; Willatts, S.; De Mendonca, A.; Bruining, H.; Reinhart, C.K.; Suter, P.M.; Thijs, L.G. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996, 22, 707–710. [Google Scholar] [CrossRef]
- Kądziołka, I.; Świstek, R.; Borowska, K.; Tyszecki, P.; Serednicki, W. Validation of APACHE II and SAPS II scales at the intensive care unit along with assessment of SOFA scale at the admission as an isolated risk of death predictor. Anaesthesiol. Intensive Ther. 2019, 51, 107–111. [Google Scholar] [CrossRef]
- Mahdavi, M.; Choubdar, H.; Zabeh, E.; Rieder, M.; Safavi-Naeini, S.; Jobbagy, Z.; Ghorbani, A.; Abedini, A.; Kiani, A.; Khanlarzadeh, V.; et al. A machine learning based exploration of COVID-19 mortality risk. PLoS ONE 2021, 16, e0252384. [Google Scholar] [CrossRef]
- Kim, H.J.; Han, D.; Kim, J.H.; Kim, D.; Ha, B.; Seog, W.; Lee, Y.K.; Lim, D.; Hong, S.O.; Park, M.J.; et al. An Easy-to-Use Machine Learning Model to Predict the Prognosis of Patients With COVID-19: Retrospective Cohort Study. J. Med. Internet Res. 2020, 22, e24225. [Google Scholar] [CrossRef]
- Jakob, C.E.M.; Mahajan, U.M.; Oswald, M.; Stecher, M.; Schons, M.; Mayerle, J.; Rieg, S.; Pletz, M.; Merle, U.; Wille, K.; et al. Prediction of COVID-19 deterioration in high-risk patients at diagnosis: An early warning score for advanced COVID-19 developed by machine learning. Infection 2022, 50, 359–370. [Google Scholar] [CrossRef]
- Singhal, L.; Garg, Y.; Yang, P.; Tabaie, A.; Wong, A.I.; Mohammed, A.; Chinthala, L.; Kadaria, D.; Sodhi, A.; Holder, A.L.; et al. eARDS: A multi-center validation of an interpretable machine learning algorithm of early onset Acute Respiratory Distress Syndrome (ARDS) among critically ill adults with COVID-19. PLoS ONE 2021, 16, e0257056. [Google Scholar] [CrossRef]
- Liang, W.; Yao, J.; Chen, A.; Lv, Q.; Zanin, M.; Liu, J.; Wong, S.; Li, Y.; Lu, J.; Liang, H.; et al. Early triage of critically ill COVID-19 patients using deep learning. Nat. Commun. 2020, 11, 3543. [Google Scholar] [CrossRef]
- Alafif, T.; Tehame, A.M.; Bajaba, S.; Barnawi, A.; Zia, S. Machine and Deep Learning towards COVID-19 Diagnosis and Treatment: Survey, Challenges, and Future Directions. Int. J. Environ. Res. Public Health 2021, 18, 1117. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, B.J.; Bucholz, E.M.; Desai, N.R.; Huang, C.; Curtis, J.P.; Masoudi, F.A.; Shaw, R.E.; Negahban, S.N.; Krumholz, H.M. Comparison of Machine Learning Methods with National Cardiovascular Data Registry Models for Prediction of Risk of Bleeding After Percutaneous Coronary Intervention. JAMA Netw. Open 2019, 2, e196835. [Google Scholar] [CrossRef] [PubMed]
- Wainberg, M.; Merico, D.; Delong, A.; Frey, B.J. Deep learning in biomedicine. Nat. Biotechnol. 2018, 36, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. Introduction to machine learning: K-nearest neighbors. Ann. Transl. Med. 2016, 4, 218. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Xie, R.; Wang, J.; Leier, A.; Marquez-Lago, T.T.; Akutsu, T.; Webb, G.I.; Chou, K.C.; Song, J. Computational analysis and prediction of lysine malonylation sites by exploiting informative features in an integrative machine-learning framework. Brief. Bioinform. 2019, 20, 2185–2199. [Google Scholar] [CrossRef]
- Yaman, E.; Subasi, A. Comparison of Bagging and Boosting Ensemble Machine Learning Methods for Automated EMG Signal Classification. BioMed Res. Int. 2019, 2019, 9152506. [Google Scholar] [CrossRef]
- Sarica, A.; Cerasa, A.; Quattrone, A. Random Forest Algorithm for the Classification of Neuroimaging Data in Alzheimer’s Disease: A Systematic Review. Front. Aging Neurosci. 2017, 9, 329. [Google Scholar] [CrossRef]
