Detecting Abnormal Axillary Lymph Nodes on Mammograms Using a Deep Convolutional Neural Network
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Data Preparation
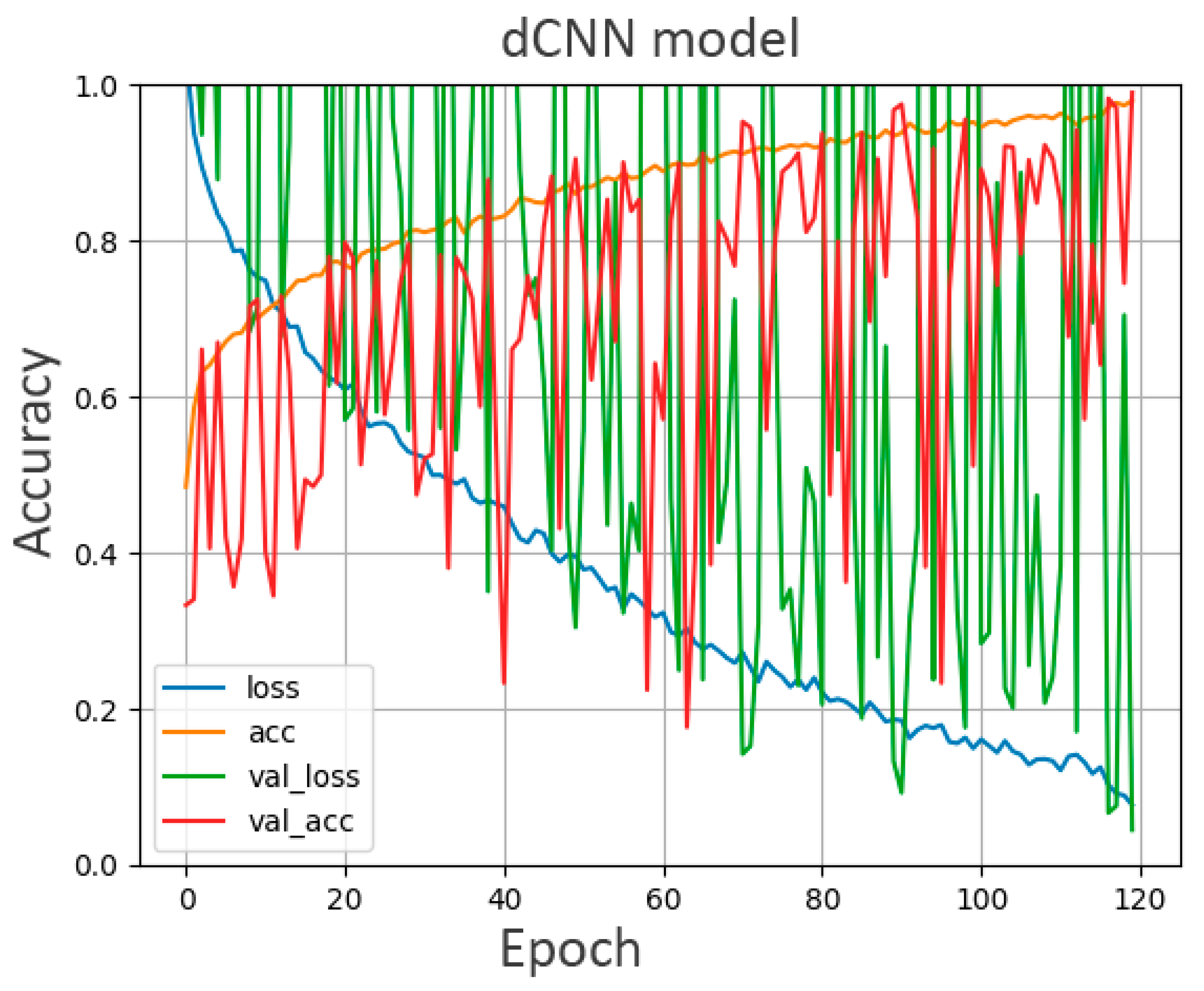
2.3. Training of dCNN Model
2.4. Human Readout on “Real World” Data
2.5. Computation of Colored Probability Maps
2.6. Statistical Analysis
3. Results
3.1. Accuracy of the dCNN Model
3.2. Human Readout
3.3. Colored Probability Map
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Dibden, A.; Offman, J.; Duffy, S.W.; Gabe, R. Worldwide Review and Meta-Analysis of Cohort Studies Measuring the Effect of Mammography Screening Programmes on Incidence-Based Breast Cancer Mortality. Cancers 2020, 12, 976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duijm, L.E.M.; Louwman, M.W.J.; Groenewoud, J.H.; Van De Poll-Franse, L.V.; Fracheboud, J.; Coebergh, J.W. Inter-observer variability in mammography screening and effect of type and number of readers on screening outcome. Br. J. Cancer 2009, 100, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Heywang-Köbrunner, S.H.; Hacker, A.; Sedlacek, S. Advantages and Disadvantages of Mammography Screening. Breast Care 2011, 6, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, R.A.; Kerlikowske, K.; Flowers, C.I.; Yankaskas, B.C.; Zhu, W.; Miglioretti, D.L. Cumulative Probability of False-Positive Recall or Biopsy Recommendation After 10 Years of Screening Mammography: A cohort study. Ann. Intern. Med. 2011, 155, 481–492. [Google Scholar] [CrossRef]
- Bird, R.E.; Wallace, T.W.; Yankaskas, B.C. Analysis of cancers missed at screening mammography. Radiology 1992, 184, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Weber, R.J.; van Bommel, R.M.; Louwman, M.W.; Nederend, J.; Voogd, A.C.; Jansen, F.H.; Tjan-Heijnen, V.C.; Duijm, L.E. Characteristics and prognosis of interval cancers after biennial screen-film or full-field digital screening mammography. Breast Cancer Res. Treat. 2016, 158, 471–483. [Google Scholar] [CrossRef]
