Simple Summary
Recently, there has been increasing interest in identifying NTRK fusions in various tumors, as they are therapeutically targetable driver mutations. In tumor types with low-frequency NTRK fusions, recent recommendations on NTRK testing recommend pan-Trk immunohistochemistry (IHC) as the initial screening test to validate pan-Trk expression cases with next- generation sequencing (NGS) assays. This retrospective study was conducted on 1113 solid tumor samples (510 non-small cell lung cancers, 503 colorectal cancers, and 100 inflammatory myofibroblastic tumors) to evaluate using pan-Trk IHC assay, and TRK expression cases were followed by validation with NGS. We investigated the accuracy of an IHC assay in detecting NTRK fusions and characterizing the clinicopathological and molecular features of NTRK-rearranged common tumors. Despite its rarity, this study confirms the importance of identifying potential target groups based on the pathological and immunohistochemical characteristics of NTRK fusion-driven solid tumors for effective targeted therapy.
Abstract
Most NTRK fusions occur at very low frequencies in various common cancers. Recent recommendations on NTRK testing recommend immunohistochemistry (IHC) as the initial test for tumor types with a low frequency of NTRK fusions. This study investigated the accuracy of an IHC assay to detect NTRK fusions and characterize the clinicopathological and molecular features of NTRK-rearranged tumors. This retrospective study was conducted on 1113 solid tumor samples known to harbor no oncogenic driver alterations, including 510 non-small cell lung cancers (NSCLC), 503 colorectal cancers (CRC), and 79 inflammatory myofibroblastic tumors (IMT). Additionally, 21 ALK expression-positive cases were included. TRK expression was evaluated using a pan-Trk IHC assay, and positive cases were validated using NGS. TRK expression was observed in three NSCLCs (0.6%), six CRCs (1.2%), and six IMTs (6%). NTRK fusions were finally detected in two NSCLCs (0.4%), six CRCs (1.2%), and one IMT (1%). In NSCLC and CRC, the majority of NTRK fusions were readily discernible due to diffuse moderate-to-strong cytoplasmic staining on pan-Trk IHC. In IMT, focal weak nuclear staining indicated the presence of NTRK fusion. Therefore, the utility of pan-Trk IHC should be assessed considering that the difference in performance depends on tumor type.
1. Introduction
Members of the tyrosine receptor kinase (TRK) family bind to neurotrophins and affect neuronal differentiation and survival, thereby playing important roles in the nervous system. NTRKs, including NTRK1/2/3, encode tropomyosin receptor kinase A/B/C (TRKA/B/C), respectively [1,2]. NTRK fusion can cause constitutive activation of TRK receptors and overexpression of TRK proteins, which can lead to oncogenesis in various types of cancers [1,3].
Recently, there is increasing interest in identifying NTRK fusions in various tumors, as they are therapeutically targetable driver mutations. Two TRK inhibitors have received FDA therapeutic approval for the treatment of NTRK fusion-positive tumors [4]. Entrectinib (Genetech, Roche) was the first drug developed against NTRK fusions, which also targets ALK and ROS1 fusion proteins, and was designated as an orphan drug for NTRK fusion-positive non-small cell lung cancer (NSCLC) and colorectal cancer (CRC) by the FDA in 2015 [5,6]. Larotrectinib (VITRAKVI, Loxo Oncology Inc., Bayer) is highly specific for NTRK fusions and was designated as a breakthrough therapy for NTRK fusion-positive solid tumors in 2016 [3]. In 2018, the FDA accepted a new drug application and granted a priority review for larotrectinib in the treatment of adult and pediatric patients with locally advanced or metastatic solid tumors harboring an NTRK fusion regardless of tumor type [7].
Tumor types where NTRK fusions are characteristic or even considered pathognomonic, such as secretory breast carcinoma and secretory carcinoma of the salivary gland, infantile fibrosarcoma, and congenital mesoblastic nephroma, are very rare [8,9,10,11]. Conversely, the majority of NTRK fusions occur at very low frequencies, with an average rate of 0.5–1% in a variety of common cancers—such as lung adenocarcinoma, colorectal and malignant melanoma, and soft tissue sarcoma [12,13,14,15]. However, these common cancer types contribute to most patients with NTRK fusions.
Therefore, it is important to identify patients who could benefit from TRK inhibitor therapy using reliable and cost-effective techniques for common cancer types that rarely harbor NTRK fusions. In tumor types with a low frequency of NTRK fusions, recent NTRK testing recommendations suggesting using pan-Trk immunohistochemistry as a screening tool to identify cases for definitive NTRK fusion detection by NGS assay. RNA-based targeted NGS assays to detect NTRK fusions can accurately characterize fusion transcripts if sufficient RNA of adequate quality is available [16].
In this study, to uncover the NTRK fusion frequency in the South Korean population with NSCLC, CRC, and inflammatory myofibroblastic tumors (IMT), we performed a pan-Trk IHC assay and confirmed the pan-Trk-positive samples with NGS assays. Furthermore, we investigated the accuracy of an IHC assay to detect NTRK fusions and characterize the clinicopathological and molecular features of NTRK-rearranged tumors.