- Maltarollo, V.G.; Gertrudes, J.C.; Oliveira, P.R.; Honorio, K.M. Applying machine learning techniques for ADME-Tox prediction: A review. Expert Opin. Drug Metab. Toxicol. 2015, 11, 259–271. [Google Scholar] [CrossRef]
- Park, D.J.; Park, M.W.; Lee, H.; Kim, Y.J.; Kim, Y.; Park, Y.H. Development of machine learning model for diagnostic disease prediction based on laboratory tests. Sci. Rep. 2021, 11, 7567. [Google Scholar] [CrossRef]
- Creatore, C.; Sabathiel, S.; Solstad, T. Learning exact enumeration and approximate estimation in deep neural network models. Cognition 2021, 215, 104815. [Google Scholar] [CrossRef]
- Yuan, X.; Ou, C.; Wang, Y.; Yang, C.; Gui, W. A Layer-Wise Data Augmentation Strategy for Deep Learning Networks and Its Soft Sensor Application in an Industrial Hydrocracking Process. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 3296–3305. [Google Scholar] [CrossRef] [PubMed]
- Jurtz, V.I.; Johansen, A.R.; Nielsen, M.; Almagro Armenteros, J.J.; Nielsen, H.; Sønderby, C.K.; Winther, O.; Sønderby, S.K. An introduction to deep learning on biological sequence data: Examples and solutions. Bioinformatics 2017, 33, 3685–3690. [Google Scholar] [CrossRef] [PubMed]
- Min, S.; Lee, B.; Yoon, S. Deep learning in bioinformatics. Brief. Bioinform. 2017, 18, 851–869. [Google Scholar] [CrossRef] [Green Version]
- Laino, M.E.; Ammirabile, A.; Posa, A.; Cancian, P.; Shalaby, S.; Savevski, V.; Neri, E. The Applications of Artificial Intelligence in Chest Imaging of COVID-19 Patients: A Literature Review. Diagnostics 2021, 11, 1317. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Lee, E.H.; Zheng, J.; Zhang, W.; Halabi, S.; Liu, C.; Deng, K.; Song, J.; Yeom, K.W. Decoding COVID-19 pneumonia: Comparison of deep learning and radiomics CT image signatures. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1478–1486. [Google Scholar] [CrossRef]
- Cao, C.; Liu, F.; Tan, H.; Song, D.; Shu, W.; Li, W.; Zhou, Y.; Bo, X.; Xie, Z. Deep Learning and Its Applications in Biomedicine. Genom. Proteom. Bioinform. 2018, 16, 17–32. [Google Scholar] [CrossRef]
- Khan, S.H.; Hayat, M.; Porikli, F. Regularization of deep neural networks with spectral dropout. Neural Netw. 2019, 110, 82–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Yu, M.K.; Fong, S.; Ono, K.; Sage, E.; Demchak, B.; Sharan, R.; Ideker, T. Using deep learning to model the hierarchical structure and function of a cell. Nat. Methods 2018, 15, 290–298. [Google Scholar] [CrossRef]
- Chen, C.; Shi, H.; Jiang, Z.; Salhi, A.; Chen, R.; Cui, X.; Yu, B. DNN-DTIs: Improved drug-target interactions prediction using XGBoost feature selection and deep neural network. Comput. Biol. Med. 2021, 136, 104676. [Google Scholar] [CrossRef]
- Poernomo, A.; Kang, D.K. Biased Dropout and Crossmap Dropout: Learning towards effective Dropout regularization in convolutional neural network. Neural Netw. 2018, 104, 60–67. [Google Scholar] [CrossRef]
- Li, H.; Weng, J.; Mao, Y.; Wang, Y.; Zhan, Y.; Cai, Q.; Gu, W. Adaptive Dropout Method Based on Biological Principles. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 4267–4276. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, C.; Liu, S.; Guan, H.; Zhang, Y. Age Prediction by DNA Methylation in Neural Networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 3, 1393–1402. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Ma, Z.; Lei, J.; Zhang, G.; Xue, J.H.; Tan, Z.H.; Guo, J. Advanced Dropout: A Model-free Methodology for Bayesian Dropout Optimization. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 1. [Google Scholar] [CrossRef] [PubMed]
- Consistent Individualized Feature Attribution for Tree Ensembles. arXive 2019, arXiv:1802.03888v3.
