A Novel ARMC5 Germline Variant in Primary Macronodular Adrenal Hyperplasia Using Whole-Exome Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Ethics Statement
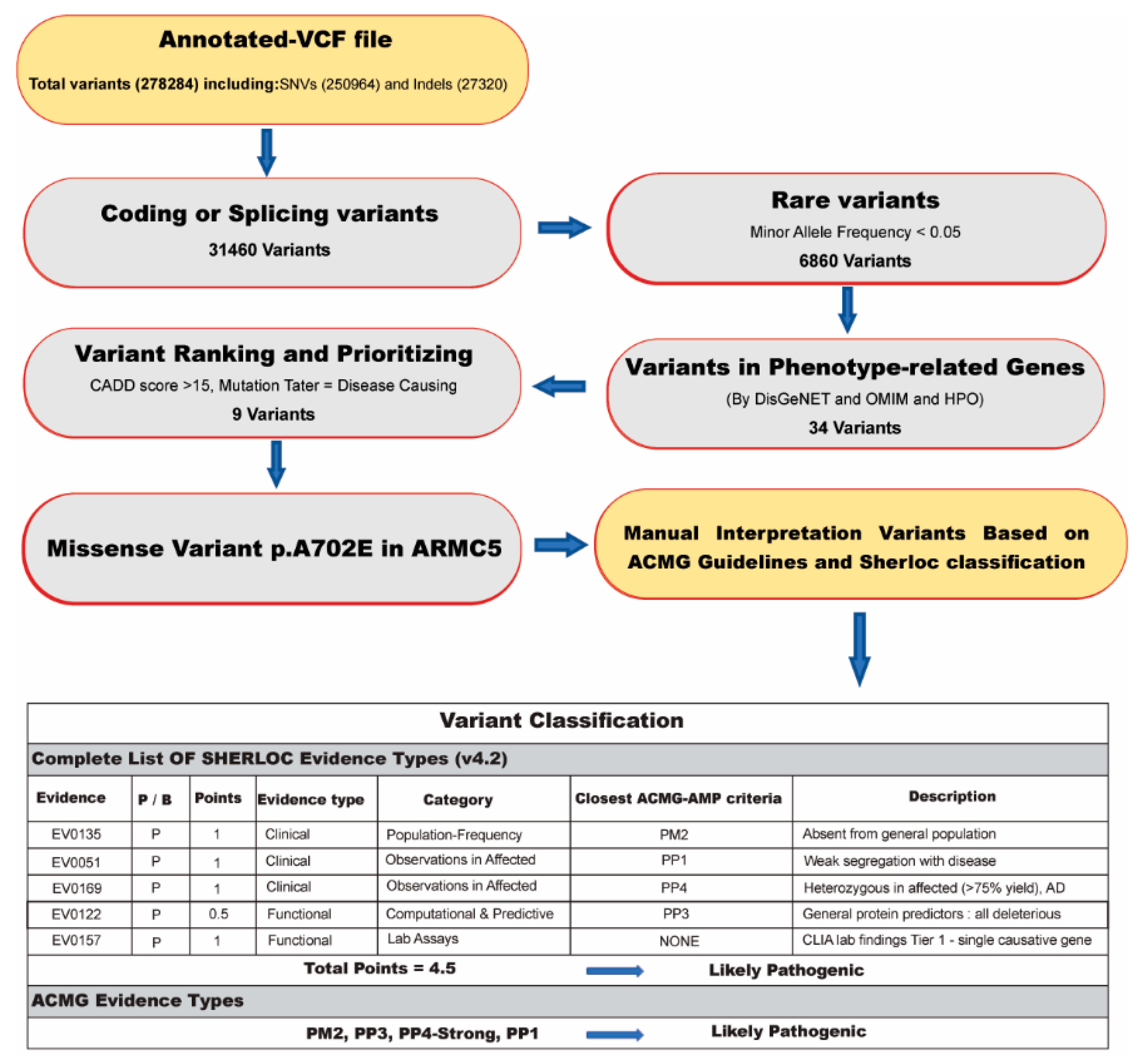
2.2. Whole-Exome Sequencing and Bioinformatics Analysis
- (1)
- Including and selecting known/unknown missense variants and loss-of-function (LOF) variants.
