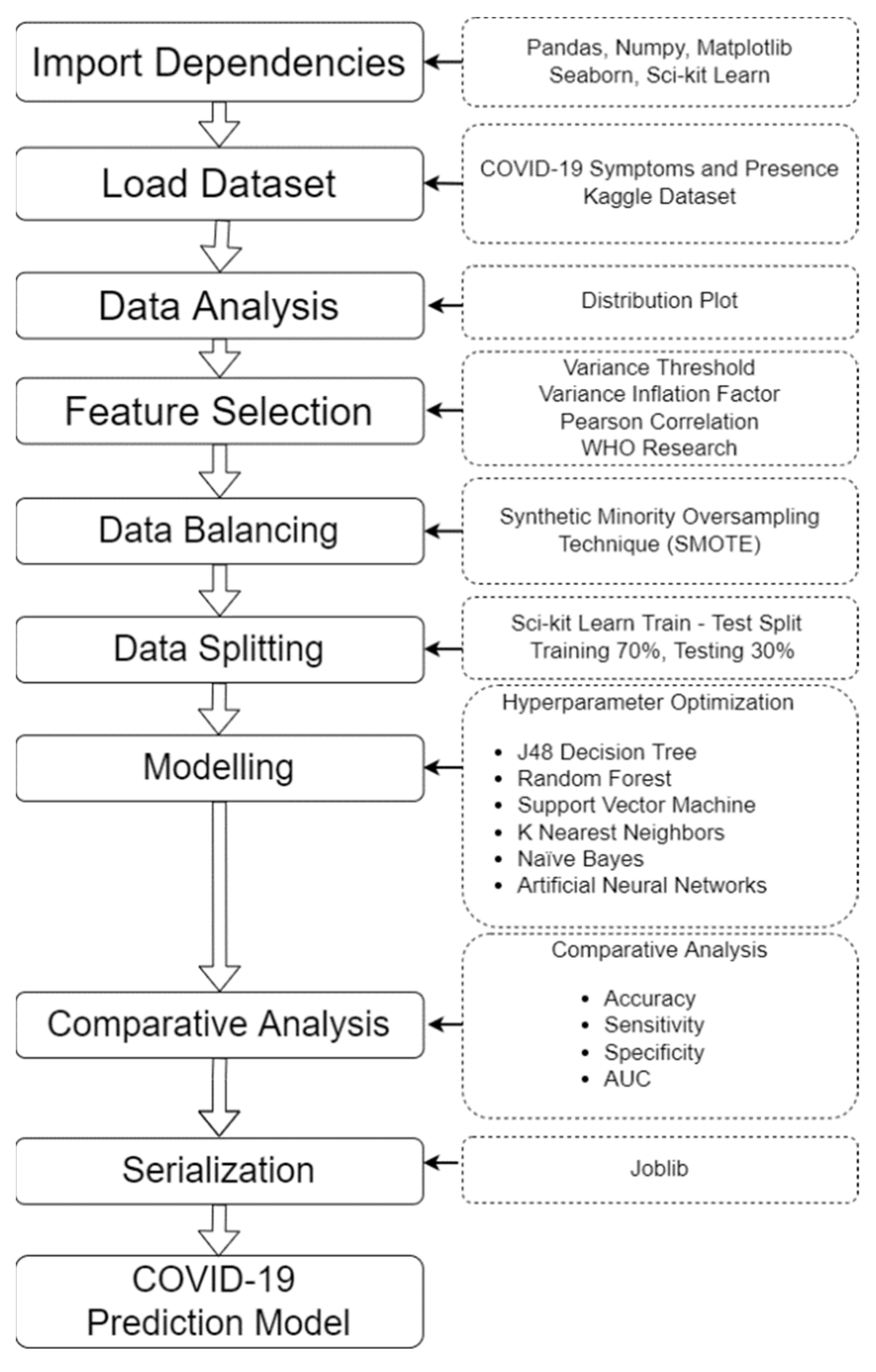
2.2. Data Analysis and Feature Selection
To be able to depict the distribution of data across the whole dataset, the displot function of the Seaborn package was used along with the Matplotlib. This function plots the data by a histogram combined with a line on it, representing the univariate distribution of a variable against the density distribution [
17]. The distplot of the collected dataset can be seen in
Figure 2.
