Noninvasive Classification of Glioma Subtypes Using Multiparametric MRI to Improve Deep Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Pathological Analysis
2.3. MRI Protocols
2.4. Dataset
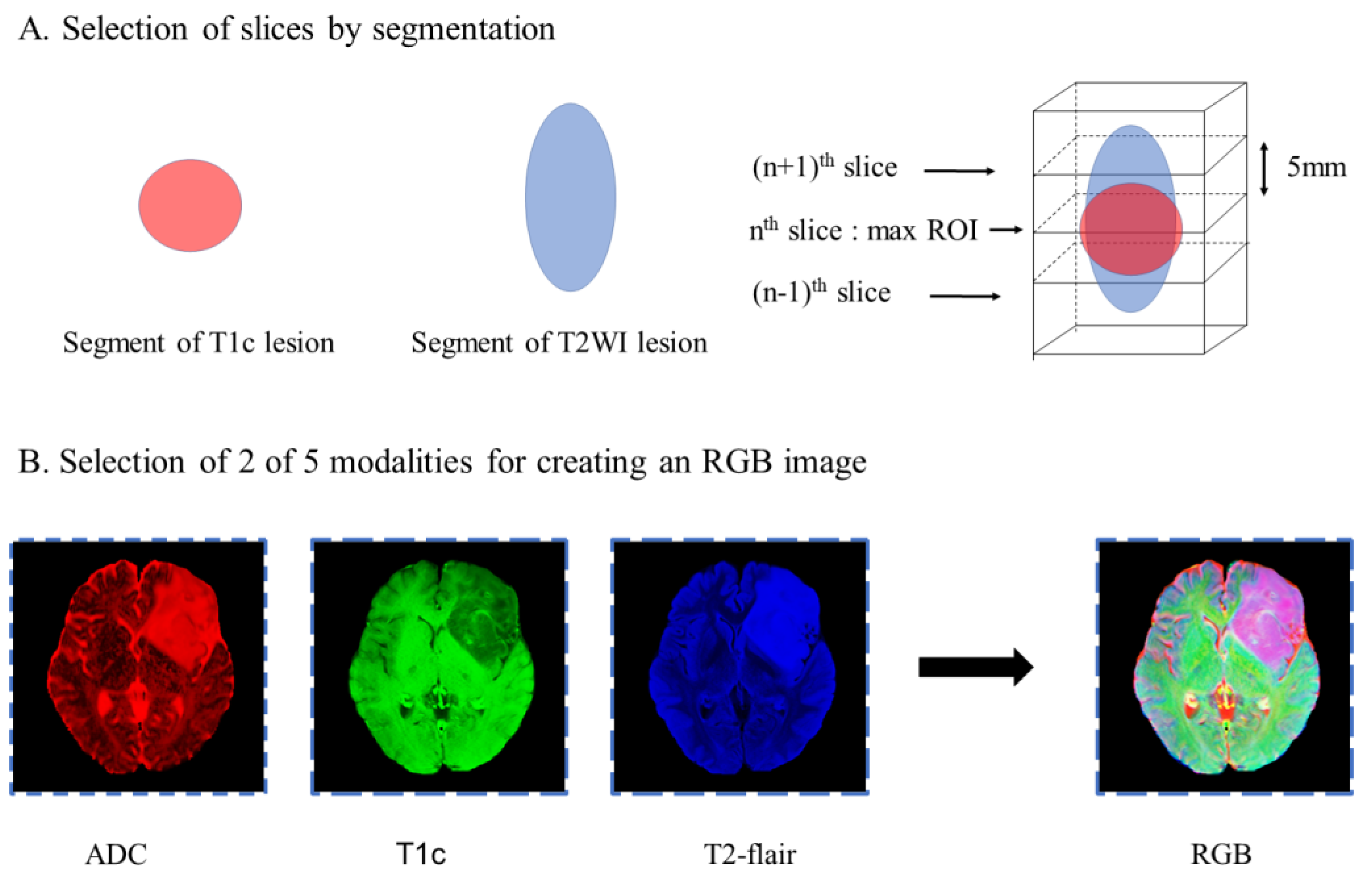
2.5. Image Postprocessing
2.6. Numeric Data
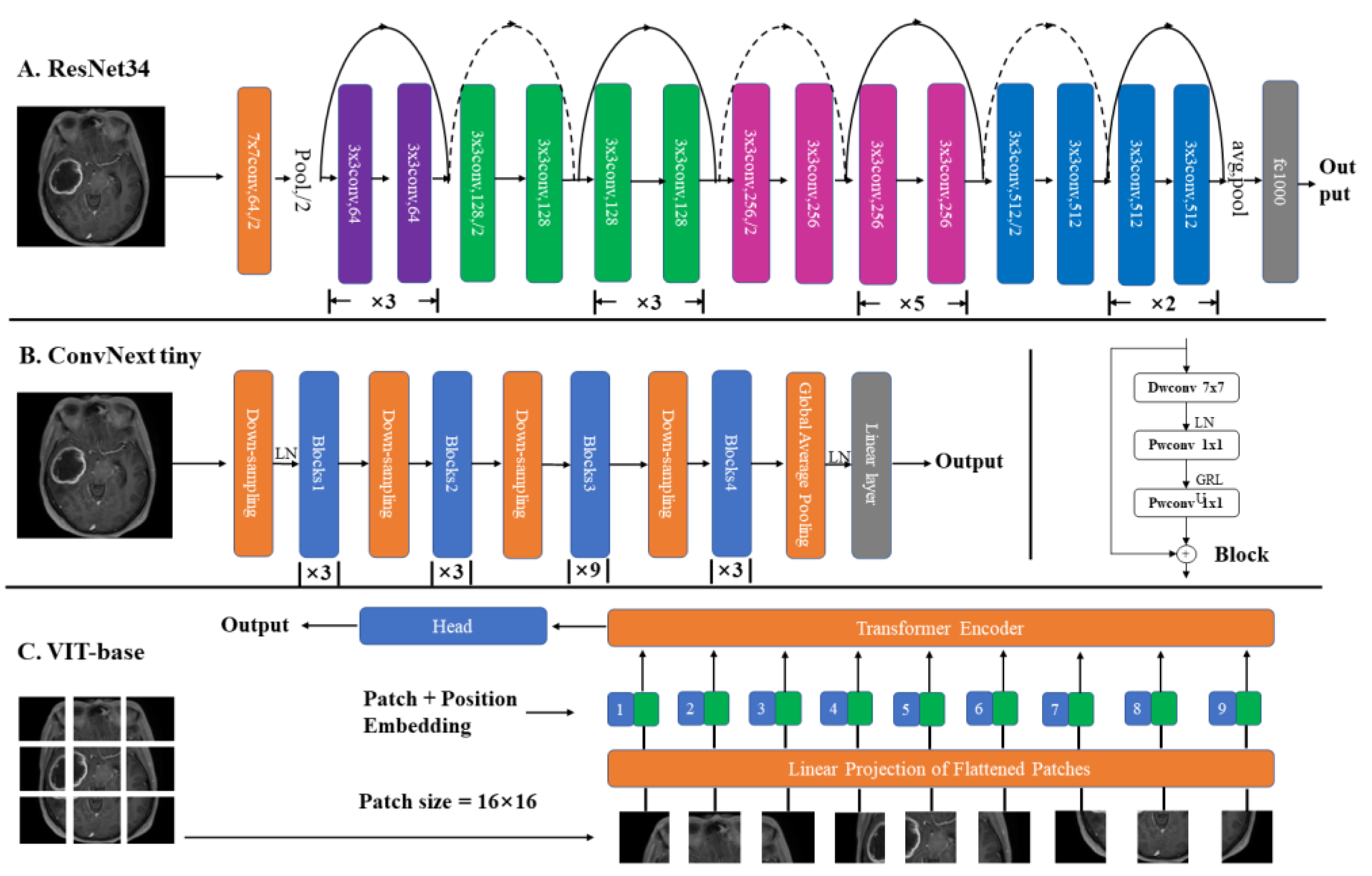
2.7. Model Details
2.7.1. Slice Preprocessing Strategies
2.7.2. Structures
2.7.3. Model Explanation
2.7.4. Model Evaluation
3. Results
3.1. Patient Characteristics
3.2. Model Comparison
3.2.1. Addition of ADC to Models
3.2.2. Different Slice Preprocessing Strategies
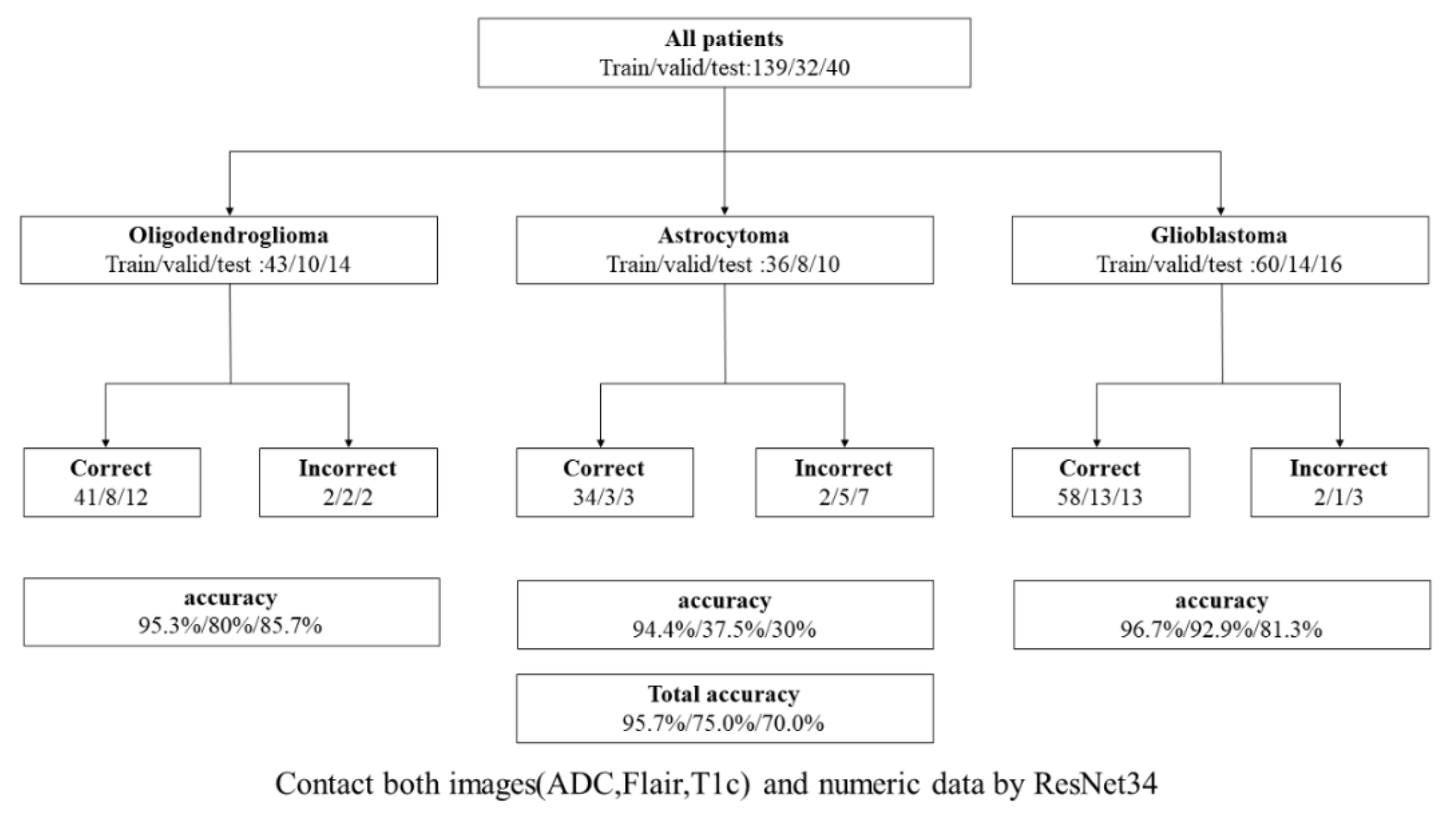
3.2.3. Benefits of Using Numeric Data
3.2.4. Comparison of Different Networks
3.2.5. Visualization and GradCAM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van den Bent, M.J. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: A clinician’s perspective. Acta Neuropathol. 2010, 120, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Brat, D.J.; Verhaak, R.G.; Aldape, K.D.; Yung, W.K.; Salama, S.R.; Cooper, L.A.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N. Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; van den Bent, M.; Tonn, J.C.; Stupp, R.; Preusser, M.; Cohen-Jonathan-Moyal, E.; Henriksson, R.; Le Rhun, E.; Balana, C.; Chinot, O.; et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017, 18, e315–e329. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, X.; Zhang, K.; Fung, K.M.; Thai, T.C.; Moore, K.; Mannel, R.S.; Liu, H.; Zheng, B.; Qiu, Y. Recent advances and clinical applications of deep learning in medical image analysis. Med. Image Anal. 2022, 79, 102444. [Google Scholar] [CrossRef]
- Kadry, S.; Nam, Y.; Rauf, H.T.; Rajinikanth, V.; Lawal, I.A. Automated detection of brain abnormality using deep-learning-scheme: A study. In Proceedings of the 2021 Seventh International Conference on Bio Signals, Images, and Instrumentation (ICBSII), Chennai, India, 25–27 March 2021; pp. 1–5. [Google Scholar]
