The DendrisCHIP® Technology as a New, Rapid and Reliable Molecular Method for the Diagnosis of Osteoarticular Infections
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial, Clinical Specimens, and DNA
2.1.1. Bacterial and Clinical Specimens Provision
2.1.2. Microbiological Cultures for Bacteria Identification
2.2. Bacterial, Probes Design, and Manufacture of DendrisCHIP®OA
2.3. Process Flow for DendrisCHIP®OA Validation
2.3.1. DNA Extraction and PCR
2.3.2. Semiautomated Hybridization Process and Reading of the DendrisCHIP®OA
2.3.3. Data Treatment Using Machine-Learning Methods and Statistical Analysis
2.4. Process Flow with Next-Generation Sequencing (NGS) Targeted on 16S rRNA Gene
3. Results
3.1. Construction and Validation of the DendrisCHIP®OA
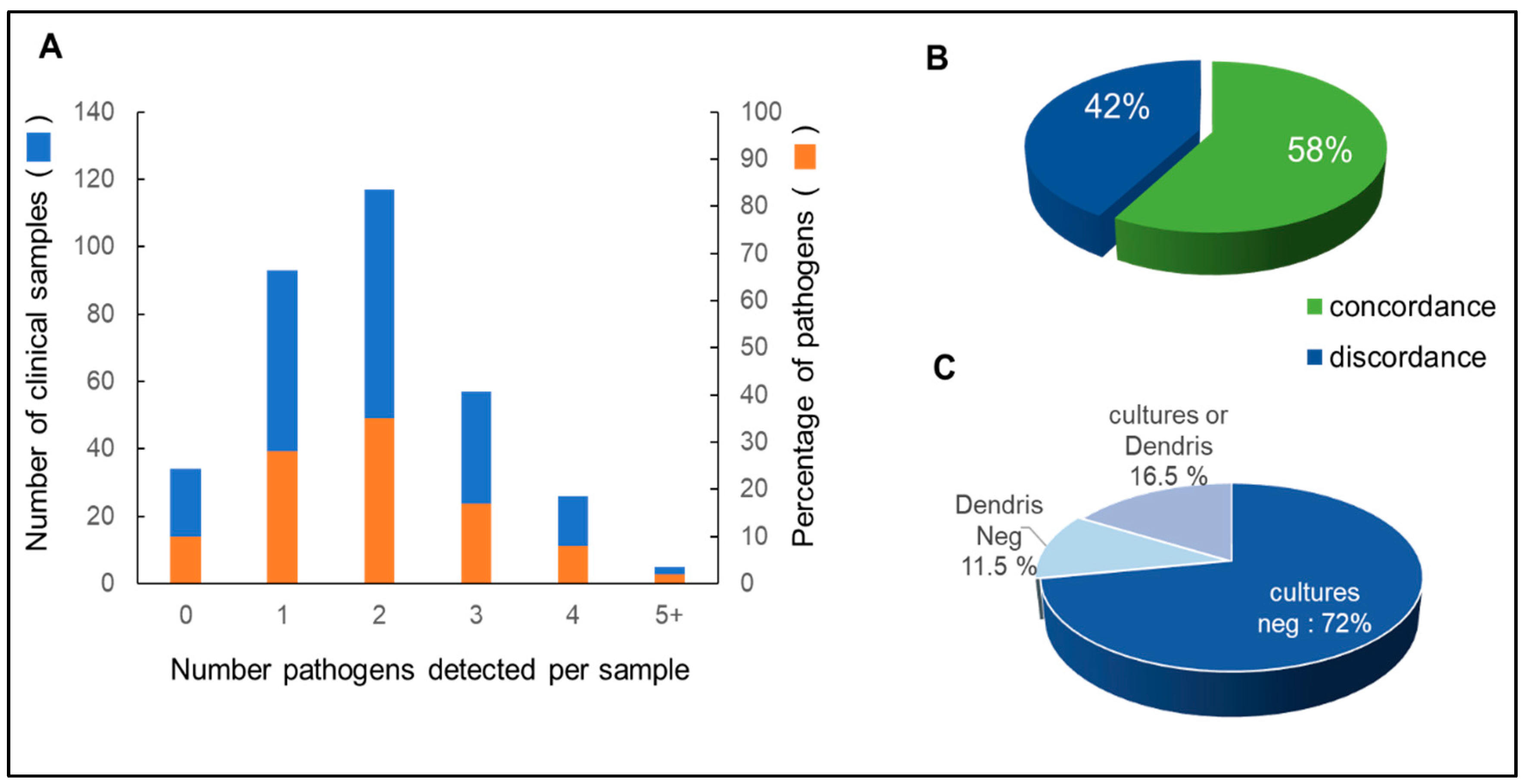
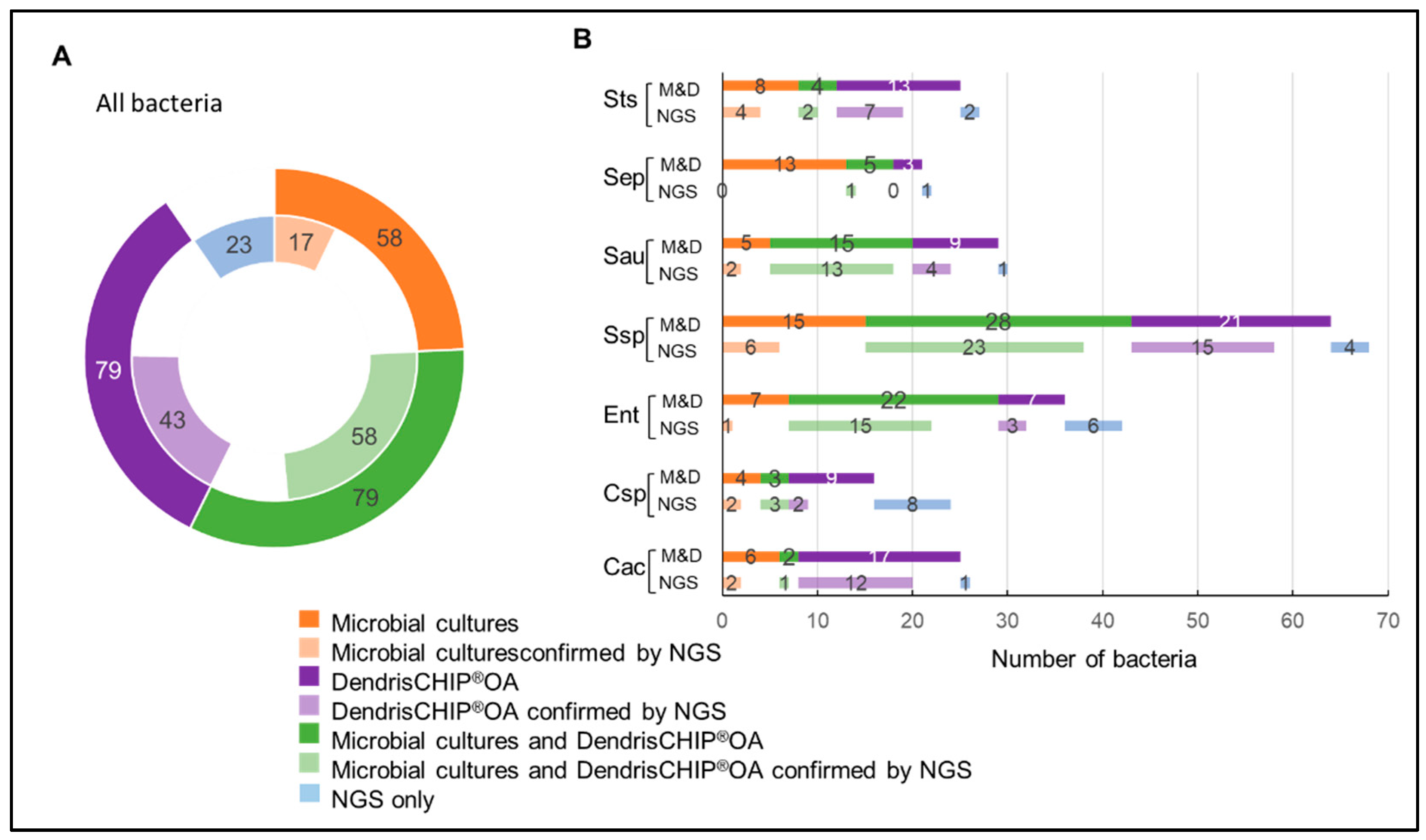
3.2. Isolates Analyzed by DendrisCHIP®OA and Comparison with Microbial Cultures
3.3. Reliability of DendrisCHIP®OA Technology in Diagnosing Osteoarticular Infection by Comparison with NGS
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Zimmerli, W. Infection and musculoskeletal conditions: Prosthetic-joint-associated infections. Best Pract. Res. Clin. Rheumatol. 2006, 20, 1045–1063. [Google Scholar] [CrossRef] [PubMed]
- Tande, A.J.; Patel, R. Prosthetic joint infection. Clin. Microbiol. Rev. 2014, 27, 302–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurtz, S.; Ong, K.; Lau, E.; Mowat, F.; Halpern, M. Projections of primary and revision hip and knee arthroplasty in the united states from 2005 to 2030. J. Bone Jt. Surg. Am. 2007, 89, 780–785. [Google Scholar] [CrossRef]
- Parvizi, J.; Erkocak, O.F.; Della Valle, C.J. Culture-negative periprosthetic joint infection. J. Bone Jt. Surg. Am. 2014, 96, 430–436. [Google Scholar] [CrossRef]
- Laurent, E.; Gras, G.; Druon, J.; Rosset, P.; Baron, S.; Le-Louarn, A.; Rusch, E.; Bernard, L.; Grammatico-Guillon, L. Key features of bone and joint infections following the implementation of reference centers in france. Med. Mal. Infect. 2018, 48, 256–262. [Google Scholar] [CrossRef]
- Chauvelot, P.; Ferry, T.; Tafani, V.; Diot, A.; Tasse, J.; Conrad, A.; Chidiac, C.; Braun, E.; Lustig, S.; Laurent, F.; et al. Bone and joint infection involving corynebacterium spp.: From clinical features to pathophysiological pathways. Front. Med. 2020, 7, 539501. [Google Scholar] [CrossRef]
- McNally, M.; Sousa, R.; Wouthuyzen-Bakker, M.; Chen, A.F.; Soriano, A.; Vogely, H.C.; Clauss, M.; Higuera, C.A.; Trebše, R. The ebjis definition of periprosthetic joint infection. Bone Jt. J. 2021, 103, 18–25. [Google Scholar] [CrossRef]