- Krizhevsky, I.S.A.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Adv. Neural Inf. Processing Syst. 2012, 25. [Google Scholar] [CrossRef]
- Becker, A.; Marcon, M.; Ghafoor, S.; Wurnig, M.C.; Frauenfelder, T.; Boss, A. Deep Learning in Mammography: Diagnostic Accuracy of a Multipurpose Image Analysis Software in the Detection of Breast Cancer. Investig. Radiol. 2017, 52, 434–440. [Google Scholar] [CrossRef]
- Ciritsis, A.; Rossi, C.; De Martini, I.V.; Eberhard, M.; Marcon, M.; Becker, A.S.; Berger, N.; Boss, A. Determination of mammographic breast density using a deep convolutional neural network. Br. J. Radiol. 2019, 92, 20180691. [Google Scholar] [CrossRef]
- Schönenberger, C.; Hejduk, P.; Ciritsis, A.; Marcon, M.; Rossi, C.; Boss, A. Classification of Mammographic Breast Microcalcifications Using a Deep Convolutional Neural Network: A BI-RADS-Based Approach. Investig. Radiol. 2020, 56, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Augasta, M.G.; Kathirvalavakumar, T. Reverse Engineering the Neural Networks for Rule Extraction in Classification Prob-lems. Neural Processing Lett. 2011, 35, 131–150. [Google Scholar] [CrossRef]
- Walsh, R.; Kornguth, P.J.; Soo, M.S.; Bentley, R.; Delong, D.M. Axillary lymph nodes: Mammographic, pathologic, and clinical correlation. Am. J. Roentgenol. 1997, 168, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Bergkvist, L.; Frodis, E.; Hedborg-Mellander, C.; Hansen, J. Management of accidentally found pathological lymph nodes on routine screening mammography. Eur. J. Surg. Oncol. 1996, 22, 250–253. [Google Scholar] [CrossRef]
