Correlation Analysis of Anti-SARS-CoV-2 RBD IgG and Neutralizing Antibody against SARS-CoV-2 Omicron Variants after Vaccination
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Anti-Receptor Binding Domain (RBD) IgG Assay
2.3. Neutralizing Antibody (NAb) Assay
2.4. Statistical Analysis
3. Results
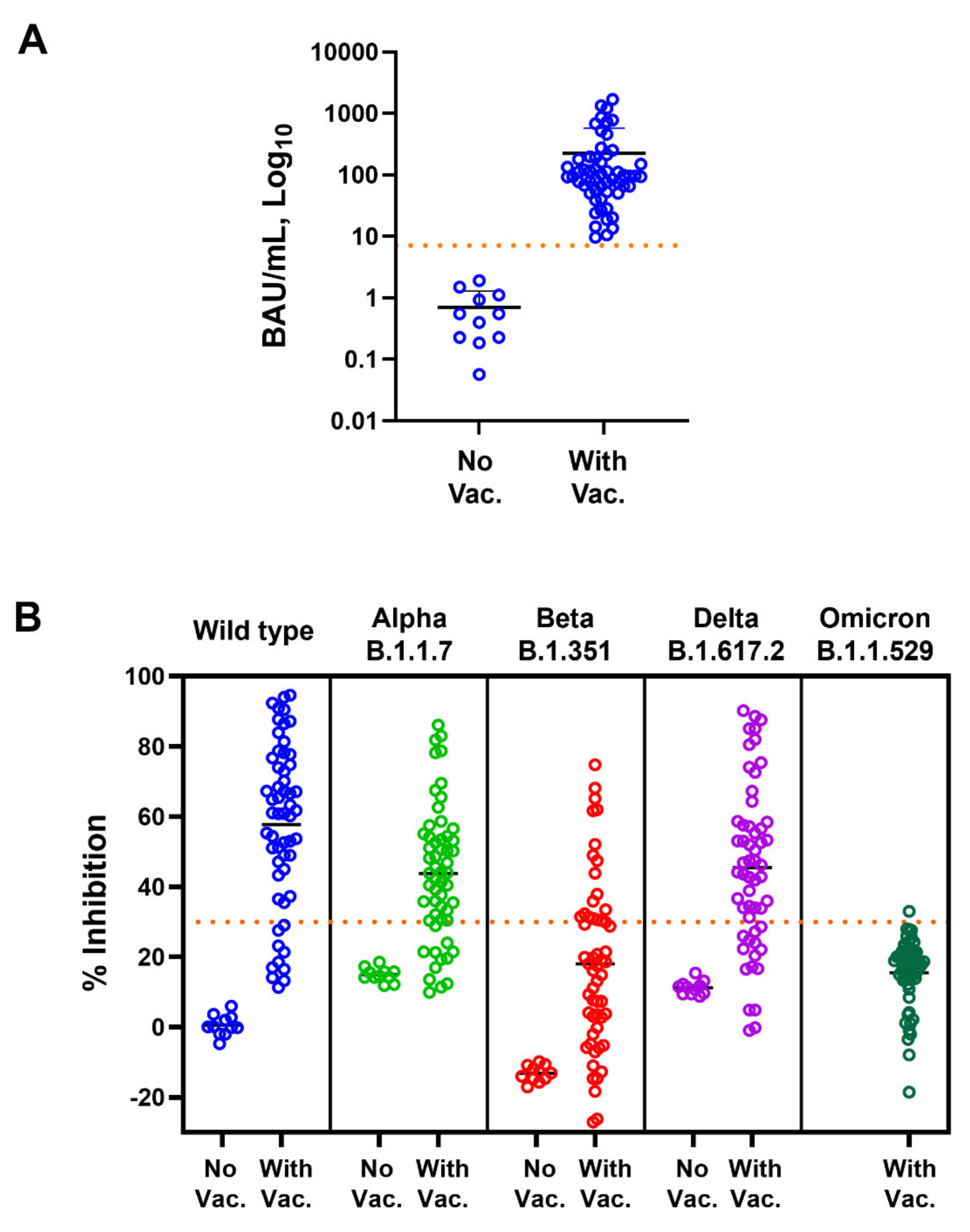
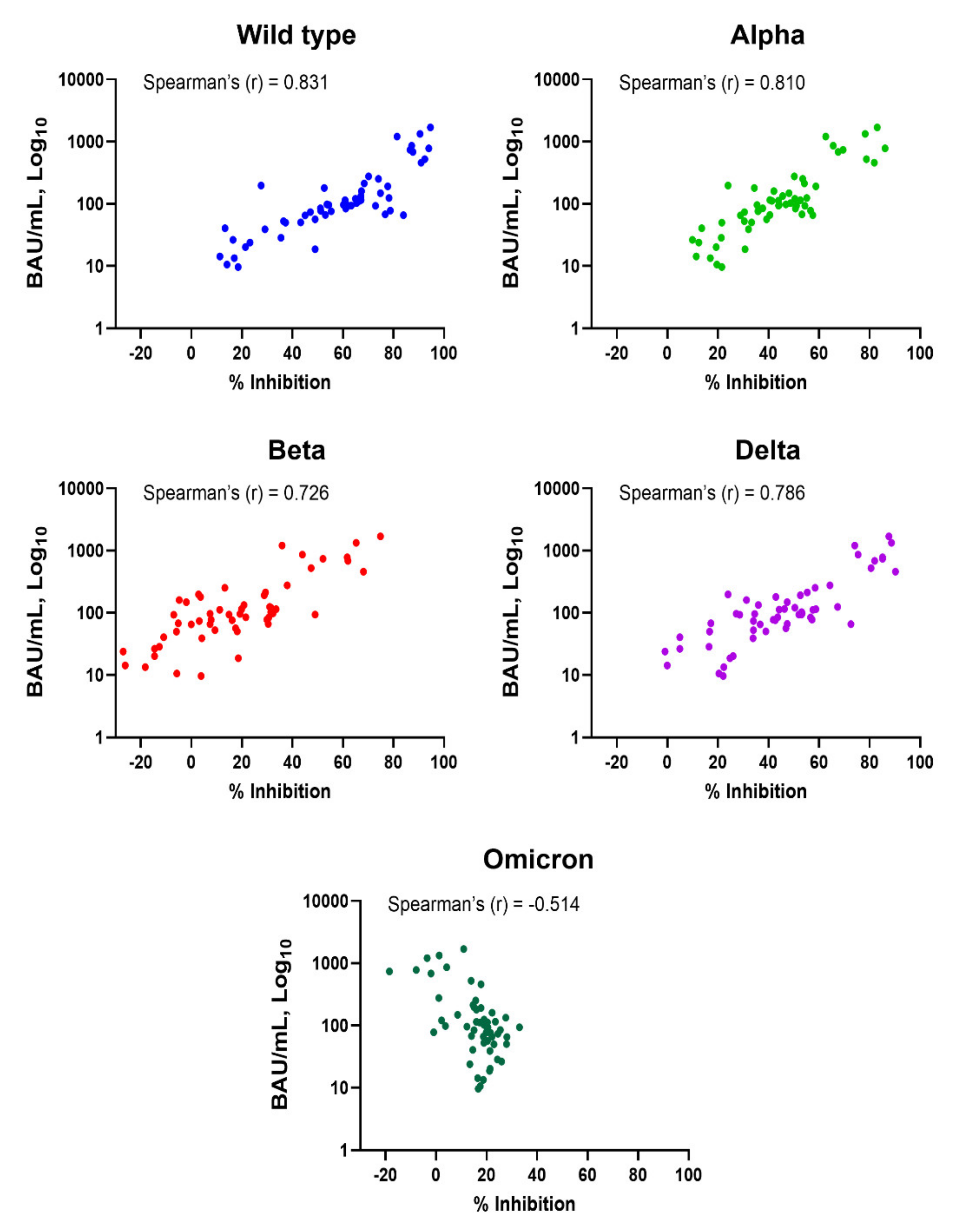
3.1. Correlation of Anti-SARS-CoV-2 RBD IgG and Neutralizing Antibodies against SARS-CoV-2 Variants
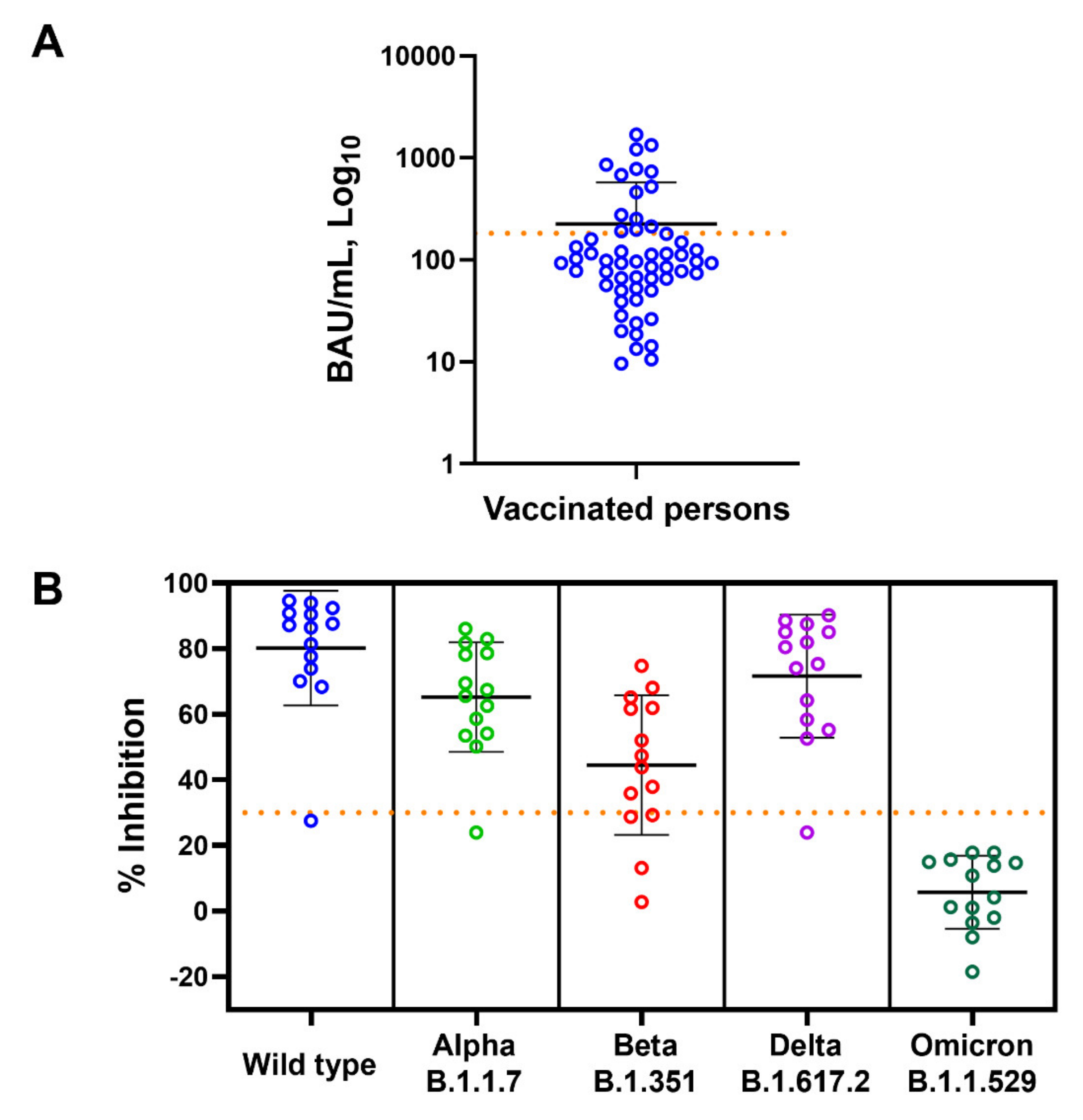
3.2. Vaccine Induced Anti-RBD IgG Response but Not Neutralizing Antibody against SARS-CoV-2 Omicron Variant
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nagy, A.; Alhatlani, B. An overview of current COVID-19 vaccine platforms. Comput. Struct. Biotechnol. J. 2021, 19, 2508–2517. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Vaccine Tracker and Landscape. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 11 April 2022).