- Kaiming, H.; Xiangyu, Z.; Shaoqing, R.; Jian, S. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Kim, J.W.; Park, C.-K.; Park, S.-H.; Kim, Y.H.; Han, J.H.; Kim, C.-Y.; Sohn, C.-H.; Chang, K.-H.; Jung, H.-W. Relationship between radiological characteristics and combined 1p and 19q deletion in World Health Organization grade III oligodendroglial tumours. J. Neurol. Neurosurg. Psychiatry 2011, 82, 224–227. [Google Scholar] [CrossRef]
- Decuyper, M.; Bonte, S.; Deblaere, K.; Van Holen, R. Automated MRI based pipeline for segmentation and prediction of grade, IDH mutation and 1p19q co-deletion in glioma. Comput Med. Imaging Graph. 2021, 88, 101831. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Niu, C.; Wang, G. Unsupervised contrastive learning based transformer for lung nodule detection. Phys. Med. Biol. 2022, 67, 204001. [Google Scholar] [CrossRef]
- Wu, Y.; Qi, S.; Sun, Y.; Xia, S.; Yao, Y.; Qian, W. A vision transformer for emphysema classification using CT images. Phys. Med. Biol. 2021, 66, 245016. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, G.; Oh, Y.; Seo, J.B.; Lee, S.M.; Kim, J.H.; Moon, S.; Lim, J.K.; Park, C.M.; Ye, J.C. Self-evolving vision transformer for chest X-ray diagnosis through knowledge distillation. Nat. Commun. 2022, 13, 3848. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, H.; Wu, C.-Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 19–20 June 2022; pp. 11976–11986. [Google Scholar]
- Tian, G.; Wang, Z.; Wang, C.; Chen, J.; Liu, G.; Xu, H.; Lu, Y.; Han, Z.; Zhao, Y.; Li, Z.; et al. A deep ensemble learning-based automated detection of COVID-19 using lung CT images and Vision Transformer and ConvNeXt. Front. Microbiol 2022, 13, 1024104. [Google Scholar] [CrossRef] [PubMed]
- Cluceru, J.; Interian, Y.; Phillips, J.J.; Molinaro, A.M.; Luks, T.L.; Alcaide-Leon, P.; Olson, M.P.; Nair, D.; LaFontaine, M.; Shai, A.; et al. Improving the noninvasive classification of glioma genetic subtype with deep learning and diffusion-weighted imaging. Neuro Oncol. 2022, 24, 639–652. [Google Scholar] [CrossRef]
