End-to-End Calcification Distribution Pattern Recognition for Mammograms: An Interpretable Approach with GNN
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Datasets
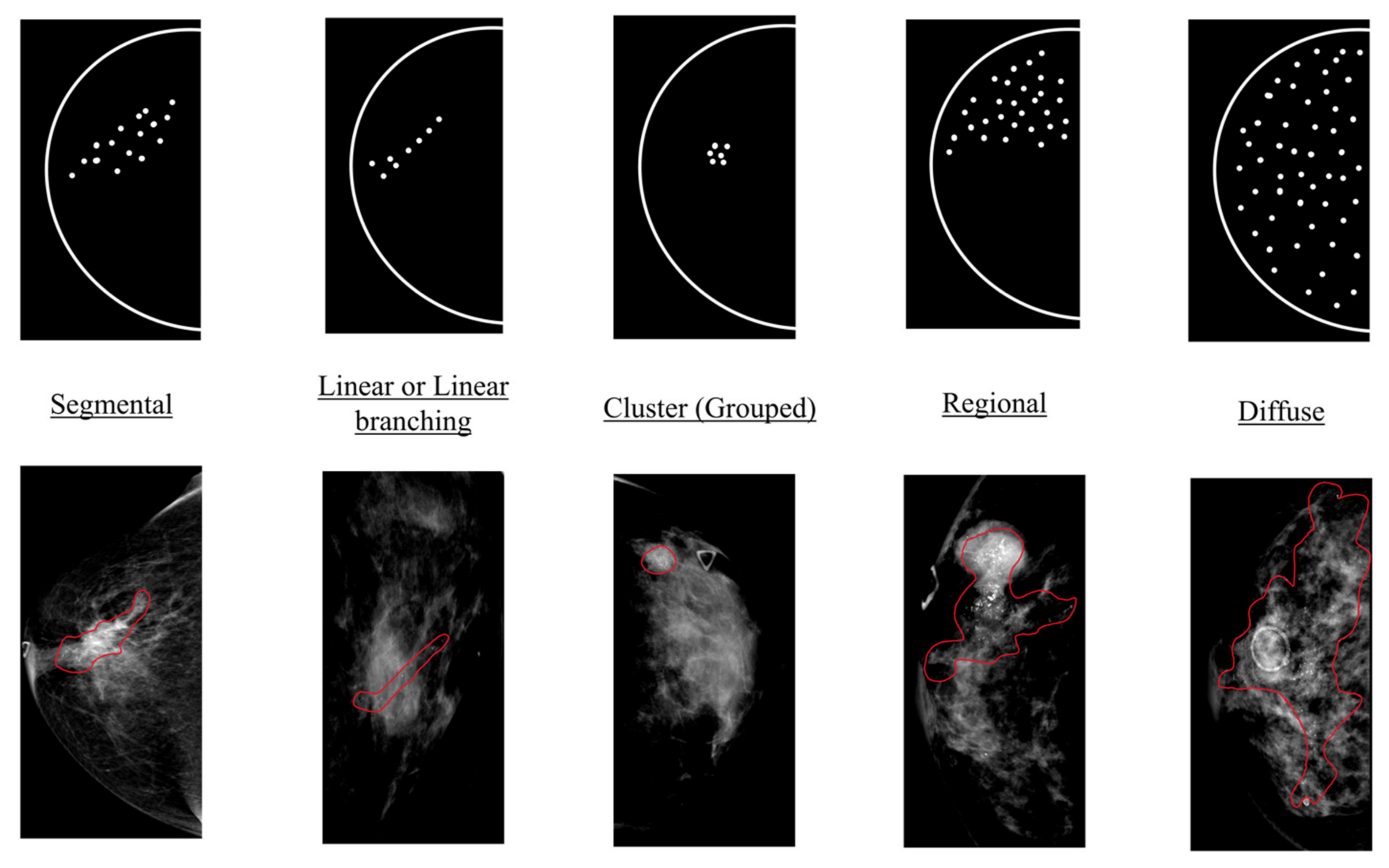
2.2. Annotation and Ground Truth
2.3. Study Design
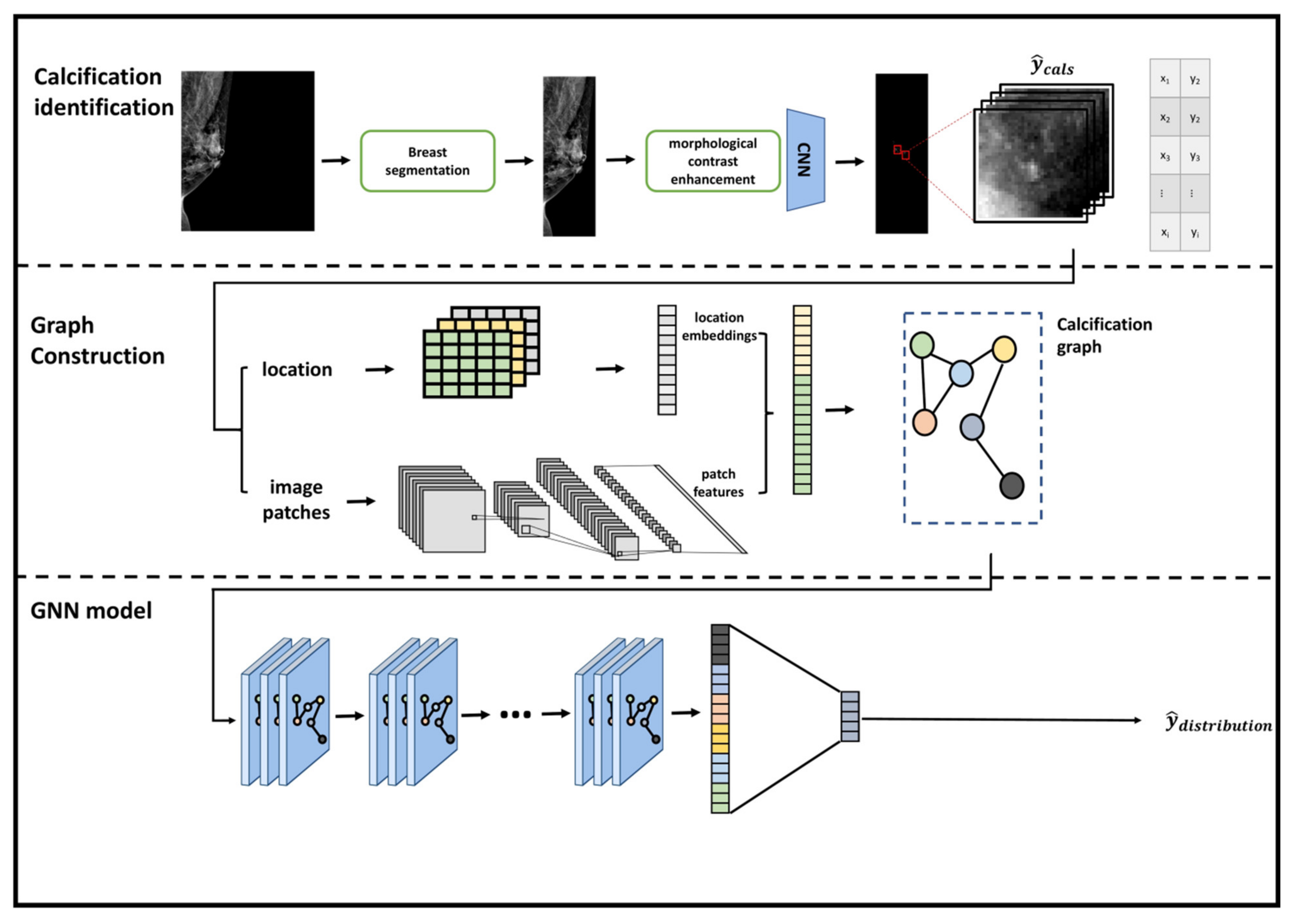
- We designed a graph construction module which detects calcifications and transforms them into nodes in graphs. We defined node features and an adjacency matrix to represent the calcification graph. Node features were represented by deep feature maps from a convolutional neural network which was trained on patches that were centered at proposed calcifications. Adjacency matrix of nodes was defined according to the spatial relationship among calcifications.
