Clinical Validation of a Rapid Variant-Proof RT-RPA Assay for the Detection of SARS-CoV-2
Abstract
:1. Introduction
2. Methods
2.1. Clinical Samples
2.2. Real-Time RT-PCR
2.3. Sequencing
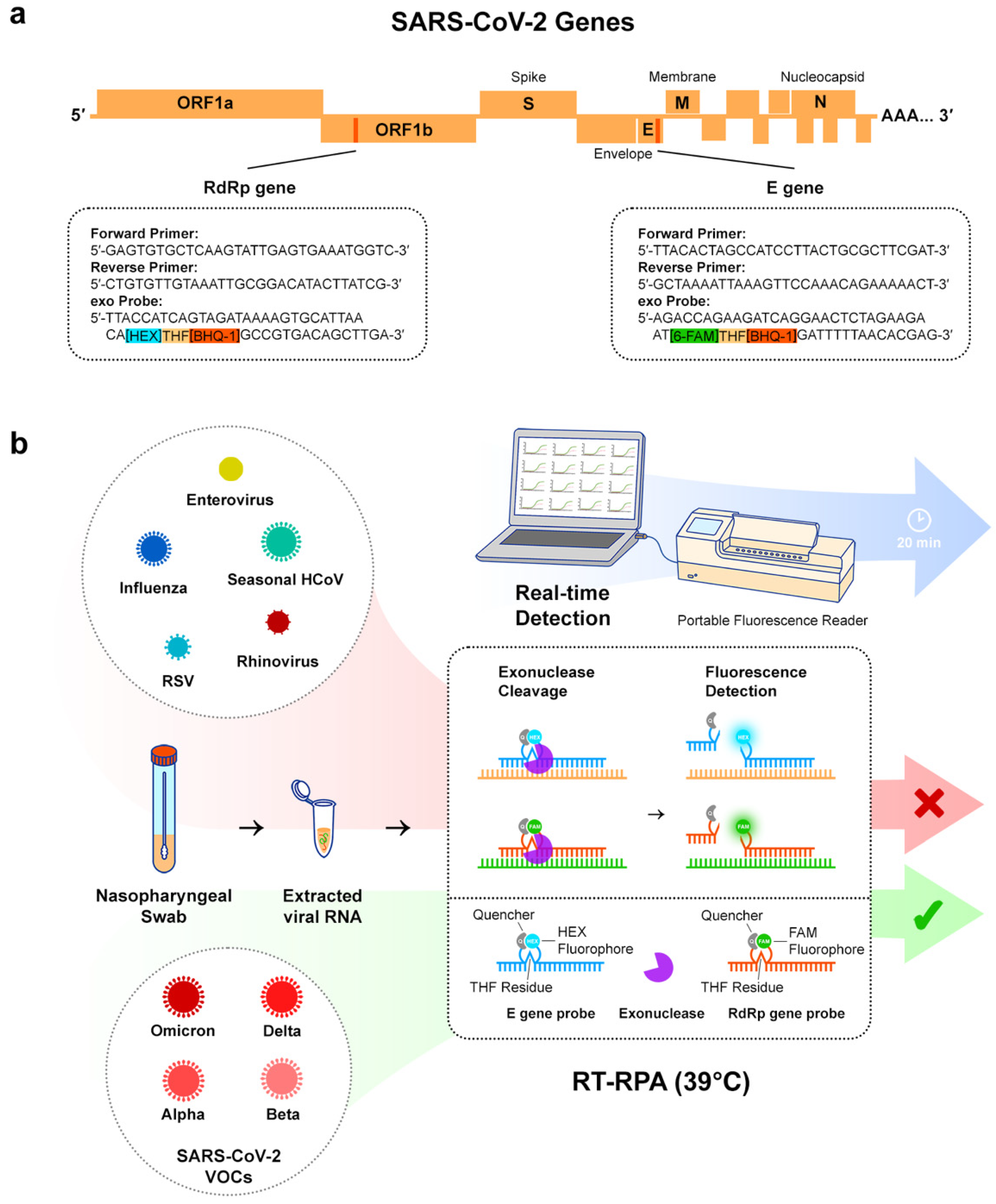
2.4. Multi-Gene RT-RPA
2.5. Correlation between Cq and TT
3. Results