- A Unified Approach to Interpreting Model Predictions. arXive 2017, arXiv:1705.07874v2.
- Rhodes, N.J.; O’Donnell, J.N.; Lizza, B.D.; McLaughlin, M.M.; Esterly, J.S.; Scheetz, M.H. Tree-Based Models for Predicting Mortality in Gram-Negative Bacteremia: Avoid Putting the CART before the Horse. Antimicrob. Agents Chemother. 2016, 60, 838–844. [Google Scholar] [CrossRef] [Green Version]
- Parikh, S.A.; Gomez, R.; Thirugnanasambandam, M.; Chauhan, S.S.; De Oliveira, V.; Muluk, S.C.; Eskandari, M.K.; Finol, E.A. Decision Tree Based Classification of Abdominal Aortic Aneurysms Using Geometry Quantification Measures. Ann. Biomed. Eng. 2018, 46, 2135–2147. [Google Scholar] [CrossRef]
- Freeman, C.; Kuli, D.; Basir, O. Feature-selected tree-based classification. IEEE Trans. Cybern. 2013, 43, 1990–2004. [Google Scholar] [CrossRef]
- Vaid, A.; Somani, S.; Russak, A.J.; De Freitas, J.K.; Chaudhry, F.F.; Paranjpe, I.; Johnson, K.W.; Lee, S.J.; Miotto, R.; Richter, F.; et al. Machine Learning to Predict Mortality and Critical Events in a Cohort of Patients With COVID-19 in New York City: Model Development and Validation. J. Med. Internet Res. 2020, 22, e24018. [Google Scholar] [CrossRef]
- Li, X.; Ge, P.; Zhu, J.; Li, H.; Graham, J.; Singer, A.; Richman, P.S.; Duong, T.Q. Deep learning prediction of likelihood of ICU admission and mortality in COVID-19 patients using clinical variables. PeerJ 2020, 8, e10337. [Google Scholar] [CrossRef]
- Wernly, B.; Heramvand, N.; Masyuk, M.; Rezar, R.; Bruno, R.R.; Kelm, M.; Niederseer, D.; Lichtenauer, M.; Hoppe, U.C.; Bakker, J.; et al. Acidosis predicts mortality independently from hyperlactatemia in patients with sepsis. Eur. J. Intern. Med. 2020, 76, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Karon, B.S. Comparison of measured and calculated bicarbonate values. Clin. Chem. 2008, 54, 1586–1587. [Google Scholar] [CrossRef] [PubMed]
- Pourbagheri-Sigaroodi, A.; Bashash, D.; Fateh, F.; Abolghasemi, H. Laboratory findings in COVID-19 diagnosis and prognosis. Clin. Chim. Acta 2020, 510, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.; Rhodes, A.; Alhazzani, W.; Antonelli, M.; Coopersmith, C.M.; French, C.; Machado, F.R.; McIntyre, L.; Ostermann, M.; Prescott, H.C.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock 2021. Crit. Care Med. 2021, 49, e1063–e1143. [Google Scholar] [CrossRef] [PubMed]



| Variable | Total (n = 203) | Non-Survival (n = 49) | Survival (n = 154) | p |
|---|---|---|---|---|
| Male sex (%) | 98 (48.3) | 24 (49.0) | 74 (48.1) | 0.910 |
| Age (years) | 67.5 ± 16.16 | 75.7 ± 11.58 | 64.9 ± 16.57 | <0.05 |
| Hospitalization period | 13.4 ± 10.20 | 16.8 ± 12.81 | 12.4 ± 9.00 | <0.05 |
| Comorbidities (%) | ||||
| Hypertension | 98 (48.3) | 30 (61.2) | 68 (44.2) | 0.055 |
| Diabetes mellitus | 56 (27.6) | 20 (40.8) | 36 (23.4) | <0.05 |