- We developed a graph convolutional neural network to fuse the node characteristics of the calcification graph and the spatial topological relationship to perform the graph classification task. The graph convolutional neural network was trained to fuse the features and topological structures from neighboring nodes and extract the most correlated information for the classification task.
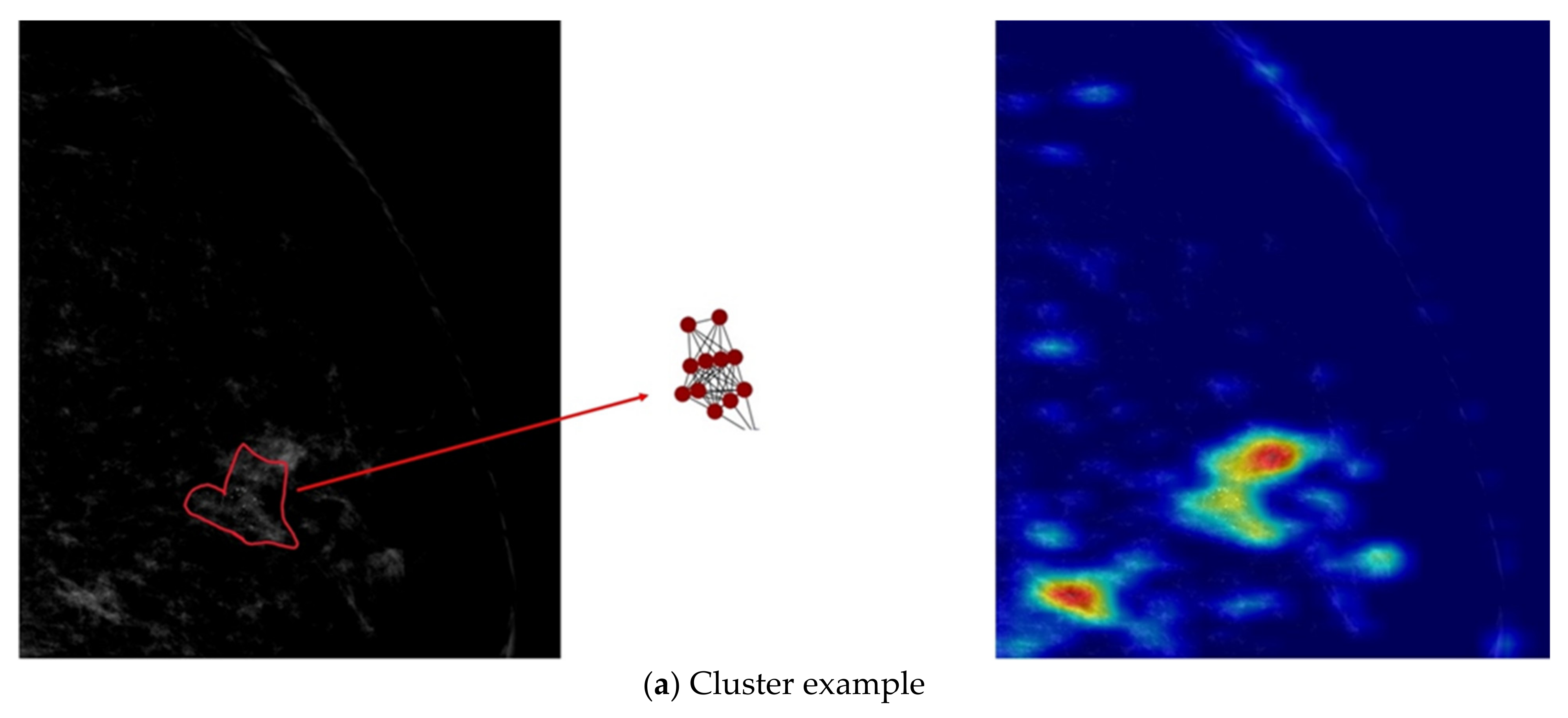
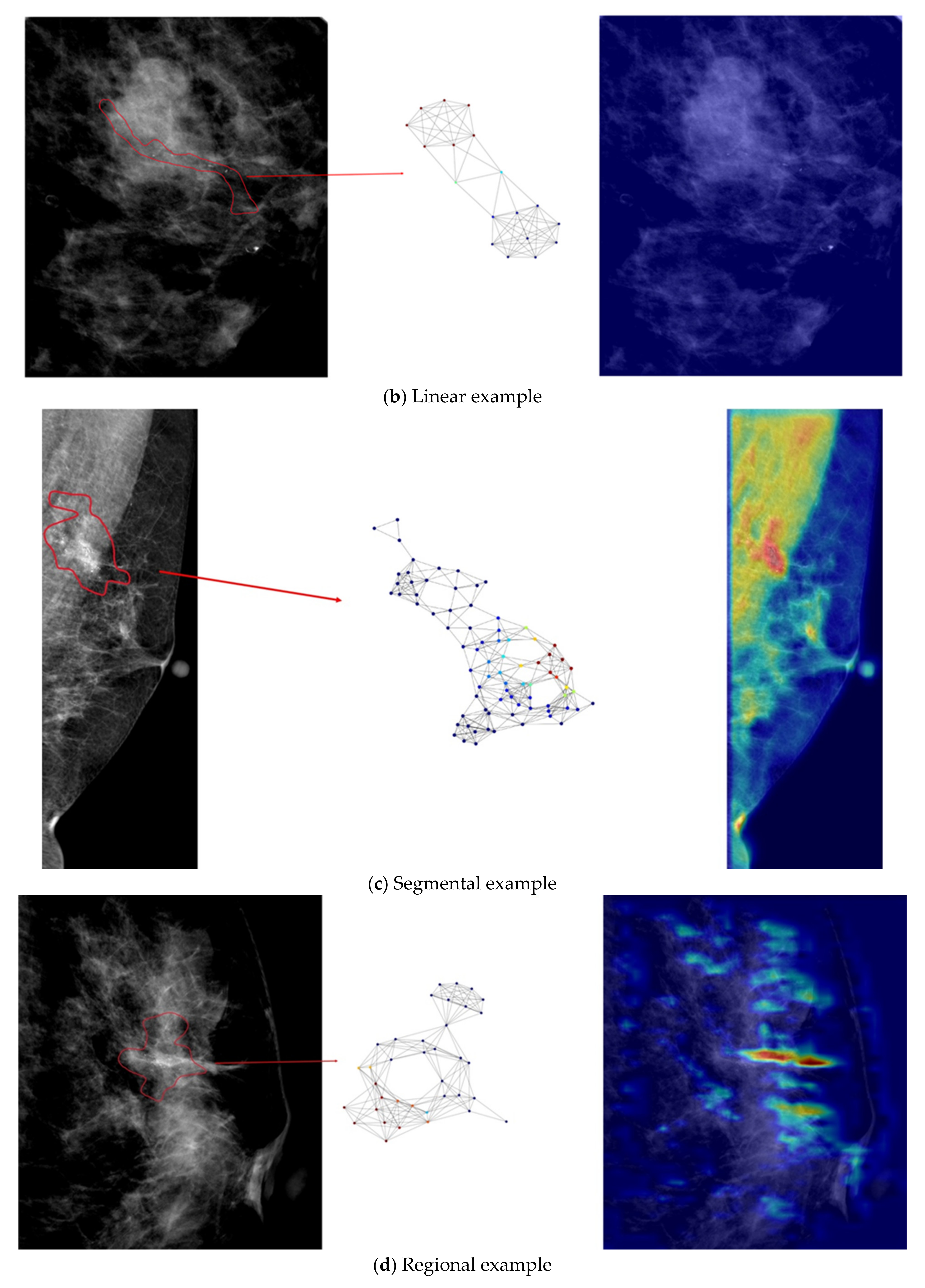
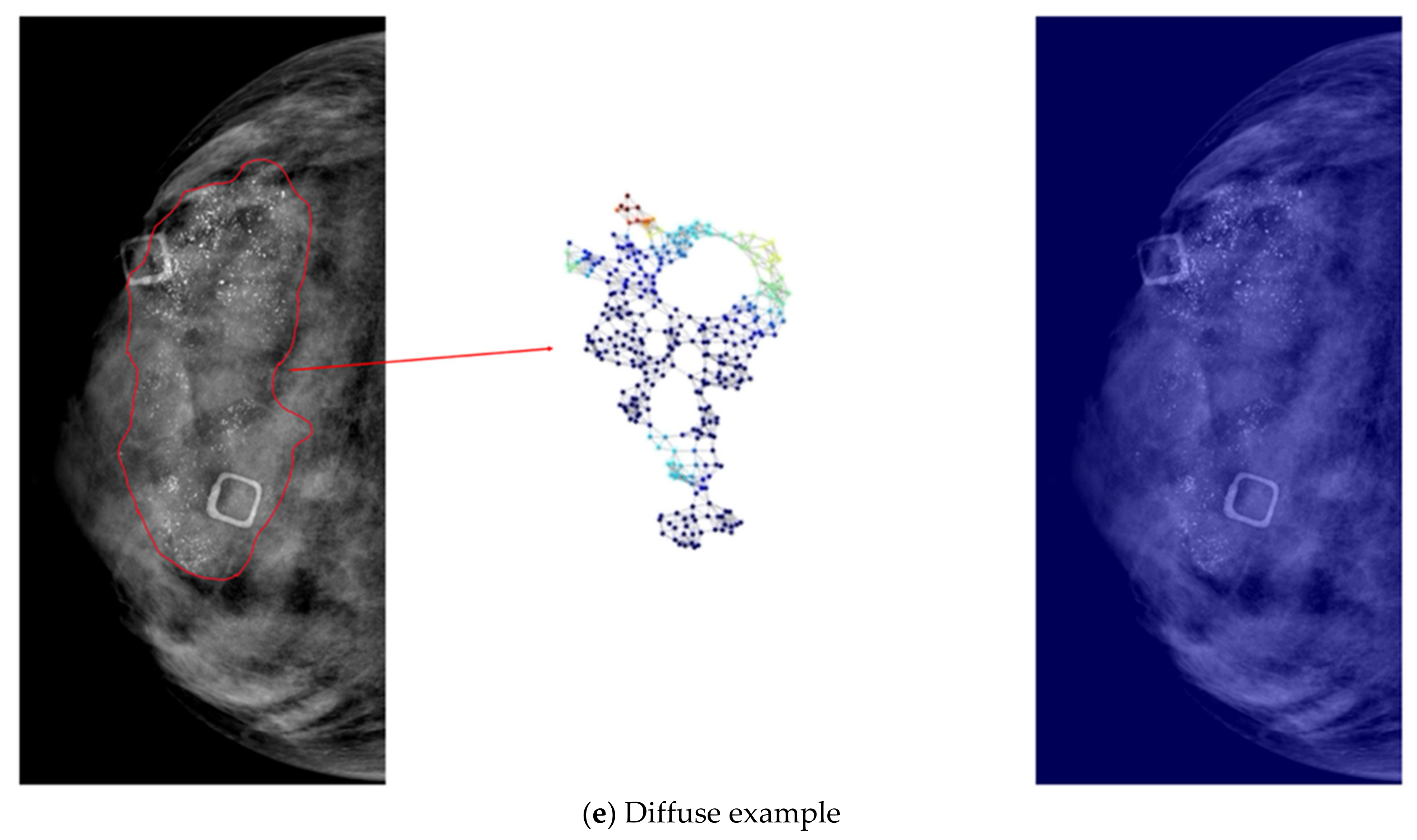
- Our developed model is interpretable by highlighting important nodes in graphs. For each distribution descriptor, the highlighted nodes are consistent with the clinical descriptions.
2.4. Calcification Identification
2.5. Calcification Graph Construction
2.5.1. Learning Feature Representations and Spatial Embeddings of Calcification Patches