- Kolarik, D.; Pecha, V.; Skovajsova, M.; Zahumensky, J.; Trnková, M.; Petruzelka, L.; Halaska, M.; Sottner, O.; Otcenasek, M.; Kolarova, H. Predicting axillary sentinel node status in patients with primary breast cancer. Neoplasma 2013, 60, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Woods, R.W.; Camp, M.S.; Durr, N.J.; Harvey, S.C. A Review of Options for Localization of Axillary Lymph Nodes in the Treatment of Invasive Breast Cancer. Acad. Radiol. 2019, 26, 805–819. [Google Scholar] [CrossRef] [PubMed]
- Choschzick, M.; Alyahiaoui, M.; Ciritsis, A.; Rossi, C.; Gut, A.; Hejduk, P.; Boss, A. Deep learning for the standardized classification of Ki-67 in vulva carcinoma: A feasibility study. Heliyon 2021, 7, e07577. [Google Scholar] [CrossRef]
- Hejduk, P.; Marcon, M.; Unkelbach, J.; Ciritsis, A.; Rossi, C.; Borkowski, K.; Boss, A. Fully automatic classification of automated breast ultrasound (ABUS) imaging according to BI-RADS using a deep convolutional neural network. Eur. Radiol. 2022, 1–11. [Google Scholar] [CrossRef]
- Landsmann, A.; Wieler, J.; Hejduk, P.; Ciritsis, A.; Borkowski, K.; Rossi, C.; Boss, A. Applied Machine Learning in Spiral Breast-CT: Can We Train a Deep Convolutional Neural Network for Automatic, Standardized and Observer Independent Classification of Breast Density? Diagnostics 2022, 12, 181. [Google Scholar] [CrossRef]
- Bartko, J.J. The Intraclass Correlation Coefficient as a Measure of Reliability. Psychol. Rep. 1966, 19, 3–11. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Lyman, G.H.; Giuliano, A.E.; Somerfield, M.R.; Benson, A.B.; Bodurka, D.C.; Burstein, H.J.; Cochran, A.J.; Cody, H.S.; Edge, S.B.; Galper, S.; et al. American Society of Clinical Oncology Guideline Recommendations for Sentinel Lymph Node Biopsy in Early-Stage Breast Cancer. J. Clin. Oncol. 2005, 23, 7703–7720. [Google Scholar] [CrossRef] [Green Version]