- Johnson, D.R.; Diehn, F.E.; Giannini, C.; Jenkins, R.B.; Jenkins, S.M.; Parney, I.F.; Kaufmann, T.J. Genetically Defined Oligodendroglioma Is Characterized by Indistinct Tumor Borders at MRI. Am. J. Neuroradiol. 2017, 38, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Bai, H.X.; Zhou, H.; Su, C.; Bi, W.L.; Agbodza, E.; Kavouridis, V.K.; Senders, J.T.; Boaro, A.; Beers, A.; et al. Residual Convolutional Neural Network for the Determination of IDH Status in Low- and High-Grade Gliomas from MR Imaging. Clin. Cancer Res. 2018, 24, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Bien, N.; Rajpurkar, P.; Ball, R.L.; Irvin, J.; Park, A.; Jones, E.; Bereket, M.; Patel, B.N.; Yeom, K.W.; Shpanskaya, K.; et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PLoS Med. 2018, 15, e1002699. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE conference on computer vision and pattern recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE international conference on computer vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Yebasse, M.; Shimelis, B.; Warku, H.; Ko, J.; Cheoi, K.J. Coffee Disease Visualization and Classification. Plants (Basel) 2021, 10, 1257. [Google Scholar] [CrossRef]
- Matsui, Y.; Maruyama, T.; Nitta, M.; Saito, T.; Tsuzuki, S.; Tamura, M.; Kusuda, K.; Fukuya, Y.; Asano, H.; Kawamata, T.; et al. Prediction of lower-grade glioma molecular subtypes using deep learning. J. Neurooncol. 2020, 146, 321–327. [Google Scholar] [CrossRef]
- Wu, S.; Meng, J.; Yu, Q.; Li, P.; Fu, S. Radiomics-based machine learning methods for isocitrate dehydrogenase genotype prediction of diffuse gliomas. J. Cancer Res. Clin. Oncol. 2019, 145, 543–550. [Google Scholar] [CrossRef]
- Tummala, S.; Kadry, S.; Bukhari, S.A.C.; Rauf, H.T. Classification of Brain Tumor from Magnetic Resonance Imaging Using Vision Transformers Ensembling. Curr. Oncol. 2022, 29, 7498–7511. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Ashraf, I.; Alhaisoni, M.; Damaševičius, R.; Scherer, R.; Rehman, A.; Bukhari, S.A.C. Multimodal brain tumor classification using deep learning and robust feature selection: A machine learning application for radiologists. Diagnostics 2020, 10, 565. [Google Scholar] [CrossRef] [PubMed]