2. Materials and Methods
2.1. Case Selection
A total of 1113 patients with solid tumors who underwent surgical resection or biopsy at Samsung Medical Center between January 2010 and April 2020 were selected. These included 510 NSCLCs, 503 CRCs, and 100 IMTs. All cases were pathologically confirmed. Patients with NSCLC and CRC were excluded if they had known driver mutations, such as ALK, ROS1, BRAF, and EGFR mutations in NSCLC and KRAS, NRAS, and EGFR mutations in CRC. Of the total 503 CRCs, 333 cases had microsatellite instability (MSI) analysis information, of which 14 cases were MSI-H (high-level MSI). The IMT included 21 ALK expression-positive cases and 79 ALK expression-negative cases. EGFR alteration was detected by either real-time PCR with PNA-clamping methods, direct sequencing, or both methods. For the ALK fusion, ALK IHC or fluorescence in situ hybridization (FISH) was performed, and ROS1 fusion was detected by RT-PCR. BRAF, KRAS, and NRAS alterations were detected by RT-PCR or NGS assay. Patients with inadequate tumor specimens for molecular analysis were excluded. Clinical data on sex, age at surgery, smoking history, tumor histology, and tumor size were extracted from electronic medical records. This study was reviewed and approved by the Institutional Review Board of Samsung Medical Center (#2020-04-105-003).
2.2. Tissue Microarrays (TMA)
TMAs were constructed to include two 3 mm cores of representative tumors in formalin-fixed paraffin-embedded (FFPE) tissues from 1113 cases. IHC using TMAs was reviewed by two pathologists (HB and SEL).
2.3. IHC Assay
The FFPE samples used in this study were tissues from NSCLC, CRC, and IMT patients diagnosed between 2013 and 2020. FFPE TMA blocks were cut into 4 μm thick sections and placed on slides. We used a commercially available pan-Trk assay (rabbit monoclonal antibody, clone EPR17341 assay, ready to use (RTU), Roche, Ventana, Oro Valley, AZ, USA) to screen for TRK expression in FFPE specimens. In the case of the pan-Trk IHC assay, there is no scoring algorithm or criteria for determining IHC positivity. Tumors were considered positive when tumor cells exhibited staining at any intensity if ≥1% of tumor cells. In addition, different subcellular staining patterns (nuclear, cytoplasmic, membranous, etc.) were considered positive.
2.4. NGS Analysis
A total of 15 IHC-positive cases with available FFPE material were analyzed by NGS to confirm NTRK fusion status and identify possible fusion partners. NGS was conducted using the TruSightTM Oncology (TSO) 500 assay (Illumina) according to the manufacturer’s recommendations. The positive samples were further validated by an additional hybridization capture-based targeted RNA panel (SOLIDaccuTest RNA), which includes all exons of NTRK1/2/3.
3. Results
3.1. Prevalence of NTRK Fusions in 1113 Solid Tumors
Of the 1113 solid tumor samples screened with pan-Trk IHC, 15 cases (1.3% of the entire cohort) showed TRK expression. RNA-based NGS assay identified 15 cases, of which nine (0.8% of the entire cohort) had an NTRK fusion. The clinicopathological characteristics of the 15 TRK expression cases (three NSCLCs, six CRCs, and six IMTs) are summarized in Table 1.

Table 1.
Clinicopathological characteristics of 15 TRK expression cases.
The pan-Trk immunohistochemical and molecular characteristics of the 15 TRK expression cases are summarized in Table 2. NTRK1 fusion was the most common (n = 6), followed by NTRK3 (n = 3). Fusion was not observed in NTRK2. Two NTRKs were involved in the fusion with five different partner genes: TPM3–NTRK1, LMNA–NTRK1, CD74–NTRK1, ETV6–NTRK3, and SQSTM1–NTRK3. These five types of NTRK fusion have been previously reported in various tumors. Strong and uniform expression with pan-Trk IHC identified 5/6 NTRK1 fused cases, and all NTRK3 fusion cases showed moderate staining intensity. Of the 15 TRK expression positive cases, six NTRK fusion-negative, namely false-positive pan-Trk IHC cases, showed weak and moderate staining intensity but not strong staining intensity. The subcellular distribution of immunohistochemical staining differed depending on the fusion partner.

Table 2.
Pan-Trk immunohistochemical and molecular characteristic of 15 TRK expression cases.
3.2. Non-Small Cell Lung Cancer
Of the 510 patients diagnosed with NSCLC, 341 (66.9%) were male, and the median age was 64.6 years (range, 32–84 years).
All cases were immunohistochemically analyzed using VENTANA pan-Trk IHC, and TRK expression was observed in 3 of 510 cases (0.6%); (L099, L347, L491) (Figure 1). Pan-Trk IHC staining showed cytoplasmic and membranous staining in the tumor cells of all three cases but with different intensity (1–3). The three positive cases were further validated using RNA-based targeted NGS assay (TSO500). Immunohistochemical analysis of pan-Trk was concordant with the TSO500 assay in two of three cases. Finally, two NSCLC cases harbored NTRK fusion among 510 NSCLCs (0.4%). In the L347 and L491 cases, NTRK fusion genes were detected with SQSTM1–NTRK3 and CD74–NTRK1, respectively. Non-NTRK-rearranged cases showed cytoplasmic staining with weak to moderate intensity for pan-Trk antibodies in IHC.