- Veronesi, U.; Paganelli, G.; Viale, G.; Luini, A.; Zurrida, S.; Galimberti, V.; Intra, M.; Veronesi, P.; Maisonneuve, P.; Gatti, G.; et al. Sentinel-lymph-node biopsy as a staging procedure in breast cancer: Update of a randomised controlled study. Lancet Oncol. 2006, 7, 983–990. [Google Scholar] [CrossRef]
- Valente, S.A.; Levine, G.M.; Silverstein, M.J.; Rayhanabad, J.A.; Weng-Grumley, J.G.; Ji, L.; Holmes, D.R.; Sposto, R.; Sener, S.F. Accuracy of Predicting Axillary Lymph Node Positivity by Physical Examination, Mammography, Ultrasonography, and Magnetic Resonance Imaging. Ann. Surg. Oncol. 2012, 19, 1825–1830. [Google Scholar] [CrossRef] [PubMed]
- Shetty, M.K.; Carpenter, W.S. Sonographic evaluation of isolated abnormal axillary lymph nodes identified on mammograms. J. Ultrasound Med. 2004, 23, 63–71. [Google Scholar] [CrossRef]
- Liu, Q.; Xing, P.; Dong, H.; Zhao, T.; Jin, F. Preoperative assessment of axillary lymph node status in breast cancer patients by ultrasonography combined with mammography: A STROBE compliant article. Medicine 2018, 97, e11441. [Google Scholar] [CrossRef]
- Kim, K.; Shim, S.-R.; Kim, S.-J. Diagnostic Values of 8 Different Imaging Modalities for Preoperative Detection of Axillary Lymph Node Metastasis of Breast Cancerr: A Bayesian Network Meta-analysis. Am. J. Clin. Oncol. 2021, 44, 331–339. [Google Scholar] [CrossRef]
- Cooper, K.L.; Meng, Y.; Harnan, S.; Ward, S.E.; Fitzgerald, P.; Papaioannou, D.; Wyld, L.; Ingram, C.; Wilkinson, I.D.; Lorenz, E. Positron emission tomography (PET) and magnetic resonance imaging (MRI) for the assessment of axillary lymph node metastases in early breast cancer: Systematic review and economic evaluation. Health Technol. Assess. 