- Li, C.; Renz, N.; Trampuz, A.; Ojeda-Thies, C. Twenty common errors in the diagnosis and treatment of periprosthetic joint infection. Int. Orthop. 2020, 44, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Parvizi, J.; Gehrke, T.; Chen, A.F. In Proceedings of the international consensus on periprosthetic joint infection. Bone Jt. J. 2013, 95, 1450–1452. [Google Scholar] [CrossRef]
- Kalbian, I.; Park, J.W.; Goswami, K.; Lee, Y.K.; Parvizi, J.; Koo, K.H. Culture-negative periprosthetic joint infection: Prevalence, aetiology, evaluation, recommendations, and treatment. Int. Orthop. 2020, 44, 1255–1261. [Google Scholar] [CrossRef]
- Tarabichi, M.; Shohat, N.; Goswami, K.; Alvand, A.; Silibovsky, R.; Belden, K.; Parvizi, J. Diagnosis of periprosthetic joint infection: The potential of next-generation sequencing. J. Bone Jt. Surg. Am. 2018, 100, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Esteban, J.; Gómez-Barrena, E. An update about molecular biology techniques to detect orthopaedic implant-related infections. EFORT Open Rev. 2021, 6, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Fihman, V.; Hannouche, D.; Bousson, V.; Bardin, T.; Liote, F.; Raskine, L.; Riahi, J.; Sanson-Le Pors, M.J.; Bercot, B. Improved diagnosis specificity in bone and joint infections using molecular techniques. J. Infect. 2007, 55, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Marin, M.; Garcia-Lechuz, J.M.; Alonso, P.; Villanueva, M.; Alcala, L.; Gimeno, M.; Cercenado, E.; Sanchez-Somolinos, M.; Radice, C.; Bouza, E. Role of universal 16s rrna gene pcr and sequencing in diagnosis of prosthetic joint infection. J. Clin. Microbiol. 2012, 50, 583–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bemer, P.; Plouzeau, C.; Tande, D.; Leger, J.; Giraudeau, B.; Valentin, A.S.; Jolivet-Gougeon, A.; Vincent, P.; Corvec, S.; Gibaud, S.; et al. Evaluation of 16s rrna gene pcr sensitivity and specificity for diagnosis of prosthetic joint infection: A prospective multicenter cross-sectional study. J. Clin. Microbiol. 2014, 52, 3583–3589. [Google Scholar] [CrossRef] [Green Version]
- Portillo, M.E.; Salvado, M.; Sorli, L.; Alier, A.; Martinez, S.; Trampuz, A.; Gomez, J.; Puig, L.; Horcajada, J.P. Multiplex pcr of sonication fluid accurately differentiates between prosthetic joint infection and aseptic failure. J. Infect. 2012, 65, 541–548. [Google Scholar] [CrossRef]
- Renz, N.; Feihl, S.; Cabric, S.; Trampuz, A. Performance of automated multiplex pcr using sonication fluid for diagnosis of periprosthetic joint infection: A prospective cohort. Infection 2017, 45, 877–884. [Google Scholar] [CrossRef]