- Le Bihan, D. The ‘wet mind’: Water and functional neuroimaging. Phys. Med. Biol. 2007, 57, R57–R90. [Google Scholar] [CrossRef] [PubMed]
- Le Bihan, D.; Johansen-Berg, H. Diffusion MRI at 25: Exploring brain tissue structure and function. Neuroimage 2012, 61, 324–341. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Cantor, J.R.; Sabatini, D.M. Cancer cell metabolism: One hallmark, many faces. Cancer Discov. 2012, 2, 881–898. [Google Scholar] [CrossRef]
- Hensley, C.T.; Wasti, A.T.; DeBerardinis, R.J. Glutamine and cancer: Cell biology, physiology, and clinical opportunities. J. Clin. Investig. 2013, 123, 3678–3684. [Google Scholar] [CrossRef]
- Dang, C.V. Glutaminolysis: Supplying carbon or nitrogen or both for cancer cells? Cell Cycle 2010, 9, 3884–3886. [Google Scholar] [CrossRef]
- Glunde, K.; Bhujwalla, Z.M.; Ronen, S.M. Choline metabolism in malignant transformation. Nat. Rev. Cancer 2011, 11, 835–848. [Google Scholar] [CrossRef]
- Kickingereder, P.; Sahm, F.; Radbruch, A.; Wick, W.; Heiland, S.; Deimling, A.; Bendszus, M.; Wiestler, B. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci. Rep. 2015, 5, 16238. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Xiong, J.; Huang, W.; Wu, J.; Zhan, S.; Geng, D. Noninvasively detecting Isocitrate dehydrogenase 1 gene status in astrocytoma by dynamic susceptibility contrast MRI. J. Magn. Reson. Imaging 2017, 45, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wei, D.; Liu, X.; Fan, X.; Wang, K.; Li, S.; Zhang, Z.; Ma, K.; Qian, T.; Jiang, T.; et al. Molecular subtyping of diffuse gliomas using magnetic resonance imaging: Comparison and correlation between radiomics and deep learning. Eur Radiol 2022, 32, 747–758. [Google Scholar] [CrossRef]