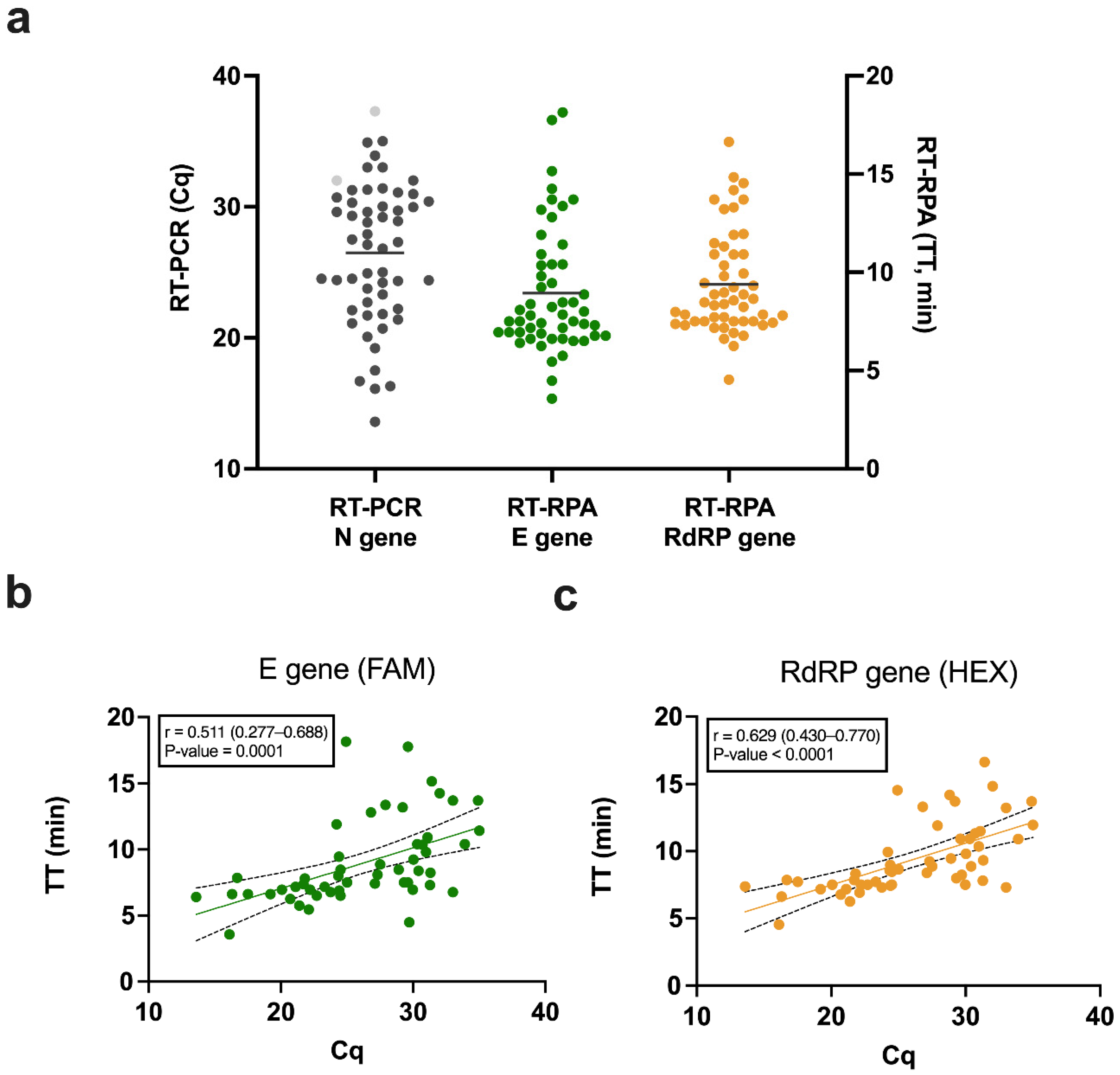
3.1. Clinical Sensitivity and Specificity
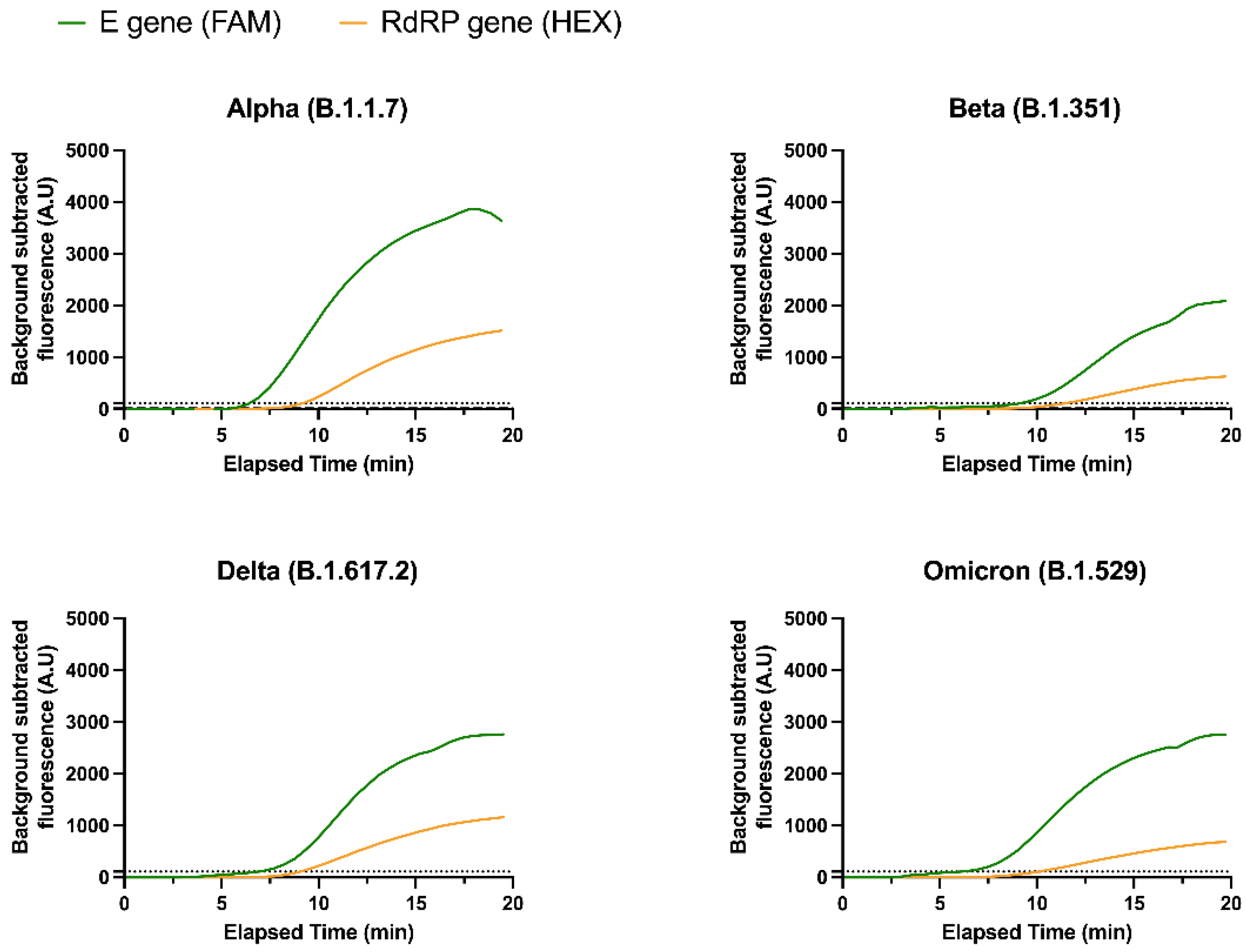
3.2. Detection of SARS-CoV-2 Variants
3.3. Correlation between Cq and TT
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 15 May 2022).
- Vandenberg, O.; Martiny, D.; Rochas, O.; van Belkum, A.; Kozlakidis, Z. Considerations for Diagnostic COVID-19 Tests. Nat. Rev. Microbiol. 2020, 19, 171–183. [Google Scholar] [CrossRef] [PubMed]
- The Foundation for Innovative New Diagnostics (FIND). SARS-CoV-2 Test Tracker. Available online: https://www.finddx.org/covid-19/test-tracker/ (accessed on 15 May 2022).