2.5.2. Building a Calcification Graph
2.5.3. Learning Spatial and Distribution Relationships Using a Graph Neural Network
2.6. Data Analysis
- ResNet was proposed by He et al. and won the ILSVRS competition in 2015. The authors proposed residual blocks with skip connections to train deep CNNs with up to 152 layers. ResNet is one of the most popular and successful methods in the computer vision community with various applications in medical imaging [32,33,34]. ResNet-50 was used in this study.
- DenseNet was proposed with dense connections between layers to reuse features and preserve the global state. DenseNet demonstrated outperforming results on small benchmarking datasets such as Cifar-10 and Cifar-100 [29]. DenseNet-121 was adopted in this study.
- MobileNet was proposed primarily with depthwise separable convolutions as an efficient model with high accuracy and low latency for mobile and embedded applications. The effectiveness of MobileNets has been demonstrated for various applications, such as object detection, traffic density estimation, and computer-aided diagnosis systems [30,35,36,37,38]. MobileNetV2 was used in this study.
- EfficientNets is a family of models proposed by Google in 2019. EfficientNets outperformed state-of-the-art accuracy with up to 10 times better efficiency. A compound scaling method was proposed in EfficientNets to expand the depth, width, and resolution of the network. EfficientNets obtained state-of-the-art capacity in various benchmark datasets while requiring less computing resource than other models. EfficientNet-B7 was used in experiments of this study.