- Wesseling, P.; van den Bent, M.; Perry, A. Oligodendroglioma: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015, 129, 809–827. [Google Scholar] [CrossRef] [PubMed]
- Giannini, C.; Burger, P.C.; Berkey, B.A.; Cairncross, J.G.; Jenkins, R.B.; Mehta, M.; Curran, W.J.; Aldape, K. Anaplastic oligodendroglial tumors: Refining the correlation among histopathology, 1p 19q deletion and clinical outcome in Intergroup Radiation Therapy Oncology Group Trial 9402. Brain Pathol. 2008, 18, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Varghese, J.; Jain, R. Adult Glioma WHO Classification Update, Genomics, and Imaging: What the Radiologists Need to Know. Top. Magn Reson Imaging 2020, 29, 71–82. [Google Scholar] [CrossRef]
- Carrillo, J.A.; Lai, A.; Nghiemphu, P.L.; Kim, H.J.; Phillips, H.S.; Kharbanda, S.; Moftakhar, P.; Lalaezari, S.; Yong, W.; Ellingson, B.M.; et al. Relationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastoma. AJNR Am. J. Neuroradiol 2012, 33, 1349–1355. [Google Scholar] [CrossRef]
- Qi, S.; Yu, L.; Li, H.; Ou, Y.; Qiu, X.; Ding, Y.; Han, H.; Zhang, X. Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol. Lett. 2014, 7, 1895–1902. [Google Scholar] [CrossRef]
- Patel, S.H.; Poisson, L.M.; Brat, D.J.; Zhou, Y.; Cooper, L.; Snuderl, M.; Thomas, C.; Franceschi, A.M.; Griffith, B.; Flanders, A.E.; et al. T2-FLAIR Mismatch, an Imaging Biomarker for IDH and 1p/19q Status in Lower-grade Gliomas: A TCGA/TCIA Project. Clin. Cancer Res. 2017, 23, 6078–6085. [Google Scholar] [CrossRef]




| All Dataset | Training | Validation | Testing | |
|---|---|---|---|---|
| All gliomas | 211 | 139 | 32 | 40 |
| Oligodendrogliomas | 67 | 43 | 10 | 14 |
| Astrocytoma | 54 | 36 | 8 | 10 |
| Glioblastoma | 90 | 60 | 14 | 16 |
| Parameter | All Gliomas | Oligodendroglioma | Astrocytoma | Glioblastoma |
|---|---|---|---|---|
| Number of patients | 211 | 67 | 54 | 90 |
| Median age (years) | 48.1 ± 11.8 | 46.9 ± 10.0 | 40.3 ± 11.5 | 53.6 ± 10.4 |
| Gender | ||||
| Male | 105 | 32 | 24 | 50 |
| Female | 106 | 35 | 30 | 51 |