- Skegg, D.; Gluckman, P.; Boulton, G.; Hackmann, H.; Karim, S.S.A.; Piot, P.; Woopen, C. Future Scenarios for the COVID-19 Pandemic. Lancet 2021, 397, 777–778. [Google Scholar] [CrossRef]
- World Health Organization. COVAX: Working for Global Equitable Access to COVID-19 Vaccines. Available online: https://www.who.int/initiatives/act-accelerator/covax (accessed on 15 May 2022).
- World Health Organization. The Access to COVID-19 Tools (ACT) Accelerator. Available online: https://www.who.int/initiatives/act-accelerator (accessed on 15 May 2022).
- World Health Organization. Recommendations for National SARS-CoV-2 Testing Strategies and Diagnostic Capacities; Interim Guidance; 25 June 2021; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Bustin, S.A.; Nolan, T. RT-QPCR Testing of SARS-CoV-2: A Primer. Int. J. Mol. Sci. 2020, 21, 3004. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, Y.; Orihara, Y.; Kawamura, R.; Imai, K.; Sakai, J.; Tarumoto, N.; Matsuoka, M.; Takeuchi, S.; Maesaki, S.; Maeda, T. Evaluation of Rapid Diagnosis of Novel Coronavirus Disease (COVID-19) Using Loop-Mediated Isothermal Amplification. J. Clin. Virol. 2020, 129, 104446. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.D.; Poirier, E.Z.; Cardoso, A.; Frederico, B.; Canton, J.; Barrell, S.; Beale, R.; Byrne, R.; Caidan, S.; Crawford, M.; et al. SARS-CoV-2 Detection by a Clinical Diagnostic RT-LAMP Assay. Wellcome Open Res. 2021, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- El Wahed, A.A.; Patel, P.; Maier, M.; Pietsch, C.; Rüster, D.; Böhlken-Fascher, S.; Kissenkötter, J.; Behrmann, O.; Frimpong, M.; Diagne, M.M.; et al. Suitcase Lab for Rapid Detection of SARS-CoV-2 Based on Recombinase Polymerase Amplification Assay. Anal. Chem. 2021, 93, 2627–2634. [Google Scholar] [CrossRef]
- Lau, Y.L.; Ismail, I.B.; Mustapa, N.I.B.; Lai, M.Y.; Tuan Soh, T.S.; Haji Hassan, A.; Peariasamy, K.M.; Lee, Y.L.; Abdul Kahar, M.K.B.; Chong, J.; et al. Development of a Reverse Transcription Recombinase Polymerase Amplification Assay for Rapid and Direct Visual Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). PLoS ONE 2021, 16, e0245164. [Google Scholar]
- Shelite, T.R.; Uscanga-Palomeque, A.C.; Castellanos-Gonzalez, A.; Melby, P.C.; Travi, B.L. Isothermal Recombinase Polymerase Amplification-Lateral Flow Detection of SARS-CoV-2, the Etiological Agent of COVID-19. J. Virol. Methods 2021, 296, 114227. [Google Scholar] [CrossRef]
- Behrmann, O.; Bachmann, I.; Spiegel, M.; Schramm, M.; Abd El Wahed, A.; Dobler, G.; Dame, G.; Hufert, F.T.; Theodor Fontane, B. Rapid Detection of SARS-CoV-2 by Low Volume Real-Time Single Tube Reverse Transcription Recombinase Polymerase Amplification Using an Exo Probe with an Internally Linked Quencher (Exo-IQ). Clin. Chem. 2020, 66, 1047–1054. [Google Scholar] [CrossRef]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical Validation of a Cas13-Based Assay for the Detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef]
- Cherkaoui, D.; Huang, D.; Miller, B.S.; Turbé, V.; McKendry, R.A. Harnessing Recombinase Polymerase Amplification for Rapid Multi-Gene Detection of SARS-CoV-2 in Resource-Limited Settings. Biosens. Bioelectron. 2021, 189, 113328. [Google Scholar] [CrossRef] [PubMed]
- University of Liverpool. Liverpool COVID-19 Community Testing Pilot; Interim Evaluation Report, 23 December 2020; University of Liverpool: Liverpool, UK, 2020. [Google Scholar]