3. Results
3.1. Implementation Details
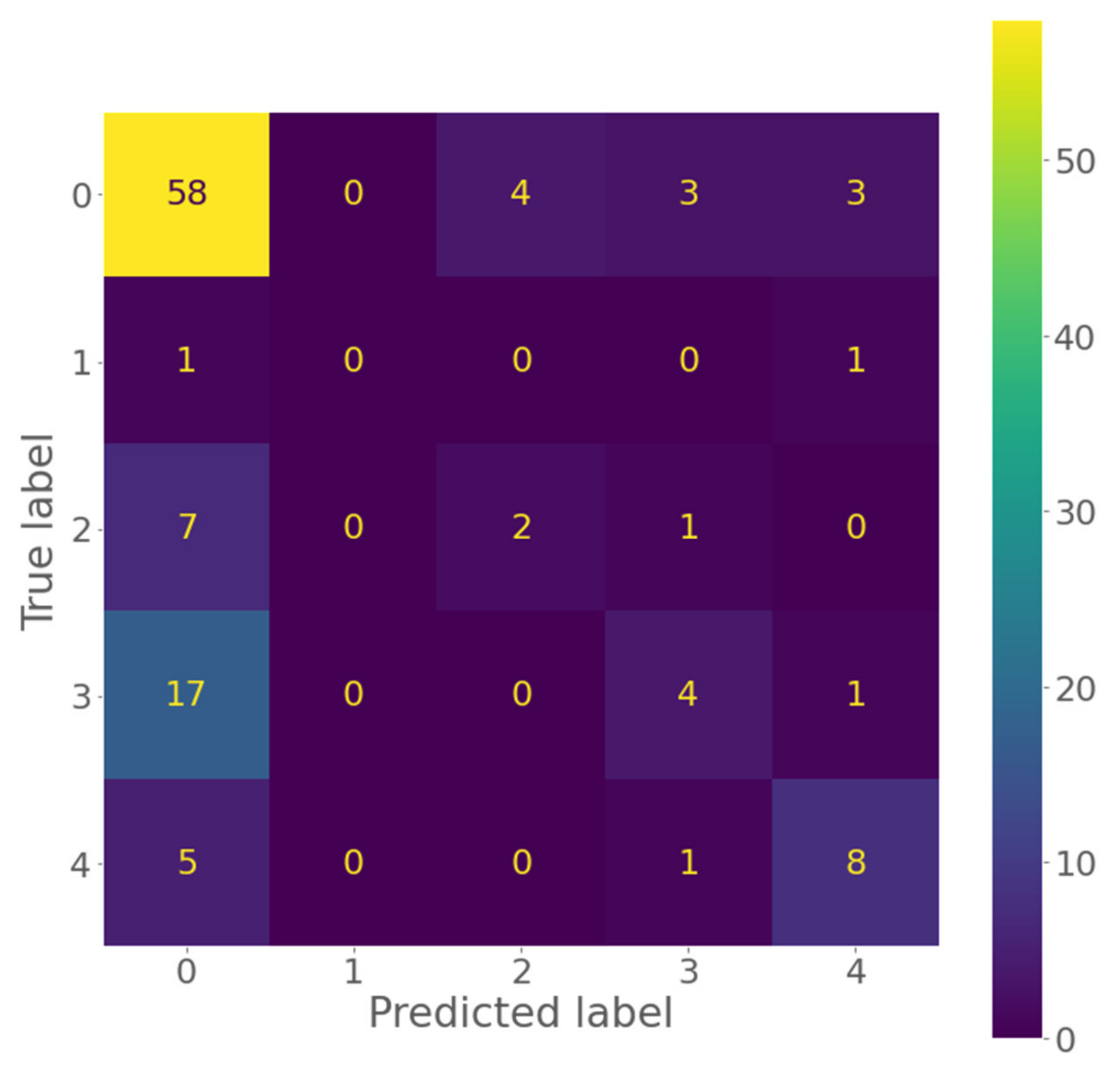
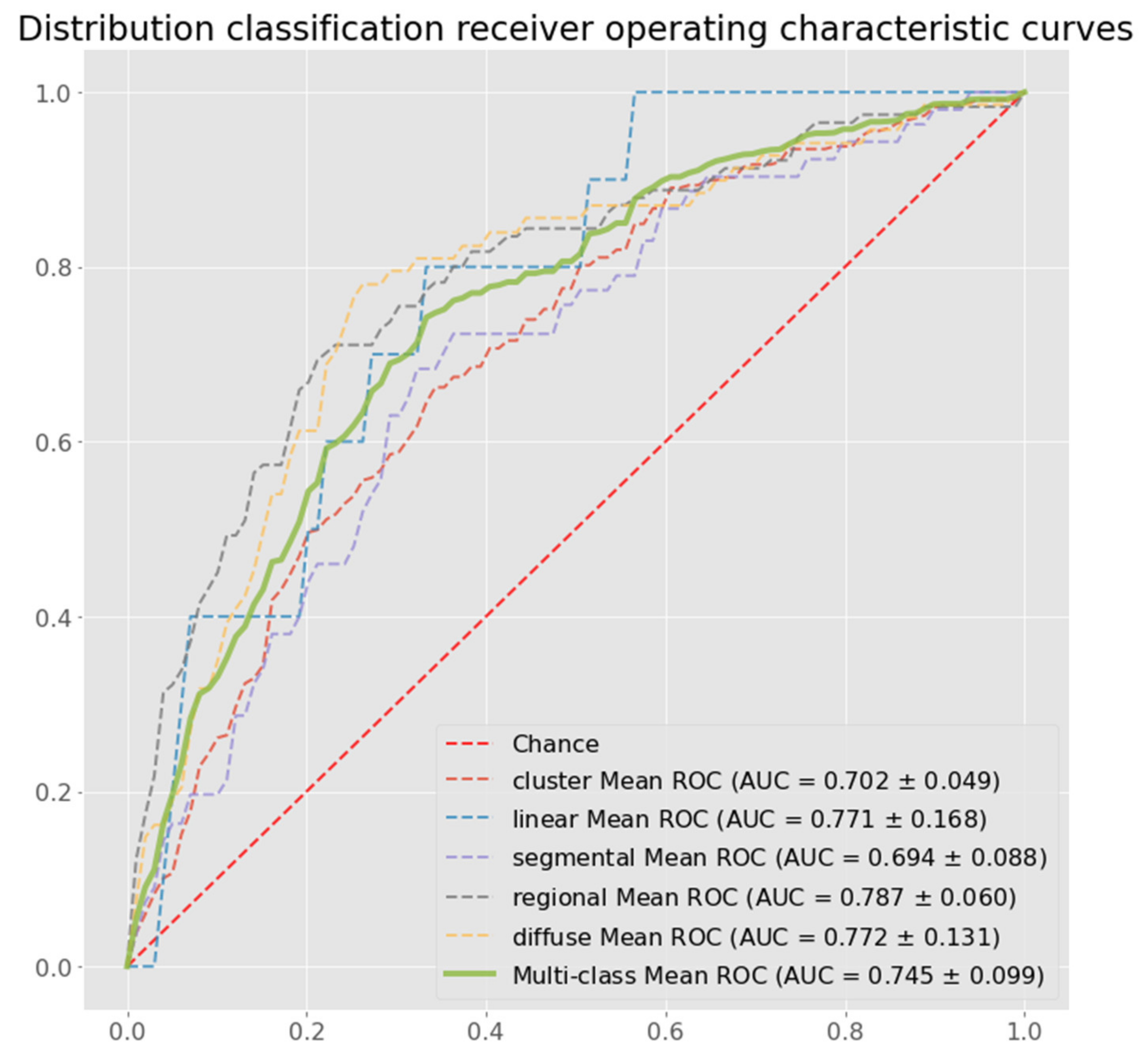
3.2. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Myers, E.R.; Moorman, P.; Gierisch, J.M.; Havrilesky, L.J.; Grimm, L.J.; Ghate, S.; Davidson, B.; Mongtomery, R.C.; Crowley, M.J.; McCrory, D.C.; et al. Benefits and Harms of Breast Cancer Screening: A Systematic Review. JAMA 2015, 314, 1615–1634. [Google Scholar] [CrossRef]
- Moorman, S.E.H.; Pujara, A.C.; Sakala, M.D.; Neal, C.H.; Maturen, K.E.; Swartz, L.; Egloff, H.; Helvie, M.A. Annual Screening Mammography Associated with Lower Stage Breast Cancer Compared with Biennial Screening. AJR Am. J. Roentgenol. 2021, 217, 40–47. [Google Scholar] [CrossRef]
- Sanderson, M.; Levine, R.S.; Fadden, M.K.; Kilbourne, B.; Pisu, M.; Cain, V.; Husaini, B.A.; Langston, M.; Gittner, L.; Zoorob, R.; et al. Mammography Screening Among the Elderly: A Research Challenge. Am. J. Med. 2015, 128, 1362.e7–1362.e14. [Google Scholar] [CrossRef]