| Enhancement category | ||||
| Nonenhancing | 76 | 37 | 37 | 2 |
| Patchy enhancing | 47 | 28 | 13 | 6 |
| Rim enhancing | 88 | 3 | 3 | 82 |
| Tumor location category | ||||
| Frontal, parietal, or occipital | 139 | 57 | 36 | 46 |
| Temporal and insular | 50 | 7 | 10 | 33 |
| Others | 22 | 3 | 8 | 11 |
| T2-FLAIR mismatch | ||||
| Present | 14 | 1 | 13 | 0 |
| Absent | 197 | 66 | 41 | 90 |
| Tumor margin | ||||
| Present | 27 | 4 | 21 | 2 |
| Absent | 184 | 63 | 33 | 88 |
| Combinations of 3 Image Modalities | n (Total) | n (Correct) | Accuracy | Subtype | Confusion Matrices | ||
|---|---|---|---|---|---|---|---|
| T1, T2, T1c | 40 | 20 | 50.0% | Oligodendroglioma | Astrocytoma | Glioblastoma | |
| Oligodendroglioma | 14 | 7 | 50.0% | Oligodendroglioma | 7 | 0 | 7 |
| Astrocytoma | 10 | 3 | 30.0% | Astrocytoma | 0 | 3 | 7 |
| Glioblastoma | 16 | 10 | 62.5% | Glioblastoma | 3 | 3 | 10 |
| T1, T2, ADC | 40 | 20 | 50.0% | ||||
| Oligodendroglioma | 14 | 4 | 28.6% | Oligodendroglioma | 4 | 0 | 10 |
| Astrocytoma | 10 | 0 | 0% | Astrocytoma | 4 | 0 | 6 |
| Glioblastoma | 16 | 16 | 100.0% | Glioblastoma | 0 | 0 | 16 |
| FLAIR, T1c, Zero* | 40 | 24 | 60.0% | ||||
| Oligodendroglioma | 14 | 11 | 78.6% | Oligodendroglioma | 11 | 2 | 1 |
| Astrocytoma | 10 | 5 | 50.0% | Astrocytoma | 5 | 3 | 2 |
| Glioblastoma | 16 | 10 | 62.5% | Glioblastoma | 5 | 1 | 10 |
| FLAIR, T1c, ADC | 40 | 27 | 67.5% | ||||
| Oligodendroglioma | 14 | 12 | 85.7% | Oligodendroglioma | 12 | 0 | 2 |
| Astrocytoma | 10 | 4 | 40.0% | Astrocytoma | 5 | 4 | 1 |
| Glioblastoma | 16 | 11 | 68.8% | Glioblastoma | 5 | 0 | 11 |
| Image Processing Strategy | n (Total) | n (Correct) | Accuracy | Subtype | Confusion Matrices | ||
|---|---|---|---|---|---|---|---|
| Skull stripping (FLAIR, ADC, T1c) | 40 | 27 | 67.5% | Oligodendroglioma | Astrocytoma | Glioblastoma | |
| Oligodendroglioma | 14 | 12 | 85.7% | Oligodendroglioma | 12 | 0 | 2 |
| Astrocytoma | 10 | 4 | 40.0% | Astrocytoma | 5 | 4 | 1 |
| Glioblastoma | 16 | 11 | 62.5% | Glioblastoma | 5 | 0 | 11 |