In
Figure 2, 20 columns in the dataset were presented, which were referred to as the COVID-19-related symptoms, namely breathing problems, fever, dry cough, sore throat, runny nose, asthma, chronic lung disease, headache, heart disease, diabetes, hypertension, fatigue, gastrointestinal, abroad travel, contact with a COVID-19 patient, attended a large gathering, visited public areas, family working in public areas, wearing masks, and sanitization of things bought from the market. The vertical labels (
y-axis) represent the density of the samples in the dataset, while the horizontal labels (
x-axis) represent the classes wherein 0 or the indicated symptom is not present in the person, while number 1 means yes or currently being experienced by the person. Based on the distribution plot, a preliminary intuition can be drawn such as the majority of the samples in the dataset were having breathing problems, fever, dry cough, sore throat, runny nose, headache, fatigue, and visited public areas. Meanwhile, some samples in the dataset do not have asthma, chronic lung disease, heart disease, diabetes, hypertension, gastrointestinal, abroad travel history, attended a large gathering, and family working in public areas. Dataset samples have a slightly close density of having contact with COVID-19 patients, while wearing masks and sanitization from the market have only one content that is 0 or no.
We used the variance threshold to perform feature selection to remove the attribute values that do not have significant variance or have the same value for all samples [
18]. We used 80% as the threshold and found out that the attributes wearing masks and sanitization from the market exceeded the threshold, since the values were the same for all samples. These columns were removed from the dataset having only 18 features left.
Another feature selection method, which is the Pearson Correlation Coefficient (PCC), was applied in the dataset. This is a statistical measure to determine the correlation of two random attributes [
19]. The formula used in this method can be seen in Equation (1).
where
r is the Pearson correlation coefficient,
is the variable’s samples,
is the sample mean,
is the samples of another variable, and
is the value of its sample. The value of
r ranges from −1 to +1. PCC was applied to measure the correlation of the attributes to the target variable, which is the COVID-19 attribute and to know which features were positively and negatively correlated to the target class. By doing this, we can have intuition on what features must be retained in training the model, and the results are displayed in
Table 3.
In
Table 3, the correlation values of each feature toward the target variable were listed. The highest correlation goes to the symptom sore throat having 0.503 correlation, next is the Dry Cough with 0.464 correlation value, and the Abroad Travel and Breathing Problems have a 0.444 correlation value. Having a positive correlation means that the variables were positively correlated: as
x increases, the value of
y also increases, and vice versa. In contrast, the negative correlation means that the variables were negatively correlated: when the value of
x decreases, the value of
y increases, and vice versa [
20]. Symptoms that were found to be negatively correlated to the target variable were gastrointestinal, runny nose, headache, fatigue, and chronic lung disease. Features that have very low correlation to the COVID-19 variable were asthma, diabetes, and heart disease, which were mostly were termed as comorbidities.
After studying the correlation of the predictors to the target, we also took into consideration the collinearity. Collinearity happens when two predictors are linearly associated or having a high correlation to each other, and both were used as predictors of the target variable [
21]. Multicollinearity may also happen, which is a situation wherein the variable has collinearity with more than one predictors in the dataset. We used the Variance Inflation Factor (VIF) to detect the collinearity of the predictors in the dataset. The VIF starts from 1 to infinity, and the value of 1 means that the features were not correlated. VIF values less than 5 are moderately correlated, while VIF values of 10 and above are highly correlated and a cause of concern [
21]. The VIF values of each predictor in the dataset can be seen in
Table 4.