- Prieto-Borja, L.; Rodriguez-Sevilla, G.; Aunon, A.; Perez-Jorge, C.; Sandoval, E.; Garcia-Canete, J.; Gadea, I.; Fernandez-Roblas, R.; Blanco, A.; Esteban, J. Evaluation of a commercial multiplex pcr (unyvero i60((r))) designed for the diagnosis of bone and joint infections using prosthetic-joint sonication. Enferm. Infecc. Microbiol. Clin. 2017, 35, 236–242. [Google Scholar] [CrossRef]
- Malandain, D.; Bemer, P.; Leroy, A.G.; Leger, J.; Plouzeau, C.; Valentin, A.S.; Jolivet-Gougeon, A.; Tande, D.; Hery-Arnaud, G.; Lemarie, C.; et al. Assessment of the automated multiplex-pcr unyvero i60 iti((r)) cartridge system to diagnose prosthetic joint infection: A multicentre study. Clin. Microbiol. Infect. 2018, 24, 83.e81–83.e86. [Google Scholar] [CrossRef] [Green Version]
- Vasoo, S.; Cunningham, S.A.; Greenwood-Quaintance, K.E.; Mandrekar, J.N.; Hanssen, A.D.; Abdel, M.P.; Osmon, D.R.; Berbari, E.F.; Patel, R. Evaluation of the filmarray blood culture id panel on biofilms dislodged from explanted arthroplasties for prosthetic joint infection diagnosis. J. Clin. Microbiol. 2015, 53, 2790–2792. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Fang, X.; Chen, Y.; Huang, Z.; Zhang, C.; Li, W.; Yang, B.; Zhang, W. Metagenomic next generation sequencing improves diagnosis of prosthetic joint infection by detecting the presence of bacteria in periprosthetic tissues. Int. J. Infect. Dis. 2020, 96, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Thoendel, M.; Jeraldo, P.; Greenwood-Quaintance, K.E.; Chia, N.; Abdel, M.P.; Steckelberg, J.M.; Osmon, D.R.; Patel, R. A novel prosthetic joint infection pathogen, mycoplasma salivarium, identified by metagenomic shotgun sequencing. Clin. Infect. Dis. 2017, 65, 332–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Indelli, P.F.; Ghirardelli, S.; Violante, B.; Amanatullah, D.F. Next generation sequencing for pathogen detection in periprosthetic joint infections. EFORT Open Rev. 2021, 6, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Boers, S.A.; Jansen, R.; Hays, J.P. Understanding and overcoming the pitfalls and biases of next-generation sequencing (ngs) methods for use in the routine clinical microbiological diagnostic laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1059–1070. [Google Scholar] [CrossRef] [Green Version]
- Senescau, A.; Kempowsky, T.; Bernard, E.; Messier, S.; Besse, P.; Fabre, R.; Francois, J.M. Innovative dendrischips((r)) technology for a syndromic approach of in vitro diagnosis: Application to the respiratory infectious diseases. Diagnostics 2018, 8, 77. [Google Scholar] [CrossRef] [Green Version]
- Call, D.R. Challenges and opportunities for pathogen detection using DNA microarrays. Crit. Rev. Microbiol. 2005, 31, 91–99. [Google Scholar] [CrossRef]