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cardenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against COVID-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef] [PubMed]
- Tregoning, J.S.; Flight, K.E.; Higham, S.L.; Wang, Z.; Pierce, B.F. Progress of the COVID-19 vaccine effort: Viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021, 21, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, A.; Robertson, C.; Taylor, B. BNT162b2 and ChAdOx1 nCoV-19 Vaccine Effectiveness against Death from the Delta Variant. N. Engl. J. Med. 2021, 385, 2195–2197. [Google Scholar] [CrossRef] [PubMed]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Addetia, A.; Crawford, K.H.D.; Dingens, A.; Zhu, H.; Roychoudhury, P.; Huang, M.L.; Jerome, K.R.; Bloom, J.D.; Greninger, A.L. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J. Clin. Microbiol. 2020, 58, e02107-20. [Google Scholar] [CrossRef]
- McMahan, K.; Yu, J.; Mercado, N.B.; Loos, C.; Tostanoski, L.H.; Chandrashekar, A.; Liu, J.; Peter, L.; Atyeo, C.; Zhu, A.; et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 2021, 590, 630–634. [Google Scholar] [CrossRef]
- Jiang, Y.; Wu, Q.; Song, P.; You, C. The Variation of SARS-CoV-2 and Advanced Research on Current Vaccines. Front. Med. 2021, 8, 806641. [Google Scholar] [CrossRef]
- Khateeb, J.; Li, Y.; Zhang, H. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Crit. Care 2021, 25, 244. [Google Scholar] [CrossRef]
- Tian, D.; Sun, Y.; Xu, H.; Ye, Q. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant. J. Med. Virol. 2022, 94, 2376–2383. [Google Scholar] [CrossRef] [PubMed]
- Waldman, S.E.; Buehring, T.; Escobar, D.J.; Gohil, S.K.; Gonzales, R.; Huang, S.S.; Olenslager, K.; Prabaker, K.K.; Sandoval, T.; Yim, J.; et al. Secondary Cases of Delta-Variant COVID-19 among Vaccinated Healthcare Workers with Breakthrough Infections is Rare. Clin. Infect. Dis. 2021, ciab916. [Google Scholar] [CrossRef] [PubMed]
- Chau, N.V.V.; Ngoc, N.M.; Nguyet, L.A.; Quang, V.M.; Ny, N.T.H.; Khoa, D.B.; Phong, N.T.; Toan, L.M.; Hong, N.T.T.; Tuyen, N.T.K.; et al. An observational study of breakthrough SARS-CoV-2 Delta variant infections among vaccinated healthcare workers in Vietnam. EClinicalMedicine 2021, 41, 101143. [Google Scholar] [CrossRef] [PubMed]
- Perez-Then, E.; Lucas, C.; Monteiro, V.S.; Miric, M.; Brache, V.; Cochon, L.; Vogels, C.B.F.; Malik, A.A.; De la Cruz, E.; Jorge, A.; et al. Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat. Med. 2022, 28, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.M.S.; Mok, C.K.P.; Leung, Y.W.Y.; Ng, S.S.; Chan, K.C.K.; Ko, F.W.; Chen, C.; Yiu, K.; Lam, B.H.S.; Lau, E.H.Y.; et al. Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat. Med. 2022, 28, 486–489. [Google Scholar] [CrossRef]
- Muik, A.; Lui, B.G.; Wallisch, A.K.; Bacher, M.; Muhl, J.; Reinholz, J.; Ozhelvaci, O.; Beckmann, N.; Guimil Garcia, R.C.; Poran, A.; et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science 2022, 375, 678–680. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Huo, J.; Zhou, D.; Zahradnik, J.; Supasa, P.; Liu, C.; Duyvesteyn, H.M.E.; Ginn, H.M.; Mentzer, A.J.; Tuekprakhon, A.; et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022, 185, 467–484.e15. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, P.; Wang, N.; Wang, L.; Fan, K.; Zhu, Q.; Wang, K.; Chen, R.; Feng, R.; Jia, Z.; et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell 2022, 185, 860–871.e13. [Google Scholar] [CrossRef]
- Enhancing Response to Omicron SARS-CoV-2 Variant. Available online: https://www.who.int/publications/m/item/enhancing-readiness-for-omicron-(b.1.1.529)-technical-brief-and-priority-actions-for-member-states (accessed on 11 April 2022).
- Premkumar, L.; Segovia-Chumbez, B.; Jadi, R.; Martinez, D.R.; Raut, R.; Markmann, A.; Cornaby, C.; Bartelt, L.; Weiss, S.; Park, Y.; et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020, 5, eabc8413. [Google Scholar] [CrossRef]
- US Food and Drug Administration. Convalescent Plasma EUA Letter of Authorization. 28 December 2021. Available online: https://www.fda.gov/media/141477/download (accessed on 11 April 2022).