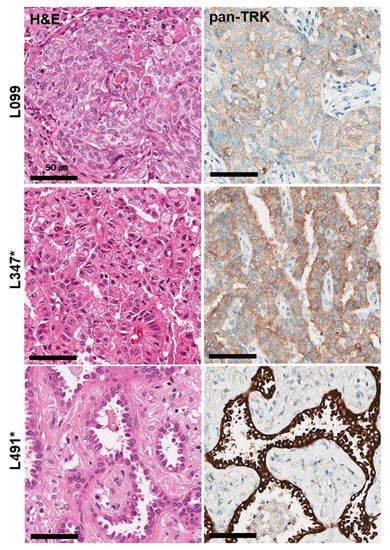
Figure 1.
Histological and immunohistochemical findings of three TRK expression cases observed in 510 NSCLC samples. Pan-TrK IHC with moderate cytoplasmic and membranous staining in NSCLC with a SQSTM1-NTRK3 fusion (L347 case). Pan-TrK IHC with strong cytoplasmic and membranous staining in NSCLC with a CD74-NTRK1 fusion (L491 case). * Confirmed cases of NTRK gene fusions in NGS assays.
The average age of NTRK-rearranged NSCLC patients was 48 years, which was younger than that of all NSCLC patients (64.6 years old). The two NTRK-rearranged NSCLCs were moderately differentiated adenocarcinomas showing papillary and acinar patterns in a 42-year-old male and a 54-year-old female, respectively.
3.3. Colorectal Cancer
In 503 patients with CRC, the median age at diagnosis was 58.2 years (range, 17–87 years), and 299 (59.4%) were male.
From pan-Trk IHC assays, a total of six cases (1.2%) were found to express TRK proteins (C175, C178, C216, C421, C478, C503) (Figure 2). NTRK fusions were detected in all five cases. (1.2%). Three partner genes were identified: four cases of TPM3–NTRK1 and one case each of LMNA–NTRK1 and ETV6–NTRK3. Immunohistochemical analysis of pan-Trk was concordant with the TSO500 assay. Pan-Trk IHC staining showed cytoplasmic and membranous staining in the four TPM3–NTRK1 cases, nuclear membranous staining in the LMNA–NTRK1 case, and nuclear staining in the ETV6–NTRK3 case (Table 2). Immunoreactivity for TRK was easily identifiable, as the majority of positive CRC cases showed strong, uniform intensity staining.
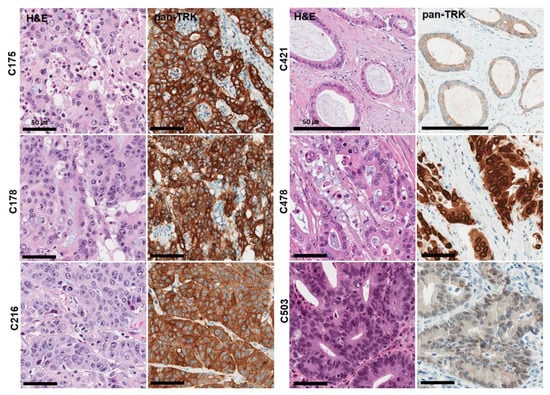
Figure 2.
Histological and immunohistochemical findings of six TRK expression cases observed in 503 CRC samples (Confirmed cases of NTRK gene fusions in NGS assays). Pan-TrK IHC with strong cytoplasmic and membranous staining in CRC with a TPM3-NTRK1 fusion (C175, C178, C216 case). Pan-TrK IHC with weak to moderate cytoplasmic and membranous staining in CRC with a TPM3-NTRK1 fusion (C421 case). Pan-TrK IHC with strong nuclear membranous staining in CRC with a LMNA-NTRK1 fusion (C478 case). Pan-TrK IHC with moderate nuclear staining in CRC with an ETV6-NTRK3 fusion (C503 case).
Interestingly, all six NTRK-rearranged CRCs were MSI-H tumors. The average age of NTRK-rearranged CRC patients was 69.8 years old, which was older than that of all CRC patients (58.2 years old). Histologically, all NTRK fusion cases were adenocarcinomas, and no characteristic histological features were identified.
3.4. Inflammatory Myofibroblastic Tumor
In 100 patients with IMT, the median age was 45.3 years (range, 1–81 years), and 43 (43.0%) were female. Six cases (6%) expressed TRK in pan-Trk IHC assays, including five cases of weak to moderate cytoplasmic staining and one case of moderate nuclear staining (Figure 3).