- Barbulovic-Nad, I.; Lucente, M.; Sun, Y.; Zhang, M.; Wheeler, A.R.; Bussmann, M. Bio-microarray fabrication techniques—A review. Crit. Rev. Biotechnol. 2006, 26, 237–259. [Google Scholar] [CrossRef] [Green Version]
- Vlacich, G.; Roe, C.; Webb, G.C. Technology insight: Microarrays—Research and clinical applications. Nat. Clin. Pract. Endocrinol. Metab. 2007, 3, 594–605. [Google Scholar] [CrossRef]
- Beaudet, A.L.; Belmont, J.W. Array-based DNA diagnostics: Let the revolution begin. Annu. Rev. Med. 2008, 59, 113–129. [Google Scholar] [CrossRef]
- Trevisiol, E.; Le Berre-Anton, V.; Leclaire, J.; Pratviel, G.; Caminade, A.-M.; Majoral, J.P.; François, J.M.; Meunier, B. Dendrislides, dendrichips: A simple chemical functionalization of glass slides with phosphorus dendrimers as an effective means for the preparation of biochips. New J. Chem. 2003, 27, 1713–1719. [Google Scholar] [CrossRef]
- Le Berre, V.; Trevisiol, E.; Dagkessamanskaia, A.; Sokol, S.; Caminade, A.M.; Majoral, J.P.; Meunier, B.; Francois, J. Dendrimeric coating of glass slides for sensitive DNA microarrays analysis. Nucleic Acids Res. 2003, 31, e88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A. Primer3plus, an enhanced web interface to primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Tsoumakas, G.; Katakis, I.; Vlahavas, I. Effective and efficient multilabel classification in domains with large number of labels. In Workshop on Mining Multidimensional Data; 2008; p. 30. Available online: http://www.ecmlpkdd2008.org/files/pdf/workshops/mmd/4.pdf (accessed on 20 April 2022).
- Fei, T.; Hemant, I. Random forest missng data algorithm. Stat. Anal. Data Min. ASA Data Sci. J. 2017, 10, 366–377. [Google Scholar]
- Rivolli, A. Utiml: Utilities for Multi-Label Learning, R Package version 0.1.7; 2021. Available online: https://cran.r-project.org/web/packages/utiml/index.html (accessed on 30 November 2021).
- Hwang, S.M.; Kim, M.S.; Park, K.U.; Song, J.; Kim, E.C. Tuf gene sequence analysis has greater discriminatory power than 16s rrna sequence analysis in identification of clinical isolates of coagulase-negative staphylococci. J. Clin. Microbiol. 2011, 49, 4142–4149. [Google Scholar] [CrossRef] [Green Version]
- Coros, A.; DeConno, E.; Derbyshire, K.M. Is6110, a mycobacterium tuberculosis complex-specific insertion sequence, is also present in the genome of mycobacterium smegmatis, suggestive of lateral gene transfer among mycobacterial species. J. Bacteriol. 2008, 190, 3408–3410. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.H.; Kim, D.E.; Sung, H.; Park, B.M.; Cho, M.J.; Yoon, O.J.; Lee, D.H. Simple detection of the is6110 sequence of mycobacterium tuberculosis complex in sputum, based on pcr with graphene oxide. PLoS ONE 2015, 10, e0136954. [Google Scholar] [CrossRef]