2011, 15, iii–iv. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.-Q.; Wu, X.-L.; Huang, S.-Y.; Wu, G.-G.; Ye, H.-R.; Wei, Q.; Bao, L.-Y.; Deng, Y.-B.; Li, X.-R.; Cui, X.-W.; et al. Lymph Node Metastasis Prediction from Primary Breast Cancer US Images Using Deep Learning. Radiology 2019, 294, 19–28. [Google Scholar] [CrossRef]
- Sun, S.; Mutasa, S.; Liu, M.Z.; Nemer, J.; Sun, M.; Siddique, M.; Desperito, E.; Jambawalikar, S.; Ha, R.S. Deep learning prediction of axillary lymph node status using ultrasound images. Comput. Biol. Med. 2022, 143, 105250. [Google Scholar] [CrossRef] [PubMed]
- Ha, R.; Chang, P.; Karcich, J.; Mutasa, S.; Fardanesh, R.; Wynn, R.T.; Liu, M.Z.; Jambawalikar, S. Axillary Lymph Node Evaluation Utilizing Convolutional Neural Networks Using MRI Dataset. J. Digit. Imaging 2018, 31, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Cattell, R.; Duanmu, H.; Huang, P.; Li, H.; Vanguri, R.; Liu, M.Z.; Jambawalikar, S.; Ha, R.; Wang, F.; et al. Convolutional Neural Network Detection of Axillary Lymph Node Metastasis Using Standard Clinical Breast MRI. Clin. Breast Cancer 2020, 20, e301–e308. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ruiz, A.; Lång, K.; Gubern-Merida, A.; Broeders, M.; Gennaro, G.; Clauser, P.; Helbich, T.H.; Chevalier, M.; Tan, T.; Mertelmeier, T.; et al. Stand-Alone Artificial Intelligence for Breast Cancer Detection in Mammography: Comparison with 101 Radiologists. J. Natl. Cancer Inst. 2019, 111, 916–922. [Google Scholar] [CrossRef]
- Lee, J.; Nishikawa, R.M. Automated mammographic breast density estimation using a fully convolutional network. Med. Phys. 2018, 45, 1178–1190. [Google Scholar] [CrossRef]
- Ha, R.; Chang, P.; Karcich, J.; Mutasa, S.; Van Sant, E.P.; Liu, M.Z.; Jambawalikar, S. Convolutional Neural Network Based Breast Cancer Risk Stratification Using a Mammographic Dataset. Acad. Radiol. 2019, 26, 544–549. [Google Scholar] [CrossRef]