Table 4 displays the VIF of each predictor in the dataset. The highest is dry cough at 5.38, which is not surprising, since it is one of the most common symptoms of COVID-19 [
22]. Predictors such as fever, sore throat, and breathing problems attained the next highest VIF having scores lower than 5. A VIF of 1 to 5 means that the predictors were not correlated and can be considered in building the COVID-19 prediction model. However, we still consider including the dry cough as a predictor in the building of the COVID-19 prediction model even if it has a VIF of 5.38, since this symptom can contribute to predicting COVID-19 in a person, and it is included in the most common symptoms declared by the World Health Organization (WHO).
Aside from the variance threshold, PCC, and VIF, we also used the WHO website to determine the common symptoms of the disease because it has been validated by the experts in the medical field and is updated regularly.
Table 5 shows the most common, less common, and serious symptoms of COVID-19.
Table 5 displays the list of symptoms from the WHO’s website. According to the WHO, people who suffer from serious symptoms may cause danger to human lives, so it is necessary to go the nearest hospital to seek immediate medical attention. People who experience mild symptoms but are still healthy may manage themselves at home [
22]. It is necessary to undergo self-isolation immediately to prevent the virus from spreading and to prevent possible transmission until tested COVID-19 negative through a Polymerase Chain Reaction (PCR) test.
Based on the findings in the feature selection process, the feature combination that can be used in building the prediction model was devised, which is composed of the positive correlation features. Negatively correlated symptoms were also included but with respect to the symptoms declared by the WHO. The features that will be included in the training process were the sore throat, dry cough, abroad travel, breathing problems, attended a large gathering, contact with COVID-19 patient, fever, family working in public, visited public exposed places, hypertension, asthma, diabetes, heart disease, runny nose, headache, and fatigue.
2.4. Modeling of the COVID-19 Prediction Model Using Supervised Machine Learning Algorithms
The modeling phase of this study discusses the process of how to develop a COVID-19 prediction model including the hyperparameter optimization, training, testing and evaluation. After data processing using the variance threshold, PCC, VIF, feature selection techniques, and finally, the SMOTE data balancing technique, several models were built using the Google colab research utilizing different supervised machine learning algorithms, namely, J48 DT, RF, SVM, k-NN, NB, and ANN.
The first algorithm we included in this study is the DT, which is an algorithm that creates a tree-like plot wherein nodes represent a condition for any classification task. A generated tree starts with a root node that tests a given sample; an example of a test condition is whether the person has a dry cough or not. After testing the samples using a condition, branches will be generated that separate the samples into their respective classes. J48 DT is a kind of decision tree algorithm primarily utilized in classification tasks.
To attain the highest possible performance of the J48 DT algorithm, we performed hyperparameter tuning using GridSearchCV and 10-fold cross validation for all the experiments. The following hyperparameters were tuned: For the maximum depth of the decision tree, we used the values 2, 3, 5, 10, and 20. In terms of the minimum samples required to be in the leaf node, we used 5, 10, 20, 50, and 100. Lastly, to measure the information gain or the purity of the nodes of the decision tree, the criterion we used were the Gini index and entropy. An innovation of this algorithm was made using several decision trees known as random forest.
- 2.
Random Forest
The RF algorithm provides a significant amount of improvement in the classification accuracy of a model because it is capable of producing multiple decision trees. Each decision tree will generate a result about the sample, and the final result will be generated according to the results of the majority of the decision trees [
25]. To obtain the highest possible performance of the RF algorithm, we performed hyperparameter tuning using GridSearchCV and 10-fold cross validation for all the experiments. We also tuned same set of hyperparameters as what we tuned in the DT wherein the maximum depth values of the decision trees to be created are 2, 3, 5, 10, and 20, and the minimum samples required to be in the leaf node, we used 5, 10, 20, 50, and 100. To measure the quality of the split of each node, the criterion we used were the Gini index and entropy. Since the RF algorithm produces several decision trees, we used the number of estimators to set how many trees to be created in the forest. The values used for this hyperparameter were 100, 200, and 300. Each decision tree learns from random sets of samples from the dataset and using bootstrap means that the samples were drawn with replacement [
26]. The hyperparameter bootstrap was also tuned with the values either True or False; the entire dataset was used to build a decision tree if the bootstrap parameter was set to False. The next algorithm used in this study is the SVM.