- Petersen, I.; Crozier, A.; Buchan, I.; Mina, M.; Bartlett, J.W. Recalibrating SARS-CoV-2 Antigen Rapid Lateral Flow Test Relative Sensitivity from Validation Studies to Absolute Sensitivity for Detecting Individuals with Live Virus. Clin. Epidemiol. 2021, 13, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J.; von Stetten, F. Review: A Comprehensive Summary of a Decade Development of the Recombinase Polymerase Amplification. Analyst 2019, 144, 31–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabaan, A.A.; Tirupathi, R.; Sule, A.A.; Aldali, J.; Mutair, A.A.; Alhumaid, S.; Gupta, N.; Koritala, T.; Adhikari, R.; Bilal, M.; et al. Viral dynamics and real-time RT-PCR Ct values correlation with disease severity in COVID-19. Diagnostics 2021, 11, 1091. [Google Scholar] [CrossRef]
- Kawasuji, H.; Takegoshi, Y.; Kaneda, M.; Ueno, A.; Miyajima, Y.; Kawago, K.; Fukui, Y.; Yoshida, Y.; Kimura, M.; Yamada, H.; et al. Transmissibility of COVID-19 Depends on the Viral Load around Onset in Adult and Symptomatic Patients. PLoS ONE 2020, 15, e0243597. [Google Scholar] [CrossRef]
- He, X.; Lau, E.H.Y.; Wu, P.; Deng, X.; Wang, J.; Hao, X.; Lau, Y.C.; Wong, J.Y.; Guan, Y.; Tan, X.; et al. Temporal Dynamics in Viral Shedding and Transmissibility of COVID-19. Nat. Med. 2020, 26, 672–675. [Google Scholar] [CrossRef] [Green Version]
- Frampton, D.; Rampling, T.; Cross, A.; Bailey, H.; Heaney, J.; Byott, M.; Scott, R.; Sconza, R.; Price, J.; Margaritis, M.; et al. Genomic Characteristics and Clinical Effect of the Emergent SARS-CoV-2 B.1.1.7 Lineage in London, UK: A Whole-Genome Sequencing and Hospital-Based Cohort Study. Lancet Infect. Dis. 2021, 21, 1246–1256. [Google Scholar] [CrossRef]
- Pujadas, E.; Chaudhry, F.; McBride, R.; Richter, F.; Zhao, S.; Wajnberg, A.; Nadkarni, G.; Glicksberg, B.S.; Houldsworth, J.; Cordon-Cardo, C. SARS-CoV-2 Viral Load Predicts COVID-19 Mortality. Lancet Respir. Med. 2020, 8, e70. [Google Scholar] [CrossRef]
- Yu, F.; Yan, L.; Wang, N.; Yang, S.; Wang, L.; Tang, Y.; Gao, G.; Wang, S.; Ma, C.; Xie, R.; et al. Quantitative Detection and Viral Load Analysis of SARS-CoV-2 in Infected Patients. Clin. Infect. Dis. 2020, 71, 793–798. [Google Scholar] [CrossRef] [Green Version]
- U.S. Food and Drug Administration. SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests. Available online: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests (accessed on 15 May 2022).
- World Health Organization. Tracking SARS-CoV-2 Variants; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Nexstrain. Genomic Epidemiology of Novel Coronavirus—Global Subsampling. Available online: https://nextstrain.org/ncov/gisaid/global (accessed on 28 January 2022).
- Onwuamah, C.K.; Kanteh, A.; Abimbola, B.S.; Ahmed, R.A.; Okoli, C.L.; Shaibu, J.O.; James, A.B.; Ajibaye, O.; Okwurauwe, A.P.; Fowora, M.; et al. SARS-CoV-2 sequencing collaboration in west Africa shows best practices. Lancet Glob. Health 2021, 9, e1499–e1500. [Google Scholar] [CrossRef]
- Grant, P.R.; Turner, M.A.; Yen Shin, G.; Nastouli, E.; Levett, L.J. Extraction-Free COVID-19 (SARS-CoV-2) Diagnosis by RT-PCR to Increase Capacity for National Testing Programmes during a Pandemic. BioRxiv 2020. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. 2019-Novel Coronavirus (2019-nCoV) Real-Time rRT-PCR Panel Primers and Probes. Available online: https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-primer-probes.pdf (accessed on 15 May 2022).