| Heart disease | 6 (3.0) | 6 (12.2) | 10 (6.5) | 0.319 |
| Lung diasease | 12 (5.9) | 3 (6.1) | 9 (5.8) | 0.943 |
| Liver disease | 5 (2.5) | 0 (0) | 5 (3.2) | 0.454 |
| Kidney disease | 5 (2.5) | 2 (4.1) | 3 (1.9) | 0.756 |
| Brain disease | 27 (13.3) | 6 (12.2) | 21 (13.6) | 0.993 |
| Malignant disease | 28 (13.8) | 7 (14.3) | 21 (13.6) | 0.909 |
| Vital signs at hospital admission | ||||
| Systolic blood pressure (mmHg) | 133.4 ± 22.08 | 132.9 ± 25.12 | 133.6 ± 21.10 | 0.866 |
| Diastolic blood pressure (mmHg) | 79.3 ± 14.76 | 75.2 ± 14.22 | 80.6 ± 14.74 | <0.05 |
| Pulse rate (PR, bpm) | 84.4 ± 17.79 | 84.3 ± 23.84 | 84.5 ± 15.53 | 0.962 |
| Respiratory rate (RR, bpm) | 22.4 ± 10.03 | 25.6 ± 17.10 | 21.3 ± 6.17 | 0.095 |
| Body temperature (°C) | 37.1 ± 0.71 | 36.9 ± 0.82 | 37.1 ± 0.65 | <0.05 |
| Pulse pressure (mmHg) | 54.1± 17.88 | 57.7 ± 20.23 | 53.0 ± 16.99 | 0.147 |
| PR/RR | 4.1 ± 1.34 | 3.9 ± 1.74 | 4.2 ± 1.18 | 0.316 |
| Classifier | AUC | Accuracy | F1-Score | Precision | Recall |
|---|---|---|---|---|---|
| XGboost | 0.8616 | 0.82 | 0.75 | 0.8 | 0.73 |
| LGBM | 0.8318 | 0.83 | 0.71 | 0.83 | 0.68 |
| RF | 0.8560 | 0.83 | 0.74 | 0.80 | 0.71 |
| KNN | 0.7631 | 0.79 | 0.59 | 0.77 | 0.58 |
| SVM | 0.8158 | 0.81 | 0.72 | 0.74 | 0.71 |
| DL | 0.8721 | 0.84 | 0.76 | 0.79 | 0.74 |
| Ensemble model * | 0.8811 | 0.85 | 0.77 | 0.81 | 0.75 |
| Classifier | AUC | Accuracy | F1-Score | Precision | Recall |
|---|---|---|---|---|---|
| XGboost | 0.8331 | 0.83 | 0.77 | 0.76 | 0.77 |
| LGBM | 0.8318 | 0.85 | 0.75 | 0.84 | 0.72 |
| RF | 0.8560 | 0.84 | 0.78 | 0.78 | 0.77 |
| KNN | 0.7631 | 0.79 | 0.72 | 0.72 | 0.74 |
| SVM | 0.8158 | 0.82 | 0.76 | 0.76 | 0.76 |
| DL | 0.8614 | 0.83 | 0.78 | 0.77 | 0.79 |
| Ensemble model * | 0.8631 | 0.85 | 0.80 | 0.80 | 0.80 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baik, S.-M.; Lee, M.; Hong, K.-S.; Park, D.-J. Development of Machine-Learning Model to Predict COVID-19 Mortality: Application of Ensemble Model and Regarding Feature Impacts. Diagnostics 2022, 12, 1464. https://doi.org/10.3390/diagnostics12061464
Baik S-M, Lee M, Hong K-S, Park D-J. Development of Machine-Learning Model to Predict COVID-19 Mortality: Application of Ensemble Model and Regarding Feature Impacts. Diagnostics. 2022; 12(6):1464. https://doi.org/10.3390/diagnostics12061464
Chicago/Turabian StyleBaik, Seung-Min, Miae Lee, Kyung-Sook Hong, and Dong-Jin Park. 2022. "Development of Machine-Learning Model to Predict COVID-19 Mortality: Application of Ensemble Model and Regarding Feature Impacts" Diagnostics 12, no. 6: 1464. https://doi.org/10.3390/diagnostics12061464
APA StyleBaik, S.-M., Lee, M., Hong, K.-S., & Park, D.-J. (2022). Development of Machine-Learning Model to Predict COVID-19 Mortality: Application of Ensemble Model and Regarding Feature Impacts. Diagnostics, 12(6), 1464. https://doi.org/10.3390/diagnostics12061464