- Azam, S.; Eriksson, M.; Sjölander, A.; Gabrielson, M.; Hellgren, R.; Czene, K.; Hall, P. Mammographic Microcalcifications and Risk of Breast Cancer. Br. J. Cancer 2021, 125, 759–765. [Google Scholar] [CrossRef]
- Spak, D.A.; Plaxco, J.S.; Santiago, L.; Dryden, M.J.; Dogan, B.E. BI-RADS® Fifth Edition: A Summary of Changes. Diagn. Interv. Imaging 2017, 98, 179–190. [Google Scholar] [CrossRef]
- Sickles, E.A.; D’Orsi, C.J.; Bassett, L.W.; Appleton, C.M.; Berg, W.A.; Burnside, E.S. 2013 ACR BI-RADS Atlas: Breast Imaging Reporting and Data System; American College of Radiology: Reston, VA, USA, 2014; ISBN 978-1-55903-016-8. [Google Scholar]
- D’Orsi, C.; Bassett, L.; Feig, S. Breast Imaging Reporting & Data System (BI-RADS). In Breast Imaging Atlas; American College of Radiology: Reston, VA, USA, 2018. [Google Scholar]
- Hernández, P.L.A.; Estrada, T.; Pizarro, A.; Cisternas, M.L.D.; Tapia, C. Breast Calcifications: Description and Classification According to BI-RADS, 5th ed. Available online: https://www.semanticscholar.org/paper/Breast-calcifications-%3A-description-and-according-5-Hern%C3%A1ndez-Estrada/12330a09d0667bede067cdff8f14a7f8bae73dd7 (accessed on 12 May 2022).