- Miragaia, M. Factors contributing to the evolution of meca-mediated β-lactam resistance in staphylococci: Update and new insights from whole genome sequencing (wgs). Front. Microbiol. 2018, 9, 2723. [Google Scholar] [CrossRef] [Green Version]
- Uchida, K.; Yayama, T.; Kokubo, Y.; Miyazaki, T.; Nakajima, H.; Negoro, K.; Takeno, K.; Mwaka, E.S.; Orwotho, N.T.; Shimadzu, M.; et al. Direct detection of pathogens in osteoarticular infections by polymerase chain reaction amplification and microarray hybridization. J. Orthop. Sci. 2009, 14, 471–483. [Google Scholar] [CrossRef]
- Plouzeau, C.; Bemer, P.; Valentin, A.S.; Hery-Arnaud, G.; Tande, D.; Jolivet-Gougeon, A.; Vincent, P.; Kempf, M.; Lemarie, C.; Guinard, J.; et al. First experience of a multicenter external quality assessment of molecular 16s rrna gene detection in bone and joint infections. J. Clin. Microbiol. 2015, 53, 419–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safari, S.; Baratloo, A.; Elfil, M.; Negida, A. Evidence based emergency medicine part 2: Positive and negative predictive values of diagnostic tests. Emergency 2015, 3, 87–88. [Google Scholar] [PubMed]
- Gray, J.; Coupland, L.J. The increasing application of multiplex nucleic acid detection tests to the diagnosis of syndromic infections. Epidemiol. Infect. 2014, 142, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ramanan, P.; Bryson, A.L.; Binnicker, M.J.; Pritt, B.S.; Patel, R. Syndromic panel-based testing in clinical microbiology. Clin. Microbiol. Rev. 2018, 31, e00024-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanella, M.C.; Meylan, P.; Kaiser, L. Syndromic panels or ‘panel syndrome’? A perspective through the lens of respiratory tract infections. Clin. Microbiol. Infect. 2020, 26, 665–668. [Google Scholar] [CrossRef]
- Lamret, F.; Colin, M.; Mongaret, C.; Gangloff, S.C.; Reffuveille, F. Antibiotic tolerance of staphylococcus aureus biofilm in periprosthetic joint infections and antibiofilm strategies. Antibiotics 2020, 9, 547. [Google Scholar] [CrossRef]
- Eisenach, K.D.; Cave, M.D.; Bates, J.H.; Crawford, J.T. Polymerase chain reaction amplification of a repetitive DNA sequence specific for mycobacterium tuberculosis. J. Infect. Dis. 1990, 161, 977–981. [Google Scholar] [CrossRef]
- Martineau, F.; Picard, F.J.; Ke, D.; Paradis, S.; Roy, P.H.; Ouellette, M.; Bergeron, M.G. Development of a pcr assay for identification of staphylococci at genus and species levels. J. Clin. Microbiol. 2001, 39, 2541–2547. [Google Scholar] [CrossRef] [Green Version]