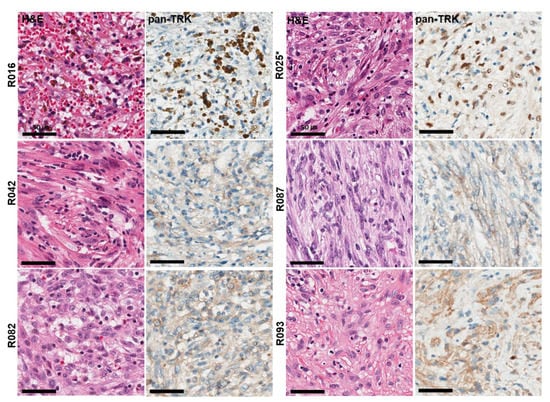
Figure 3.
Histological and immunohistochemical findings of six TRK expression cases observed in 100 IMT samples. Pan-TrK IHC with moderate nuclear staining in IMT with an ETV6-NTRK3 fusion (R025 case). * Confirmed cases of NTRK gene fusions in NGS assays.
Of the six cases of TRK-expressing IMT, NTRK fusion was detected in only one case (R025), which showed moderate nuclear staining in pan-Trk IHC assays. NTRK fusion transcripts were identified with ETV6–NTRK3 (exon 5 of ETV6 fused with exon 15 of NTRK3) using NGS. Non-NTRK rearranged cases showed cytoplasmic but not nuclear staining for pan-Trk antibody. Finally, only one case harbored NTRK fusion in 100 IMTs (1%).
The NTRK-rearranged IMT was identified in a 41-year-old female who presented with a 5.5 cm solitary, well-defined lung mass with no metastatic lesions identified at presentation. Histologically, two patterns, including a cellular area with cytologically bland spindle cells and a prominent lymphoplasma cells infiltrate and a less cellular myxoid area with spindled myofibroblasts showing vesicular nuclei, small nucleoli, and eosinophilic cytoplasm, were observed in the R025 case. TRK immunoreactivity showed heterogeneous expression and easily identifiable moderate nuclear staining intensity in less cellular myxoid areas.
4. Discussion
Identifying patients harboring NTRK fusions is very important in routine diagnostic practice. It is critical to have a good discovery strategy because the incidence of NTRK fusions is extremely low. First, it is essential to verify the prevalence of NTRK fusion in common solid tumors reported in other studies. Common cancer types, such as lung and colon cancer, harbor NTRK fusions with a prevalence <1%. In this study, the prevalence of NTRK fusion in NSCLC and CRC was 0.4% (2/510) and 1.2% (6/503), respectively. The prevalence of NTRK fusion in CRCs was higher compared with previous studies, which reported a prevalence of 0.23% [17]. These findings could be explained by the fact that our cohort was narrowed down by excluding cases harboring known driver mutations, such as KRAS, NRAS, and BRAF mutations in CRC.
Unfortunately, all nine NTRK-rearranged tumors (two NSCLCs, six CRCs, and one IMT) showed no characteristic histological features that could be useful morphological clues for the presence of NTRK fusion.
In this study, five different partner genes were identified. The majority of NTRK fusions are TPM3–NTRK1 rearrangements, which are recurring events in CRCs and are associated with tumor sensitivity to TRKA kinase inhibition [18,19]. As expected, four out of the nine NTRK fusions were TPM3–NTRK1, followed by ETV6–NTRK3 (n = 2), LMNA–NTRK1 (n = 1), SQSTM1–NTRK3 (n = 1), and CD74–NTRK1 (n = 1).
Immunoreactivity for TRK was easily identifiable, as the majority of the positive CRC and NSCLC cases showed diffuse moderate to strong intensity staining except for one case harboring ETV6–NTRK3 fusion. The ETV6–NTRK3 fusion case showed weak to intermediate nuclear staining. Weak pan-Trk IHC expression was commonly observed in various tumors, including CRCs harboring an ETV6–NTRK3 fusion, and it was recently demonstrated that the lower sensitivity of pan-Trk IHC was caused by NTRK3 fusion, especially for ETV6–NTRK3 fusion [20,21,22,23]. Notably, no false-positive CRC cases were identified in the pan-Trk IHC in this study. These findings are consistent with previous reports that pan-Trk IHC has 100% specificity for CRCs [19,22].
The six NTRK-rearranged CRCs were all MSI-H tumors, a significantly higher proportion than the 8% proportion of MSI-H in the entire CRC population. As in previous reports [19,20,24], we confirmed once again that NTRK-positive CRCs demonstrated a higher frequency of MSI. Therefore, a subset of CRCs harboring no known driver mutations and exhibiting MSI-H should be tested using pan-Trk IHC and further validated using RNA-based targeted NGS.
NTRK fusions are highly enriched in several rare specific tumor types, such as secretory carcinomas of the salivary gland and breast, congenital mesoblastic nephroma, pediatric melanoma, and infantile fibrosarcoma [1,4,25]. In our study, efforts to discover NTRK-rearranged tumors in tumors known to rarely harbor NTRK fusions led to perform the pan-Trk IHC in IMTs. IMT is a distinctive, rarely metastasizing neoplasm composed of myofibroblastic and fibroblastic spindle cells accompanied by an infiltration of lymphoplasma cells [26]. IMTs are genetically heterogeneous, and most of them harbor gene rearrangements of receptor tyrosine kinases, including ALK (approximately 50–60%), ROS1 (approximately 5–10%), and NTRK3 (approximately 5%) but rarely RET and PDGFRβ fusion [27,28,29,30,31]. Recent translational studies provided evidence of the potential activity of TKIs in sarcoma, including larotrectenib [32,33,34]
However, until recently, there have been few analyses to identify NTRK fusions in a large number of IMTs. To the best of our knowledge, this study evaluated NTRK fusion in the largest number of IMTs. In this study, NTRK fusion transcript was identified with ETV6–NTRK3 in only one IMT (1%). The lower-than-expected frequency may be due to the analysis of IMT samples, including 21 ALK-rearranged IMTs.