- Heinz, F.X.; Stiasny, K. Distinguishing features of current COVID-19 vaccines: Knowns and unknowns of antigen presentation and modes of action. NPJ Vaccines 2021, 6, 104. [Google Scholar] [CrossRef]
- Lombardi, A.; Bozzi, G.; Ungaro, R.; Villa, S.; Castelli, V.; Mangioni, D.; Muscatello, A.; Gori, A.; Bandera, A. Mini Review Immunological Consequences of Immunization with COVID-19 mRNA Vaccines: Preliminary Results. Front. Immunol. 2021, 12, 657711. [Google Scholar] [CrossRef] [PubMed]
- Guiomar, R.; Santos, A.J.; Melo, A.M.; Costa, I.; Matos, R.; Rodrigues, A.P.; Kislaya, I.; Silva, A.S.; Roque, C.; Silva, C.; et al. High correlation between binding IgG (anti-RBD/S) and neutralizing antibodies against SARS-CoV-2 six months after vaccination. medRxiv 2021. [Google Scholar] [CrossRef]
- Bosch, W.; Cowart, J.B.; Bhakta, S.; Carter, R.E.; Wadei, H.M.; Shah, S.Z.; Sanghavi, D.K.; Pollock, B.D.; Neville, M.R.; Oman, S.P.; et al. COVID-19 Vaccine-Breakthrough Infections Requiring Hospitalization in Mayo Clinic Florida through August 2021. Clin. Infect. Dis. 2021, ciab932. [Google Scholar] [CrossRef]
- Hirabara, S.M.; Serdan, T.D.A.; Gorjao, R.; Masi, L.N.; Pithon-Curi, T.C.; Covas, D.T.; Curi, R.; Durigon, E.L. SARS-CoV-2 Variants: Differences and Potential of Immune Evasion. Front. Cell. Infect. Microbiol. 2021, 11, 781429. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. Heavily mutated Omicron variant puts scientists on alert. Nature 2021, 600, 21. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Lu, L.; Choi, C.Y.; Cai, J.P.; Tsoi, H.W.; Chu, A.W.; Ip, J.D.; Chan, W.M.; Zhang, R.R.; Zhang, X.; et al. Impact of SARS-CoV-2 variant-associated RBD mutations on the susceptibility to serum antibodies elicited by COVID-19 infection or vaccination. Clin. Infect. Dis. 2021, ciab656. [Google Scholar] [CrossRef]
| SARS-CoV-2 Variants | Spearman®(r) | p-Value |
|---|---|---|
| Wild-type | 0.831 | <0.001 |
| Alpha | 0.810 | <0.001 |
| Beta | 0.726 | <0.001 |
| Delta | 0.786 | <0.001 |
| Omicron | −0.514 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takheaw, N.; Liwsrisakun, C.; Chaiwong, W.; Laopajon, W.; Pata, S.; Inchai, J.; Duangjit, P.; Pothirat, C.; Bumroongkit, C.; Deesomchok, A.; et al. Correlation Analysis of Anti-SARS-CoV-2 RBD IgG and Neutralizing Antibody against SARS-CoV-2 Omicron Variants after Vaccination. Diagnostics 2022, 12, 1315. https://doi.org/10.3390/diagnostics12061315
Takheaw N, Liwsrisakun C, Chaiwong W, Laopajon W, Pata S, Inchai J, Duangjit P, Pothirat C, Bumroongkit C, Deesomchok A, et al. Correlation Analysis of Anti-SARS-CoV-2 RBD IgG and Neutralizing Antibody against SARS-CoV-2 Omicron Variants after Vaccination. Diagnostics. 2022; 12(6):1315. https://doi.org/10.3390/diagnostics12061315
Chicago/Turabian StyleTakheaw, Nuchjira, Chalerm Liwsrisakun, Warawut Chaiwong, Witida Laopajon, Supansa Pata, Juthamas Inchai, Pilaiporn Duangjit, Chaicharn Pothirat, Chaiwat Bumroongkit, Athavudh Deesomchok, and et al. 2022. "Correlation Analysis of Anti-SARS-CoV-2 RBD IgG and Neutralizing Antibody against SARS-CoV-2 Omicron Variants after Vaccination" Diagnostics 12, no. 6: 1315. https://doi.org/10.3390/diagnostics12061315
APA StyleTakheaw, N., Liwsrisakun, C., Chaiwong, W., Laopajon, W., Pata, S., Inchai, J., Duangjit, P., Pothirat, C., Bumroongkit, C., Deesomchok, A., Theerakittikul, T., Limsukon, A., Tajarernmuang, P., Niyatiwatchanchai, N., Trongtrakul, K., & Kasinrerk, W. (2022). Correlation Analysis of Anti-SARS-CoV-2 RBD IgG and Neutralizing Antibody against SARS-CoV-2 Omicron Variants after Vaccination. Diagnostics, 12(6), 1315. https://doi.org/10.3390/diagnostics12061315