- Quick, J.; Loman, N.; ARTIC Consortium. hCoV-2019/nCoV-2019 Version 3 Amplicon Set. Available online: https://artic.network/resources/ncov/ncov-amplicon-v3.pdf (accessed on 28 January 2022).
- Artic Network. Artic-nCoV2019. Available online: https://github.com/artic-network/artic-ncov2019 (accessed on 28 January 2022).
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef] [PubMed]
- COG-UK. COG UK Mutation Explorer. Available online: https://sars2.cvr.gla.ac.uk/cog-uk/ (accessed on 28 January 2022).
- World Health Organization. COVID-19 Target Product Profiles for Priority Diagnostics to Support Response to the COVID-19 Pandemic v.1.0. Available online: https://www.who.int/publications/m/item/covid-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-covid-19-pandemic-v.0.1 (accessed on 28 January 2022).
- Dhanasekaran, S.; Doherty, T.M.; Kenneth, J. Comparison of Different Standards for Real-Time PCR-Based Absolute Quantification. J. Immunol. Methods 2010, 354, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Marks, M.; Millat-Martinez, P.; Ouchi, D.; Roberts, C.H.; Alemany, A.; Corbacho-Monné, M.; Ubals, M.; Tobias, A.; Tebé, C.; Ballana, E.; et al. Transmission of COVID-19 in 282 Clusters in Catalonia, Spain: A Cohort Study. Lancet Infect. Dis. 2021, 21, 629–636. [Google Scholar] [CrossRef]
- World Health Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 15 May 2022).
| RT-PCR | ||||||
|---|---|---|---|---|---|---|
| Positive | Negative | |||||
| RT-RPA | Positive | 53 | 1 | N | Sensitivity (95% CI) | Specificity (95% CI) |
| Negative | 2 | 35 | 91 | 96% (86–99) | 97% (84–100) | |
| VOCs Detection | ||||
|---|---|---|---|---|
| VOC | Alpha (B.1.1.7) | Beta (B.1.351) | Delta (B.1.617.2) | Omicron (B.1.1.529) |
| Sensitivity (95% CI) | 87% (58–98) | 100% (31–100) | 100% (77–100) | 100% (69–100) |
| Number of samples | 15 | 3 | 14 | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cherkaoui, D.; Heaney, J.; Huang, D.; Byott, M.; Miller, B.S.; Nastouli, E.; McKendry, R.A. Clinical Validation of a Rapid Variant-Proof RT-RPA Assay for the Detection of SARS-CoV-2. Diagnostics 2022, 12, 1263. https://doi.org/10.3390/diagnostics12051263
Cherkaoui D, Heaney J, Huang D, Byott M, Miller BS, Nastouli E, McKendry RA. Clinical Validation of a Rapid Variant-Proof RT-RPA Assay for the Detection of SARS-CoV-2. Diagnostics. 2022; 12(5):1263. https://doi.org/10.3390/diagnostics12051263
Chicago/Turabian StyleCherkaoui, Dounia, Judith Heaney, Da Huang, Matthew Byott, Benjamin S. Miller, Eleni Nastouli, and Rachel A. McKendry. 2022. "Clinical Validation of a Rapid Variant-Proof RT-RPA Assay for the Detection of SARS-CoV-2" Diagnostics 12, no. 5: 1263. https://doi.org/10.3390/diagnostics12051263
APA StyleCherkaoui, D., Heaney, J., Huang, D., Byott, M., Miller, B. S., Nastouli, E., & McKendry, R. A. (2022). Clinical Validation of a Rapid Variant-Proof RT-RPA Assay for the Detection of SARS-CoV-2. Diagnostics, 12(5), 1263. https://doi.org/10.3390/diagnostics12051263