| Not-cropped (FLAIR, ADC, T1c) | 40 | 24 | 60.0% | ||||
| Oligodendroglioma | 14 | 11 | 78.6% | Oligodendroglioma | 9 | 2 | 3 |
| Astrocytoma | 10 | 2 | 20.0% | Astrocytoma | 4 | 3 | 3 |
| Glioblastoma | 16 | 11 | 68.8% | Glioblastoma | 3 | 0 | 13 |
| Segment addition (T1c, T1c, se) | 40 | 24 | 60.0% | ||||
| Oligodendroglioma | 14 | 11 | 78.6% | Oligodendroglioma | 11 | 1 | 2 |
| Astrocytoma | 10 | 2 | 20.0% | Astrocytoma | 6 | 2 | 2 |
| Glioblastoma | 16 | 11 | 68.8% | Glioblastoma | 5 | 0 | 11 |
| Image only (T1c, T1c, Zero *) | 40 | 20 | 50.0% | ||||
| Oligodendroglioma | 14 | 4 | 28.6% | Oligodendroglioma | 4 | 6 | 4 |
| Astrocytoma | 10 | 2 | 20.0% | Astrocytoma | 3 | 2 | 5 |
| Glioblastoma | 16 | 14 | 87.5% | Glioblastoma | 0 | 2 | 14 |
| Slice pooling (ADC(n − 1), n, (n + 1)) | 40 | 17 | 42.5% | ||||
| Oligodendroglioma | 14 | 6 | 42.9% | Oligodendroglioma | 6 | 3 | 5 |
| Astrocytoma | 10 | 4 | 40.0% | Astrocytoma | 3 | 4 | 3 |
| Glioblastoma | 16 | 7 | 43.8% | Glioblastoma | 8 | 1 | 7 |
| Individual slice treatment (ADC, ADC, ADC) | 40 | 22 | 55.0% | ||||
| Oligodendroglioma | 14 | 8 | 57.1% | Oligodendroglioma | 8 | 2 | 4 |
| Astrocytoma | 10 | 1 | 10.0% | Astrocytoma | 6 | 1 | 3 |
| Glioblastoma | 16 | 13 | 81.3% | Glioblastoma | 3 | 0 | 13 |
| Combinations of 3 Image Modalities | n (Total) | n (Correct) | Accuracy | Subtype | Confusion Matrices | ||
|---|---|---|---|---|---|---|---|
| ResNet34 with Transfer method A (ADC, FLAIR, T1c) | 40 | 27 | 67.5% | Oligodendroglioma | Astrocytoma | Glioblastoma | |
| Oligodendroglioma | 14 | 12 | 85.7% | Oligodendroglioma | 12 | 0 | 2 |
| Astrocytoma | 10 | 4 | 40.0% | Astrocytoma | 5 | 4 | 1 |
| Glioblastoma | 16 | 11 | 68.8% | Glioblastoma | 5 | 0 | 11 |
| ResNet34 with Transfer method B (ADC, FLAIR, T1c) | 40 | 24 | 60.0% | ||||
| Oligodendroglioma | 14 | 10 | 71.4% | Oligodendroglioma | 10 | 1 | 3 |
| Astrocytoma | 10 | 2 | 20.0% | Astrocytoma | 7 | 2 | 1 |
| Glioblastoma | 16 | 12 | 75.0% | Glioblastoma | 4 | 0 | 12 |