- Ha, S.M.; Cha, J.H.; Kim, H.H.; Shin, H.J.; Chae, E.Y.; Choi, W.J. Retrospective Analysis on Malignant Calcification Previously Misdiagnosed as Benign on Screening Mammography. J. Korean Soc. Radiol. 2017, 76, 251. [Google Scholar] [CrossRef]
- Coolen, A.M.P.; Voogd, A.C.; Strobbe, L.J.; Louwman, M.W.J.; Tjan-Heijnen, V.C.G.; Duijm, L.E.M. Impact of the Second Reader on Screening Outcome at Blinded Double Reading of Digital Screening Mammograms. Br. J. Cancer 2018, 119, 503–507. [Google Scholar] [CrossRef]
- Al-Antari, M.A.; Al-Masni, M.A.; Choi, M.-T.; Han, S.-M.; Kim, T.-S. A Fully Integrated Computer-Aided Diagnosis System for Digital X-Ray Mammograms via Deep Learning Detection, Segmentation, and Classification. Int. J. Med. Inform. 2018, 117, 44–54. [Google Scholar] [CrossRef]
- Rodriguez-Ruiz, A.; Lång, K.; Gubern-Merida, A.; Teuwen, J.; Broeders, M.; Gennaro, G.; Clauser, P.; Helbich, T.H.; Chevalier, M.; Mertelmeier, T.; et al. Can We Reduce the Workload of Mammographic Screening by Automatic Identification of Normal Exams with Artificial Intelligence? A Feasibility Study. Eur. Radiol. 2019, 29, 4825–4832. [Google Scholar] [CrossRef]
- Cruz-Bernal, A.; Flores-Barranco, M.M.; Almanza-Ojeda, D.L.; Ledesma, S.; Ibarra-Manzano, M.A. Analysis of the Cluster Prominence Feature for Detecting Calcifications in Mammograms. J. Healthc. Eng. 2018, 2018, 2849567. [Google Scholar] [CrossRef]
- Wang, J.; Yang, X.; Cai, H.; Tan, W.; Jin, C.; Li, L. Discrimination of Breast Cancer with Microcalcifications on Mammography by Deep Learning. Sci. Rep. 2016, 6, 27327. [Google Scholar] [CrossRef]
- Rehman, M.A.; Ahmed, J.; Waqas, A.; Sawand, A. Intelligent System for Detection of Micro-Calcification in Breast Cancer. Int. J. Adv. Comput. Sci. Appl. (IJACSA) 2017, 8, 382–387. [Google Scholar] [CrossRef][Green Version]
- Mayo, R.C.; Kent, D.; Sen, L.C.; Kapoor, M.; Leung, J.W.T.; Watanabe, A.T. Reduction of False-Positive Markings on Mammograms: A Retrospective Comparison Study Using an Artificial Intelligence-Based CAD. J. Digit. Imaging 2019, 32, 618–624. [Google Scholar] [CrossRef]
- Cai, H.; Huang, Q.; Rong, W.; Song, Y.; Li, J.; Wang, J.; Chen, J.; Li, L. Breast Microcalcification Diagnosis Using Deep Convolutional Neural Network from Digital Mammograms. Comput. Math. Methods Med. 2019, 2019, 2717454. [Google Scholar] [CrossRef]
- Suhail, Z.; Denton, E.R.E.; Zwiggelaar, R. Classification of Micro-Calcification in Mammograms Using Scalable Linear Fisher Discriminant Analysis. Med. Biol. Eng. Comput. 2018, 56, 1475–1485. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, F.; Chen, C.; Wang, S.; Wang, Y.; Yu, Y. Act Like a Radiologist: Towards Reliable Multi-View Correspondence Reasoning for Mammogram Mass Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 1. [Google Scholar] [CrossRef]
- Li, H.; Chen, D.; Nailon, W.H.; Davies, M.E.; Laurenson, D.I. Dual Convolutional Neural Networks for Breast Mass Segmentation and Diagnosis in Mammography. IEEE Trans. Med. Imaging 2022, 41, 3–13. [Google Scholar] [CrossRef]
- Du, H.; Yao, M.M.-S.; Chen, L.; Chan, W.P.; Feng, M. Multi-Task Graph Convolutional Neural Network for Calcification Morphology and Distribution Analysis in Mammograms. arXiv 2021, arXiv:2105.06822. [Google Scholar]
- Zhang, J.; Hu, J. Image Segmentation Based on 2D Otsu Method with Histogram Analysis. In Proceedings of the 2008 International Conference on Computer Science and Software Engineering, Wuhan, China, 12–14 December 2008; IEEE: Washington, DC, USA, 2008; Volume 6, pp. 105–108. [Google Scholar]
- Quintanilla-Domínguez, J.; Ruiz-Pinales, J.; Barrón-Adame, J.M.; Guzmán-Cabrera, R.; Quintanilla-Domínguez, J.; Ruiz-Pinales, J.; Barrón-Adame, J.M.; Guzmán-Cabrera, R. Microcalcifications Detection Using Image Processing. Comput. Sist. 2018, 22, 291–300. [Google Scholar] [CrossRef]
- Dong, W.; Moses, C.; Li, K. Efficient K-Nearest Neighbor Graph Construction for Generic Similarity Measures. In Proceedings of the 20th International Conference on World Wide Web, Hyderabad, India, 28 March–1 April 2011; Association for Computing Machinery: New York, NY, USA, 2011; pp. 577–586. [Google Scholar]
- Simonovsky, M.; Komodakis, N. Dynamic Edge-Conditioned Filters in Convolutional Neural Networks on Graphs. arXiv 2017, arXiv:1704.02901. [Google Scholar]
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal Loss for Dense Object Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 318–327. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2018, arXiv:1608.06993. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; Volume 97, pp. 6105–6114. [Google Scholar]