- 3.
Support Vector Machine
Generating hyperplanes is a significant part of the SVM algorithm. Hyperplanes are utilized by SVM in separating the samples in the dataset according to their respective classes. SVM prioritizes in maximizing the distance of each group to the dividing hyperplane. Through the use of hyperplanes, we can minimize the errors in separating the instances. [
27]. The kernels used in the experiment were the linear, radial, polynomial, and sigmoid kernels; these kernels will help the SVM algorithm determine which are the best hyperplanes that can separate the dataset into COVID-19 positive and negative classes. A mathematical trick called the “Kernel trick” allows the SVM to create a higher dimensional space from a low-dimensional space dataset; specifically, a single-dimensional data will be converted into two-dimensional data according to their respective classes [
28].
A radial basis function (RBF) is one of the most popular among all the kernels, for it can be used to separate datasets that are not linearly separable by adding curves or bumps in the data points. Next is the polynomial kernel function, whose result depends on the direction of the two vectors, and it is the only kernel that has the hyperparameter named “degree”, which determines the flexibility of the decision boundary [
29]. The sigmoid kernel which is a quite popular kernel that originated in the neural networks field was also included. The selection of kernel to be used in the classifier depends on the kind of classification to be done; it is not fixed [
28]. In line with this, we included linear, radial, polynomial, and sigmoid kernels in the hyperparameter optimization process to find out which kernel will perform better.
The regularization parameter C, which is a hyperparameter common to all the kernels that regulates the misclassification of the samples and the simplicity of the decision boundaries, was tuned. The C values used in the hyperparameter optimization were 0, 0.01, 0.5, 0.1, 1, 2, 5, and 10. The gamma was also considered, which determines how much influence a training sample has. The values used for the gamma were 1, 0.1, 0.01, and 0.001. Large values of gamma means that the other examples are closer to be affected [
30]. Another supervised machine learning algorithm used is the k-NN, which uses a number of neighbors or samples surrounding a random sample in the dataset.
- 4.
K-Nearest Neighbors
k-NN uses a sample’s nearest neighbors in the dataset to determine to which class it belongs. k-NN is an old and simple supervised machine learning algorithm used to solve classification tasks [
31]. To determine a sample’s nearest neighbor, k-NN uses distance metrics, which are Euclidean and Manhattan distance. The most commonly used distance metric of k-NN is the Euclidean distance, which can be expressed in Equation (2).
where
x = (
a1,
a2,
a3…
an) is a sample in the dataset in a vector format, the number of the sample’s attributes is expressed as
n,
is the
rth attribute, the weight is expressed as
, and
and
are the two samples. Another distance metric used is the Manhattan distance. The Manhattan distance is also called “Taxicab Geometry” and “City Block Distance” [
32]. A Minkowski distance formula is used to find the Manhattan distance that works by calculating the distance between the samples in a grid-like path.
Hyperparameter optimization was used to determine the k-NN algorithm configuration that will yield the best possible performance of the model. The total number of neighbors used in the hyperparameter optimization were 3, 5, 7, 9, 11, and 13. The weights hyperparameters used were the uniform and distance. Uniform weights means that all the points in each neighborhood of the sample are weighted equally, while the distance weight allocates points by the inverse of their distance. The closer neighbors of the sample will have more influence than the other neighbors, which are more distant [
33]. The next supervised machine learning algorithm used in this study is the naïve Bayes which is based on Bayes’ theorem.
- 5.