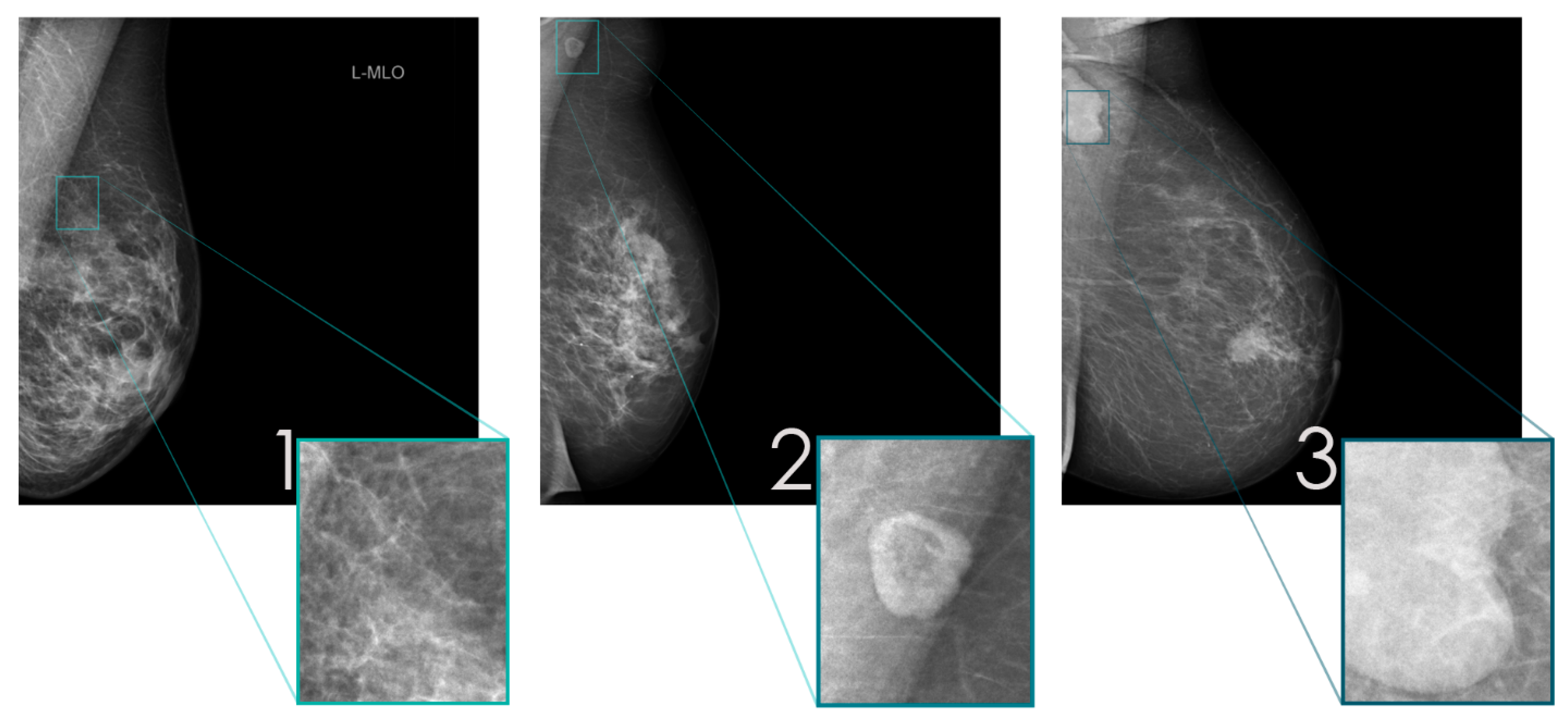
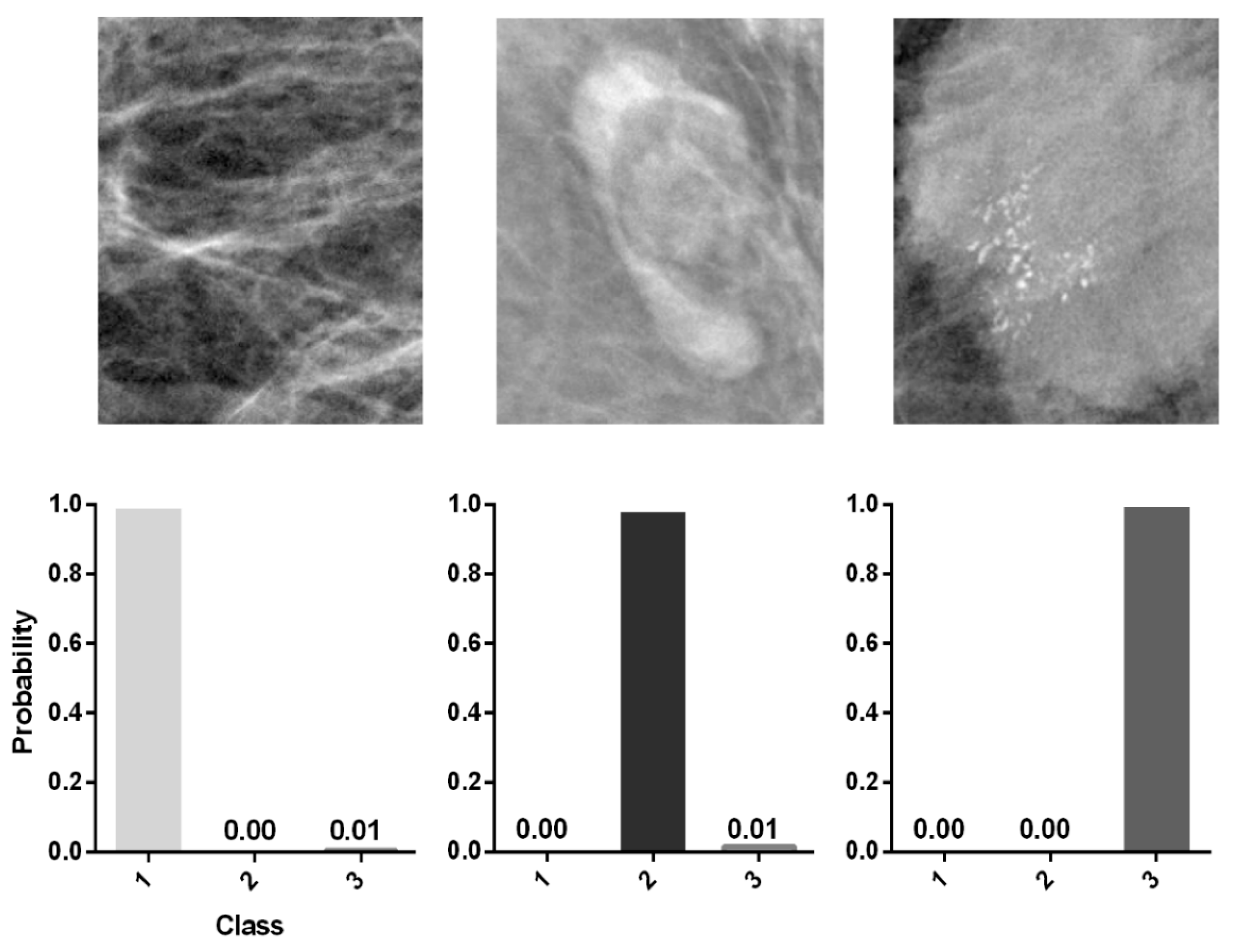


| Class | |||
|---|---|---|---|
| 1 | 2 | 3 | |
| Training data | 567 | 533 | 377 |
| Augmented | 2062 | 1926 | 1397 |
| dCNN | Reader 1 | Reader 2 | |
|---|---|---|---|
| 1 | 20 | 20 | 21 |
| 2 | 19 | 20 | 20 |
| 3 | 21 | 20 | 19 |
| Ground Truth | dCNN | Reader 1 | Reader 2 | |
|---|---|---|---|---|
| Ground Truth | 0.97 | 1 | 0.95 | |
| dCNN | 0.97 | 0.93 | ||
| Reader 1 | 0.95 | |||
| Reader 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abel, F.; Landsmann, A.; Hejduk, P.; Ruppert, C.; Borkowski, K.; Ciritsis, A.; Rossi, C.; Boss, A. Detecting Abnormal Axillary Lymph Nodes on Mammograms Using a Deep Convolutional Neural Network. Diagnostics 2022, 12, 1347. https://doi.org/10.3390/diagnostics12061347
Abel F, Landsmann A, Hejduk P, Ruppert C, Borkowski K, Ciritsis A, Rossi C, Boss A. Detecting Abnormal Axillary Lymph Nodes on Mammograms Using a Deep Convolutional Neural Network. Diagnostics. 2022; 12(6):1347. https://doi.org/10.3390/diagnostics12061347
Chicago/Turabian StyleAbel, Frederik, Anna Landsmann, Patryk Hejduk, Carlotta Ruppert, Karol Borkowski, Alexander Ciritsis, Cristina Rossi, and Andreas Boss. 2022. "Detecting Abnormal Axillary Lymph Nodes on Mammograms Using a Deep Convolutional Neural Network" Diagnostics 12, no. 6: 1347. https://doi.org/10.3390/diagnostics12061347
APA StyleAbel, F., Landsmann, A., Hejduk, P., Ruppert, C., Borkowski, K., Ciritsis, A., Rossi, C., & Boss, A. (2022). Detecting Abnormal Axillary Lymph Nodes on Mammograms Using a Deep Convolutional Neural Network. Diagnostics, 12(6), 1347. https://doi.org/10.3390/diagnostics12061347