- Ardakani, A.A.; Kanafi, A.R.; Acharya, U.R.; Khadem, N.; Mohammadi, A. Application of Deep Learning Technique to Manage COVID-19 in Routine Clinical Practice Using CT Images: Results of 10 Convolutional Neural Networks. Comput. Biol. Med. 2020, 121, 103795. [Google Scholar] [CrossRef]
- Serte, S.; Serener, A.; Al-Turjman, F. Deep Learning in Medical Imaging: A Brief Review. Trans. Emerg. Telecommun. Technol. 2020, e4080. [Google Scholar] [CrossRef]
- Xia, K.-J.; Yin, H.-S.; Zhang, Y.-D. Deep Semantic Segmentation of Kidney and Space-Occupying Lesion Area Based on SCNN and ResNet Models Combined with SIFT-Flow Algorithm. J. Med. Syst. 2018, 43, 2. [Google Scholar] [CrossRef]
- Biswas, D.; Su, H.; Wang, C.; Stevanovic, A.; Wang, W. An Automatic Traffic Density Estimation Using Single Shot Detection (SSD) and MobileNet-SSD. Phys. Chem. Earth Parts A/B/C 2019, 110, 176–184. [Google Scholar] [CrossRef]
- Velasco, J.; Pascion, C.; Alberio, J.W.; Apuang, J.; Cruz, J.S.; Gomez, M.A.; Molina, B.J.; Tuala, L.; Thio-ac, A.; Jorda, R.J. A Smartphone-Based Skin Disease Classification Using MobileNet CNN. Int. J. Adv. Trends Comput. Sci. Eng. 2019, 8, 2632–2637. [Google Scholar] [CrossRef]
- Lu, S.-Y.; Wang, S.-H.; Zhang, Y.-D. A Classification Method for Brain MRI via MobileNet and Feedforward Network with Random Weights. Pattern Recognit. Lett. 2020, 140, 252–260. [Google Scholar] [CrossRef]
- Pang, S.; Wang, S.; Rodríguez-Patón, A.; Li, P.; Wang, X. An Artificial Intelligent Diagnostic System on Mobile Android Terminals for Cholelithiasis by Lightweight Convolutional Neural Network. PLoS ONE 2019, 14, e0221720. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2017, arXiv:1412.6980. [Google Scholar]
- Pope, P.E.; Kolouri, S.; Rostami, M.; Martin, C.E.; Hoffmann, H. Explainability Methods for Graph Convolutional Neural Networks. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 10764–10773. [Google Scholar]
- Bronstein, M.M.; Bruna, J.; LeCun, Y.; Szlam, A.; Vandergheynst, P. Geometric Deep Learning: Going beyond Euclidean Data. IEEE Signal Process. Mag. 2017, 34, 18–42. [Google Scholar] [CrossRef]
- Wu, Z.; Pan, S.; Chen, F.; Long, G.; Zhang, C.; Yu, P.S. A Comprehensive Survey on Graph Neural Networks. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 4–24. [Google Scholar] [CrossRef]
- Liang, X.; Shen, X.; Feng, J.; Lin, L.; Yan, S. Semantic Object Parsing with Graph LSTM. arXiv 2016, arXiv:1603.07063. [Google Scholar]
- Liang, X.; Lin, L.; Shen, X.; Feng, J.; Yan, S.; Xing, E.P. Interpretable Structure-Evolving LSTM. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2175–2184. [Google Scholar]
- Qi, X.; Liao, R.; Jia, J.; Fidler, S.; Urtasun, R. 3D Graph Neural Networks for RGBD Semantic Segmentation. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 5209–5218. [Google Scholar]
| Precision | Sensitivity | Specificity | F1 Score | Accuracy | Multi-Class AUC | |
|---|---|---|---|---|---|---|
| ResNet | 0.388 (±0.067) | 0.594 (±0.019) | 0.810 (±0.013) | 0.459 (±0.044) | 0.594 (±0.019) | 0.672 (±0.035) |
| DenseNet | 0.388 (±0.060) | 0.590 (±0.013) | 0.808 (±0.012) | 0.451 (±0.034) | 0.590 (±0.013) | 0.657 (±0.025) |
| MobileNet | 0.507 (±0.037) | 0.607 (±0.009) | 0.816 (±0.008) | 0.481 (±0.018) | 0.607 (±0.009) | 0.695 (±0.882) |
| EfficientNet | 0.356 (±0.043) | 0.581 (±0.009) | 0.802 (±0.003) | 0.430 (±0.015) | 0.581 (±0.009) | 0.695 (±0.030) |
| Proposed Method | 0.522 (±0.028) | 0.643 (±0.017) | 0.847 (±0.009) | 0.559 (±0.018) | 0.643 (±0.017) | 0.745 (±0.030) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, M.M.-S.; Du, H.; Hartman, M.; Chan, W.P.; Feng, M. End-to-End Calcification Distribution Pattern Recognition for Mammograms: An Interpretable Approach with GNN. Diagnostics 2022, 12, 1376. https://doi.org/10.3390/diagnostics12061376
Yao MM-S, Du H, Hartman M, Chan WP, Feng M. End-to-End Calcification Distribution Pattern Recognition for Mammograms: An Interpretable Approach with GNN. Diagnostics. 2022; 12(6):1376. https://doi.org/10.3390/diagnostics12061376
Chicago/Turabian StyleYao, Melissa Min-Szu, Hao Du, Mikael Hartman, Wing P. Chan, and Mengling Feng. 2022. "End-to-End Calcification Distribution Pattern Recognition for Mammograms: An Interpretable Approach with GNN" Diagnostics 12, no. 6: 1376. https://doi.org/10.3390/diagnostics12061376
APA StyleYao, M. M.-S., Du, H., Hartman, M., Chan, W. P., & Feng, M. (2022). End-to-End Calcification Distribution Pattern Recognition for Mammograms: An Interpretable Approach with GNN. Diagnostics, 12(6), 1376. https://doi.org/10.3390/diagnostics12061376