Naïve Bayes
NB is another supervised machine learning algorithm based on the Bayes theorem by Thomas Bayes, an English mathematician, which was created in 1763 [
34]. The NB algorithm is called naïve because it does not depend on the existence of other parameters; its formula can be expressed in Equation (3).
In Equation (3), the priori probability is expressed as
P(
A), which means the probability of event
A happening. The marginal probability is expressed as
P(
B), which means the probability of the event
B happening. Then, the expression
P(
A|
B) means the posterior probability or probability of
A happening given that
B has occurred. The expression
P(
B|
A) means the likelihood probability, which is the probability of
B happening given that
A is true [
35]. Dividing the product of the likelihood and priori probability by the marginal probability will determine the posterior probability [
35].
We used the GausianNB, which is a special kind of NB algorithm, and tuned its only parameter, the variance smoothing. Variance smoothing is another form of Laplace smoothing, which is a technique that helps in the problem of zero probability in the NB. In using high values of alpha, it will push the likelihood toward a value of 0.5 [
36]. We used np.logspace(0, −9, num = 100) as the var_smothing value in the GridSearchCV hyperparameter tuning function.
- 6.
Artificial Neural Network
The last machine learning algorithm we used is the multilayer perceptron (MLP) artificial neural network, which is one of the most commonly used architectures of neural network because of its versatility in classification and regression problems [
37]. We plotted an illustration of MLP including the input layer, hidden layer, and the output layer; it can be seen in
Figure 5.
In
Figure 5, the first and the last layer are called the input layer and the output layer, respectively. The input layer introduces the MLP to the predictors given in the dataset. The output layer carries the final classification result as computed and processed by the hidden layers. The layers in between are called the hidden layers, where all the data processing and the classification of the predictors take place. Commonly, two hidden layers is enough to perform a classification task, but additional hidden layers are recommended to solve more complicated classifications and to discover deeper patterns from the predictors.
We tuned the following hyperparameters to attain the highest possible performance of the MLP ANN. As for the hidden layer sizes, first, there are 3 hidden layers with the size of 50, 50, 50; next, there are 3 hidden layers of 50, 100, and 50 sizes, and lastly, there is 1 hidden layer with the size of 100 neurons. For the activation function of the hidden layer, the “tanh” or hyperbolic tan function and relu rectified linear unit or “relu” were used. For weight optimization solver, we used “sgd” or the stochastic gradient descent. In addition, the solver adam was used, which is the default solver in the MLP classifier of sci-kit learn, which performs well in the training and validation of large datasets. Alpha values are 0.0001 and 0.05. Lastly, the learning rate is either constant or adaptive.
There are a few methods to fine-tune parameters, e.g., grid search, random search, and Bayesian optimization. Grid search requires defined values of the hyperparameters to be evaluated, which performs generally well especially in spot checking combinations [
38]. Random search defines a search space wherein random points will be evaluated; it performs great in discovering good hyperparameters combinations not particularly guessed by the developers; hence, it requires more time to execute [
38]. Lastly, the Bayesian optimization algorithm is used for more complex optimization problems such as global optimization that finds an input that determines the lowest or the highest cost of a given objective function [
39].
We aim to discover the best configuration of the supervised machine learning algorithms in developing the COVID-19 prediction model using defined values of the hyperparameters, and with that, the grid search method was selected. We make use of the GridSearchCV function from sklearn’s model_selection package. GridSearchCV will traverse through the defined hyperparameter values and use those configurations to fit the classifier onto the training set [
40]. The hyperparameter optimization will begin by creating a dictionary of the hyperparameters to be tuned including the desired values of it. Then, the GridSearchCV function will execute all the combinations of the given values and evaluate it using cross-validation. In this study, we used 10-fold cross-validation for all the experiments.
The results of this process were the accuracy or loss of each combination, and from there, we can choose the hyperparameter combination that will bring out the best possible performance of the algorithm for both the training and testing dataset [
40].