In only one NTRK-rearranged IMT case, immunoreactivity for TRK was heterogeneous and showed moderate nuclear staining intensity that was readily discernible in the less-cellular myxoid area. As shown in a recently published study, although the number of cases was very small, sensitivity and specificity were 100% for IMT [22]. Unfortunately, five false-positive cases were identified in this study, and the cytoplasmic staining pattern in the pan-Trk IHC assay was observed in all the false-positive cases. A limitation of the diagnostic utility of weak and moderate cytoplasmic pan-Trk staining, in contrast to the nuclear staining pattern, was identified. Especially in cases with mesenchymal tumors that show neural and smooth muscle differentiation, the interpretation of pan-Trk IHC should be considered due to physiological cytoplasmic expression of pan-Trk in neural and smooth muscle tissue and their malignant counterparts [14,35,36]. It is also important to note that false-positive results can be caused by the high level of surrounding background staining although the tumor cells themselves were negative. Therefore, several studies have recommended that tumors with neural and smooth muscle differentiation should not be screened using pan-Trk IHC for NTRK fusions [37,38].
However, our study showed cytoplasmic staining but not nuclear staining in the false-positive IMT cases. Nuclear staining despite focal has been described in tumors harboring ETV6-NTRK3 fusion protein [22,35,39]. It has also been demonstrated that no nuclear staining was observed in all false-positive cases of sarcoma [22], and pan-Trk nuclear staining is a highly specific diagnostic marker for secretory carcinoma harboring the ETV6–NTRK3 fusion [40].
Therefore, relevant nuclear staining in a sub-diagnostic manner (focal and weak) but not cytoplasmic staining may be meaningful in mesenchymal tumors when further validated by RNA-based targeted NGS in all advanced and metastatic ALK negative-IMTs; this can identify patients who will benefit from TRK inhibitor therapy.
Our study had several limitations. First, our study used TMA-based IHC as a primary screening tool for detecting solid tumors harboring NTRK fusions. There may be the possibility of missing positive cases due to heterogeneous staining patterns although recent studies have reported that pan-Trk IHC shows a uniform staining pattern within the same CRC section [19,41]. Another limitation of our study is that the sensitivity and specificity of the pan-Trk IHC assay could not be accurately determined, as RNA-based targeted NGS was not performed in cases with negative IHC results. Finally, this was a retrospective study, and none of our patients received a TRK inhibitor to obtain information regarding treatment response.
The caveats and limitations of pan-Trk IHC for tumor type should be considered when interpreting the IHC results. In IMT, focal, weak cytoplasmic and membranous staining for pan-Trk does not serve as a surrogate marker for NTRK fusion, whereas focal, weak nuclear staining indicates the presence of NTRK fusion. In NSCLC and CRC, the majority of NTRK fusions were readily discernible due to diffuse homogenous moderate to strong cytoplasmic staining on pan-TRK IHC. It should also be noted that focal, weak nuclear staining indicative of ETV6–NTRK3 fusion should not be overlooked.
5. Conclusions
In conclusion, the utility of pan-Trk IHC should be assessed considering the difference in the performance of pan-Trk IHC depending on the tumor type. Despite its rarity, this study confirms the importance of identifying potential target groups based on the pathological and immunohistochemical characteristics of NTRK fusion-driven solid tumors for effective targeted therapy.
Author Contributions
Conceptualization, H.B., M.-S.L., S.E.L. and Y.-L.C.; data curation, H.B., M.-S.L., S.E.L. and Y.-L.C.; formal analysis, H.B., M.-S.L., M.S., J.C., S.A., S.E.L. and Y.-L.C.; funding acquisition, M.-S.L. and Y.-L.C.; investigation, H.B., M.-S.L., S.-H.K., S.E.L. and Y.-L.C.; methodology, M.-S.L., S.E.L. and Y.-L.C.; resources, M.-S.L., M.S., J.C., S.A., S.-H.K. and Y.-L.C.; supervision, S.E.L. and Y.-L.C.; validation, M.-S.L. and Y.-L.C.; project administration, Y.-L.C.; visualization, S.E.L.; writing—original draft, H.B. and S.E.L.; writing—review and editing, Y.-L.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by grants from National Research Foundation of Korea funded by the Korean government (NRF-2016R1A5A2945889, 2021R1A2C4002158, and 2022R1A2C2006322).
Institutional Review Board Statement
This study was reviewed and approved by the Institutional Review Board of Samsung Medical Center (#2020-04-105-003) and conducted according to the guidelines of the Declaration of Helsinki.