- Murakami, K.; Minamide, W.; Wada, K.; Nakamura, E.; Teraoka, H.; Watanabe, S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 1991, 29, 2240–2244. [Google Scholar] [CrossRef] [Green Version]


| Bacteria | Abbreviation Used in This Work | Taxonomy | Gene | Accession Number |
|---|---|---|---|---|
| Enterobacteriaceae | Ent | family | 16S rRNA | / |
| Enterobacter cloacae | Ecl | species | 16S rRNA | KC990822.1 |
| Escherichia coli | Eco | species | 16S rRNA | NR024570.1 |
| Klebsiella pneumoniae | Kpn | species | 16S rRNA | KC99081717.1 |
| Proteus mirabilis | Pmi | species | 16S rRNA | MN689880.1 |
| Corynebacterium spp. | Csp | genus | 16S rRNA | LT960557.1; NR119182.1; KF564647.1 |
| Cutibacterium acnes | Cac | species | 16S rRNA | DQ672261.1 |
| Enterococcus faecalis | Efa | species | 16S rRNA | AB362602.1 |
| Mycoplasma spp. | Msp | genus | 16S rRNA | / |
| Mycoplasma pneumoniae | Mpn | species | 16S rRNA | AF132741.1 |
| Mycoplasma genitalium | Mge | species | 16S rRNA | NR026155.1 |
| Neisseria spp. | Nsp | genus | 16S rRNA | / |
| Neisseria gonorrhoeae | Ngo | species | 16S rRNA | AM921674.1 |
| Neisseria meningitidis | Nme | species | 16S rRNA | NR104946.1 |
| Mycobacterium tuberculosis | Mtu | species | IS6110 | Y14045.1 |
| Kingella kingae | Kki | species | 16S rRNA | AY628416.1 |
| Staphylococcus spp. | Ssp | genus | 16S rRNA tuf | / |
| Staphylococcus aureus | Sau | species | 16S rRNA tuf | DQ630753.1 AF298796.1 |
| Staphylococcus epidermidis | Sep | species | 16S rRNA tuf | NR036904.1 AF298800.1 |
| Staphylococcus warneri | Swa | species | 16S rRNA tuf | LN998066.1 AF298806.1 |
| Staphylococcus haemolyticus | Sha | species | 16S rRNA tuf | LN998078.1 AF298801.1 |
| Staphylococcus hominis | Sho | species | 16S rRNA tuf | HG941670.1 AF298802.1 |
| Staphylococcus lugdunensis | Slu | species | 16S rRNA tuf | NR024668.1 AF298803.1 |
| Streptococcus spp. | Sts | genus | 16S rRNA | / |
| Streptococcus agalactiae | Sag | species | 16S rRNA | LC545464.1 |
| Streptococcus pyogenes | Spy | species | 16S rRNA | NR028598.1 |
| Streptococcus pneumoniae | Spn | species | 16S rRNA | NR028665.1 |
| Bacteria | Sensitivity (%) | Specificity (%) | CI95 Sensitivity (%) | CI95 Specificity (%) |
| Cutibacterium acnes | 95.8 | 98.4 | 86–99 | 97–99 |
| Corynebacterium spp. | 82.6 | 98.1 | 69–92 | 97–99 |
| Enterobacteriacea | 96.9 | 98.4 | 92–99 | 97–99 |
| Enterobacter cloace | 60.0 | 98.8 | 36–81 | 98–100 |
| Escherichia coli | 85.2 | 99.2 | 66–96 | 98–100 |
| Klebsiella pneumoniae | 92.9 | 99.2 | 76–99 | 98–100 |
| Proteus mirabilis | 93.8 | 99.7 | 70–100 | 99–100 |