| ConvNext tiny with Transfer method B (ADC, FLAIR, T1c) | 40 | 22 | 55.0% | ||||
| Oligodendroglioma | 14 | 8 | 57.1% | Oligodendroglioma | 8 | 2 | 4 |
| Astrocytoma | 10 | 2 | 20.0% | Astrocytoma | 6 | 2 | 2 |
| Glioblastoma | 16 | 12 | 75.0% | Glioblastoma | 4 | 0 | 12 |
| VIT-base with Transfer method B (ADC, FLAIR, T1c) | 40 | 17 | 42.5% | ||||
| Oligodendroglioma | 14 | 12 | 85.7% | Oligodendroglioma | 12 | 2 | 0 |
| Astrocytoma | 10 | 2 | 20.0% | Astrocytoma | 8 | 2 | 0 |
| Glioblastoma | 16 | 0 | 0.0% | Glioblastoma | 0 | 0 | 0 |
| ResNet34 not pretrained (All images) | 40 | 20 | 50.0% | ||||
| Oligodendroglioma | 14 | 8 | 57.1% | Oligodendroglioma | 8 | 0 | 6 |
| Astrocytoma | 10 | 3 | 30.0% | Astrocytoma | 4 | 3 | 2 |
| Glioblastoma | 16 | 9 | 56.3% | Glioblastoma | 6 | 3 | 9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, D.; Ren, X.; Huang, W.; Wang, R.; Ma, L.; Gan, T.; Ai, K.; Wen, T.; Li, Y.; Wang, P.; et al. Noninvasive Classification of Glioma Subtypes Using Multiparametric MRI to Improve Deep Learning. Diagnostics 2022, 12, 3063. https://doi.org/10.3390/diagnostics12123063
Xiong D, Ren X, Huang W, Wang R, Ma L, Gan T, Ai K, Wen T, Li Y, Wang P, et al. Noninvasive Classification of Glioma Subtypes Using Multiparametric MRI to Improve Deep Learning. Diagnostics. 2022; 12(12):3063. https://doi.org/10.3390/diagnostics12123063
Chicago/Turabian StyleXiong, Diaohan, Xinying Ren, Weiting Huang, Rui Wang, Laiyang Ma, Tiejun Gan, Kai Ai, Tao Wen, Yujing Li, Pengfei Wang, and et al. 2022. "Noninvasive Classification of Glioma Subtypes Using Multiparametric MRI to Improve Deep Learning" Diagnostics 12, no. 12: 3063. https://doi.org/10.3390/diagnostics12123063
APA StyleXiong, D., Ren, X., Huang, W., Wang, R., Ma, L., Gan, T., Ai, K., Wen, T., Li, Y., Wang, P., Zhang, P., & Zhang, J. (2022). Noninvasive Classification of Glioma Subtypes Using Multiparametric MRI to Improve Deep Learning. Diagnostics, 12(12), 3063. https://doi.org/10.3390/diagnostics12123063