Informed Consent Statement
Patient consent was waived for the reason that the archival data in this study were fully anonymized before the beginning of the research.
Data Availability Statement
Not applicable.
Acknowledgments
This work was supported by Bayer.
Conflicts of Interest
This study was financially supported by Bayer. However, Bayer had no role in the study design, data collection, analysis, decision to publish, or manuscript preparation. The final decision on the content was retained exclusively by the authors.
Abbreviations
NTRK, neurotrophin kinase; TRK, tyrosine receptor kinase; NSCLC, non-small cell lung cancer; CRC, colorectal cancer; IMT, inflammatory myofibroblastic tumor; IHC, immunohistochemistry; NGS, next-generation sequencing.
References
- Vaishnavi, A.; Le, A.T.; Doebele, R.C. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015, 5, 25–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solomon, J.P.; Benayed, R.; Hechtman, J.F.; Ladanyi, M. Identifying patients with NTRK fusion cancer. Ann. Oncol. 2019, 30, viii16–viii22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cocco, E.; Scaltriti, M.; Drilon, A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 2018, 15, 731–747. [Google Scholar] [CrossRef]
- Drilon, A. TRK inhibitors in TRK fusion-positive cancers. Ann. Oncol. 2019, 30, viii23–viii30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farago, A.F.; Le, L.P.; Zheng, Z.; Muzikansky, A.; Drilon, A.; Patel, M.; Bauer, T.M.; Liu, S.V.; Ou, S.H.; Jackman, D.; et al. Durable Clinical Response to Entrectinib in NTRK1-Rearranged Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2015, 10, 1670–1674. [Google Scholar] [CrossRef] [Green Version]
- Drilon, A.; Siena, S.; Ou, S.I.; Patel, M.; Ahn, M.J.; Lee, J.; Bauer, T.M.; Farago, A.F.; Wheler, J.J.; Liu, S.V.; et al. Safety and Antitumor Activity of the Multitargeted Pan-TRK, ROS1, and ALK Inhibitor Entrectinib: Combined Results from Two Phase I Trials (ALKA-372-001 and STARTRK-1). Cancer Discov. 2017, 7, 400–409. [Google Scholar] [CrossRef] [Green Version]
- Drilon, A.; Laetsch, T.W.; Kummar, S.; DuBois, S.G.; Lassen, U.N.; Demetri, G.D.; Nathenson, M.; Doebele, R.C.; Farago, A.F.; Pappo, A.S.; et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N. Engl. J. Med. 2018, 378, 731–739. [Google Scholar] [CrossRef]
- Davis, J.L.; Lockwood, C.M.; Albert, C.M.; Tsuchiya, K.; Hawkins, D.S.; Rudzinski, E.R. Infantile NTRK-associated Mesenchymal Tumors. Pediatr. Dev. Pathol. 2018, 21, 68–78. [Google Scholar] [CrossRef] [Green Version]
- Bourgeois, J.M.; Knezevich, S.R.; Mathers, J.A.; Sorensen, P.H. Molecular detection of the ETV6-NTRK3 gene fusion differentiates congenital fibrosarcoma from other childhood spindle cell tumors. Am. J. Surg. Pathol. 2000, 24, 937–946. [Google Scholar] [CrossRef]
- Rubin, B.P.; Chen, C.J.; Morgan, T.W.; Xiao, S.; Grier, H.E.; Kozakewich, H.P.; Perez-Atayde, A.R.; Fletcher, J.A. Congenital mesoblastic nephroma t(12;15) is associated with ETV6-NTRK3 gene fusion: Cytogenetic and molecular relationship to congenital (infantile) fibrosarcoma. Am. J. Pathol. 1998, 153, 1451–1458. [Google Scholar] [CrossRef]
- Skalova, A.; Vanecek, T.; Sima, R.; Laco, J.; Weinreb, I.; Perez-Ordonez, B.; Starek, I.; Geierova, M.; Simpson, R.H.; Passador-Santos, F.; et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: A hitherto undescribed salivary gland tumor entity. Am. J. Surg. Pathol. 2010, 34, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Farago, A.F.; Taylor, M.S.; Doebele, R.C.; Zhu, V.W.; Kummar, S.; Spira, A.I.; Boyle, T.A.; Haura, E.B.; Arcila, M.E.; Benayed, R.; et al. Clinicopathologic Features of Non-Small-Cell Lung Cancer Harboring an NTRK Gene Fusion. JCO Precis Oncol. 2018, 2, 1–12. [Google Scholar] [CrossRef]