| Enterococcus faecalis | 92.1 | 99.8 | 79–98 | 99–100 |
| kingella kingae | 78.9 | 99.5 | 54–94 | 99–100 |
| Mycobacterium tuberculosis | 97.0 | 100.0 | 84–100 | 99–100 |
| Mycoplasma spp. | 75.0 | 100.0 | 59–87 | 99–100 |
| Mycoplasma genitalium | 90.0 | 99.7 | 73–98 | 99–100 |
| Mycoplasma pneumoniae | 72.7 | 100.0 | 39–94 | 99–100 |
| Neisseria spp. | 86.8 | 99.5 | 72–96 | 98–100 |
| Neisseria gonorrhoeae | 76.2 | 98.8 | 53–92 | 98–100 |
| Neisseria meningitidis | 44.4 | 99.5 | 22–69 | 99–100 |
| Pseudomonas aeruginosa | 83.3 | 99.5 | 59–96 | 99–100 |
| Staphylococcus spp. | 94.1 | 96.4 | 90–97 | 94–98 |
| Staphylococcus aureus | 93.1 | 98.0 | 87–97 | 96–99 |
| Staphylococcus epidermidis | 74.3 | 99.8 | 57–88 | 99–100 |
| Staphylococcus haemolyticus | 69.2 | 100.0 | 39–91 | 99–100 |
| Staphylococcus hominis | 91.7 | 100.0 | 62–100 | 99–100 |
| Staphylococcus lugdunensis | 81.3 | 99.8 | 54–96 | 99–100 |
| Staphylococcus warneri | 92.3 | 99.8 | 64–100 | 99–100 |
| Streptococcus spp. | 94.0 | 97.6 | 87–98 | 96–99 |
| Streptococcus agalactiae | 93.3 | 99.8 | 78–99 | 99–100 |
| Streptococcus pneumoniae | 96.0 | 99.3 | 80–100 | 98–100 |
| Streptococcus pyogenes | 76.9 | 100.0 | 46–95 | 99–100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernard, E.; Peyret, T.; Plinet, M.; Contie, Y.; Cazaudarré, T.; Rouquet, Y.; Bernier, M.; Pesant, S.; Fabre, R.; Anton, A.; et al. The DendrisCHIP® Technology as a New, Rapid and Reliable Molecular Method for the Diagnosis of Osteoarticular Infections. Diagnostics 2022, 12, 1353. https://doi.org/10.3390/diagnostics12061353
Bernard E, Peyret T, Plinet M, Contie Y, Cazaudarré T, Rouquet Y, Bernier M, Pesant S, Fabre R, Anton A, et al. The DendrisCHIP® Technology as a New, Rapid and Reliable Molecular Method for the Diagnosis of Osteoarticular Infections. Diagnostics. 2022; 12(6):1353. https://doi.org/10.3390/diagnostics12061353
Chicago/Turabian StyleBernard, Elodie, Thomas Peyret, Mathilde Plinet, Yohan Contie, Thomas Cazaudarré, Yannick Rouquet, Matthieu Bernier, Stéphanie Pesant, Richard Fabre, Aurore Anton, and et al. 2022. "The DendrisCHIP® Technology as a New, Rapid and Reliable Molecular Method for the Diagnosis of Osteoarticular Infections" Diagnostics 12, no. 6: 1353. https://doi.org/10.3390/diagnostics12061353
APA StyleBernard, E., Peyret, T., Plinet, M., Contie, Y., Cazaudarré, T., Rouquet, Y., Bernier, M., Pesant, S., Fabre, R., Anton, A., Maugis-Rabusseau, C., & François, J. M. (2022). The DendrisCHIP® Technology as a New, Rapid and Reliable Molecular Method for the Diagnosis of Osteoarticular Infections. Diagnostics, 12(6), 1353. https://doi.org/10.3390/diagnostics12061353