- Pietrantonio, F.; Di Nicolantonio, F.; Schrock, A.B.; Lee, J.; Tejpar, S.; Sartore-Bianchi, A.; Hechtman, J.F.; Christiansen, J.; Novara, L.; Tebbutt, N.; et al. ALK, ROS1, and NTRK Rearrangements in Metastatic Colorectal Cancer. J. Natl. Cancer Inst. 2017, 109, djx089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, S.; Cotzia, P.; Hyman, D.M.; Drilon, A.; Tap, W.D.; Zhang, L.; Hechtman, J.F.; Frosina, D.; Jungbluth, A.A.; Murali, R.; et al. NTRK Fusions Define a Novel Uterine Sarcoma Subtype With Features of Fibrosarcoma. Am. J. Surg. Pathol. 2018, 42, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Lezcano, C.; Shoushtari, A.N.; Ariyan, C.; Hollmann, T.J.; Busam, K.J. Primary and Metastatic Melanoma with NTRK Fusions. Am. J. Surg. Pathol. 2018, 42, 1052–1058. [Google Scholar] [CrossRef]
- Racanelli, D.; Brenca, M.; Baldazzi, D.; Goeman, F.; Casini, B.; De Angelis, B.; Guercio, M.; Milano, G.M.; Tamborini, E.; Busico, A.; et al. Next-Generation Sequencing Approaches for the Identification of Pathognomonic Fusion Transcripts in Sarcomas: The Experience of the Italian ACC Sarcoma Working Group. Front. Oncol. 2020, 10, 489. [Google Scholar] [CrossRef] [Green Version]
- Lasota, J.; Chlopek, M.; Lamoureux, J.; Christiansen, J.; Kowalik, A.; Wasag, B.; Felisiak-Golabek, A.; Agaimy, A.; Biernat, W.; Canzonieri, V.; et al. Colonic Adenocarcinomas Harboring NTRK Fusion Genes: A Clinicopathologic and Molecular Genetic Study of 16 Cases and Review of the Literature. Am. J. Surg. Pathol. 2020, 44, 162–173. [Google Scholar] [CrossRef]
- Ardini, E.; Bosotti, R.; Borgia, A.L.; De Ponti, C.; Somaschini, A.; Cammarota, R.; Amboldi, N.; Raddrizzani, L.; Milani, A.; Magnaghi, P.; et al. The TPM3-NTRK1 rearrangement is a recurring event in colorectal carcinoma and is associated with tumor sensitivity to TRKA kinase inhibition. Mol. Oncol. 2014, 8, 1495–1507. [Google Scholar] [CrossRef]
- Lasota, J.; Chlopek, M.; Wasag, B.; Kowalik, A.; Christiansen, J.; Lamoureux, J.; Kuzniacka, A.; Felisiak-Golabek, A.; Liu, Y.; Reyes, T.A.R.; et al. Colorectal Adenocarcinomas Harboring ALK Fusion Genes: A Clinicopathologic and Molecular Genetic Study of 12 Cases and Review of the Literature. Am. J. Surg. Pathol. 2020, 44, 1224–1234. [Google Scholar] [CrossRef]
- Kim, J.H.; Hong, J.H.; Choi, Y.L.; Lee, J.A.; Seo, M.K.; Lee, M.S.; An, S.B.; Sung, M.J.; Cho, N.Y.; Kim, S.S.; et al. NTRK oncogenic fusions are exclusively associated with the serrated neoplasia pathway in the colorectum and begin to occur in sessile serrated lesions. J. Pathol. 2021, 255, 399–411. [Google Scholar] [CrossRef]
- Gatalica, Z.; Xiu, J.; Swensen, J.; Vranic, S. Molecular characterization of cancers with NTRK gene fusions. Mod. Pathol. 2019, 32, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Solomon, J.P.; Linkov, I.; Rosado, A.; Mullaney, K.; Rosen, E.Y.; Frosina, D.; Jungbluth, A.A.; Zehir, A.; Benayed, R.; Drilon, A.; et al. NTRK fusion detection across multiple assays and 33,997 cases: Diagnostic implications and pitfalls. Mod. Pathol. 2020, 33, 38–46. [Google Scholar] [CrossRef]
- Koopman, B.; Kuijpers, C.; Groen, H.J.M.; Timens, W.; Schuuring, E.; Willems, S.M.; van Kempen, L.C. Detection of NTRK Fusions and TRK Expression and Performance of pan-TRK Immunohistochemistry in Routine Diagnostics: Results from a Nationwide Community-Based Cohort. Diagnostics 2022, 12, 668. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Kawazu, M.; Yamamoto, Y.; Ueno, T.; Kojima, S.; Nagae, G.; Abe, H.; Soda, M.; Oga, T.; Kohsaka, S.; et al. Fusion Kinases Identified by Genomic Analyses of Sporadic Microsatellite Instability-High Colorectal Cancers. Clin. Cancer Res. 2019, 25, 378–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amatu, A.; Sartore-Bianchi, A.; Siena, S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open 2016, 1, e000023. [Google Scholar] [CrossRef] [Green Version]
- Gleason, B.C.; Hornick, J.L. Inflammatory myofibroblastic tumours: Where are we now? J. Clin. Pathol. 2008, 61, 428–437. [Google Scholar] [CrossRef] [Green Version]
- Lovly, C.M.; Gupta, A.; Lipson, D.; Otto, G.; Brennan, T.; Chung, C.T.; Borinstein, S.C.; Ross, J.S.; Stephens, P.J.; Miller, V.A.; et al. Inflammatory myofibroblastic tumors harbor multiple potentially actionable kinase fusions. Cancer Discov. 2014, 4, 889–895. [Google Scholar] [CrossRef] [Green Version]
- Antonescu, C.R.; Suurmeijer, A.J.; Zhang, L.; Sung, Y.S.; Jungbluth, A.A.; Travis, W.D.; Al-Ahmadie, H.; Fletcher, C.D.; Alaggio, R. Molecular characterization of inflammatory myofibroblastic tumors with frequent ALK and ROS1 gene fusions and rare novel RET rearrangement. Am. J. Surg. Pathol. 2015, 39, 957–967. [Google Scholar] [CrossRef] [Green Version]
- Hornick, J.L.; Sholl, L.M.; Dal Cin, P.; Childress, M.A.; Lovly, C.M. Expression of ROS1 predicts ROS1 gene rearrangement in inflammatory myofibroblastic tumors. Mod. Pathol. 2015, 28, 732–739. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, H.; Yoshida, A.; Taguchi, K.; Kohashi, K.; Hatanaka, Y.; Yamashita, A.; Mori, D.; Oda, Y. ALK, ROS1 and NTRK3 gene rearrangements in inflammatory myofibroblastic tumours. Histopathology 2016, 69, 72–83. [Google Scholar] [CrossRef]
- Debonis, S.A.; Bongiovanni, A.; Pieri, F.; Fausti, V.; De Vita, A.; Riva, N.; Gurrieri, L.; Vanni, S.; Diano, D.; Mercatali, L.; et al. ALK-negative lung inflammatory myofibroblastic tumor in a young adult: A case report and literature review of molecular alterations. Medicine 2021, 100, e25972. [Google Scholar] [CrossRef] [PubMed]
- Recine, F.; De Vita, A.; Fausti, V.; Pieri, F.; Bongiovanni, A.; Franchini, E.; Casadei, R.; Falasconi, M.C.; Oboldi, D.; Matteucci, F.; et al. Case Report: Adult NTRK-Rearranged Spindle Cell Neoplasm: Early Tumor Shrinkage in a Case With Bone and Visceral Metastases Treated With Targeted Therapy. Front. Oncol. 2021, 11, 740676. [Google Scholar] [CrossRef] [PubMed]
- Siozopoulou, V.; Smits, E.; De Winne, K.; Marcq, E.; Pauwels, P. NTRK Fusions in Sarcomas: Diagnostic Challenges and Clinical Aspects. Diagnostics 2021, 11, 478. [Google Scholar] [CrossRef] [PubMed]
- Dang, X.; Xiang, T.; Zhao, C.; Tang, H.; Cui, P. EML4-NTRK3 Fusion Cervical Sarcoma: A Case Report and Literature Review. Front. Med. 2022, 9, 832376. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.P.; Fletcher, C.D.M.; Hornick, J.L. Evaluation of pan-TRK immunohistochemistry in infantile fibrosarcoma, lipofibromatosis-like neural tumour and histological mimics. Histopathology 2018, 73, 634–644. [Google Scholar] [CrossRef]
- Brodeur, G.M.; Minturn, J.E.; Ho, R.; Simpson, A.M.; Iyer, R.; Varela, C.R.; Light, J.E.; Kolla, V.; Evans, A.E. Trk receptor expression and inhibition in neuroblastomas. Clin. Cancer Res. 2009, 15, 3244–3250. [Google Scholar] [CrossRef] [Green Version]
- Solomon, J.P.; Hechtman, J.F. Detection of NTRK Fusions: Merits and Limitations of Current Diagnostic Platforms. Cancer Res. 2019, 79, 3163–3168. [Google Scholar] [CrossRef]
- Hsiao, S.J.; Zehir, A.; Sireci, A.N.; Aisner, D.L. Detection of Tumor NTRK Gene Fusions to Identify Patients Who May Benefit from Tyrosine Kinase (TRK) Inhibitor Therapy. J. Mol. Diagn. 2019, 21, 553–571. [Google Scholar] [CrossRef] [Green Version]
- Hechtman, J.F.; Benayed, R.; Hyman, D.M.; Drilon, A.; Zehir, A.; Frosina, D.; Arcila, M.E.; Dogan, S.; Klimstra, D.S.; Ladanyi, M.; et al. Pan-Trk Immunohistochemistry Is an Efficient and Reliable Screen for the Detection of NTRK Fusions. Am. J. Surg. Pathol. 2017, 41, 1547–1551. [Google Scholar] [CrossRef]
- Xu, B.; Haroon Al Rasheed, M.R.; Antonescu, C.R.; Alex, D.; Frosina, D.; Ghossein, R.; Jungbluth, A.A.; Katabi, N. Pan-Trk immunohistochemistry is a sensitive and specific ancillary tool for diagnosing secretory carcinoma of the salivary gland and detecting ETV6-NTRK3 fusion. Histopathology 2020, 76, 375–382. [Google Scholar] [CrossRef]
- Yamashiro, Y.; Kurihara, T.; Hayashi, T.; Suehara, Y.; Yao, T.; Kato, S.; Saito, T. NTRK fusion in Japanese colorectal adenocarcinomas. Sci. Rep. 2021, 11, 5635. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).