- (2)
- Checking variants in the Human Gene Mutation Database (HGMD) (http://www.hgmd.cf.ac.uk/ac/ access date: 22 October 2021) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar access date: 22 October 2021) to detect formerly reported mutations as pathogenic or likely pathogenic.
- (3)
- In silico analysis was directed by PolyPhen2 [17], Combined Annotation Dependent Depletion (CADD) [18], Mutation Taster [19], DANN score [20], HOPE web (https://www3.cmbi.umcn.nl/hope/ access date: 22 October 2021), and GERP score [21] to evaluate the potential pathogenicity of the variants based on the function or structure prediction. Additionally, the GERP score was used to assess the conservation score.
- (4)
- To find related variants with the patient’s clinical information, we used the Human Phenotype Ontology (HPO) as the phenotype–gene association database and the Online Mendelian Inheritance in Man (OMIM) as the gene–disease association database to discover the damaged genes related to the phenotype of the patients.
- (5)
2.3. The Variant Validation and Co-Segregation Analysis
2.4. Protein Structure Analysis
3. Results
3.1. Subjects
3.2. Whole-Exome Sequencing Results
3.3. Genetic Findings
3.4. Protein Assessment Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PMAH | Primary macronodular adrenocortical hyperplasia |
| WES | Whole-exome sequencing |
| AIMAH | ACTH-independent macronodular adrenal hyperplasia |
| CS | Cushing’s syndrome |
| LOH | Loss of heterozygosity |
| SNP | Single-nucleotide polymorphism |
| ARMC5 | Armadillo repeat-containing protein 5 |
| APC | Adenomatous polyposis coli gene |
| GNAS | Guanine nucleotide-binding protein alpha-stimulating activity polypeptide |
| PRKACA | cAMP-dependent protein kinase catalytic subunit alpha |
| PDE11A | phosphodiesterase 11A |
| PDE8B | phosphodiesterase 8B |
| SAM | Sequence Alignment/Map |
| SNVs | Single nucleotide variants |
| IGV | Integrated genome viewer |
| VCF | Variant call format |
| Indels | Insertions or deletions |
| LOF | Loss-of-function |
| HGMD | Human Gene Mutation Database |
| ESP | Exon Sequencing Projects |
| CADD | Combined Annotation Dependent Depletion |
| HPO | Human Phenotype Ontology |
| OMIM | Online Mendelian Inheritance in Man |
| ACMG | American College of Medical Genetics and Genomics guideline |
| Sherloc | Semiquantitative, hierarchical evidence-based rules for locus interpretation |
| SMR | SWISS-MODEL Repository |
| CVD | Cardiovascular disorder |
| HTN | Hypertension |
| DM | Diabetes mellitus |
| CNS | Central nervous system |
| BMI | Body mass index |
| ARR | Aldosterone renin ratio |
| LDDS | Low-Dose Dexamethasone-Suppression |
| MAF | Minor allele frequency |
| PGD | Preimplantation genetic diagnosis |
References
- Vélayoudom-Céphise, F.L.; Haissaguerre, M.; Tabarin, A. Etiopathogeny of primary adrenal hypercortisolism. Front. Horm. Res. 2016, 46, 39–53. [Google Scholar] [PubMed]
- Newell-Price, J. Cushing’s Syndrome. Clin. Med. 2008, 8, 204–208. [Google Scholar] [CrossRef]
- Kyo, C.; Usui, T.; Kosugi, R.; Torii, M.; Yonemoto, T.; Ogawa, T.; Kotani, M.; Tamura, N.; Yamamoto, Y.; Katabami, T.; et al. Armc5 Alterations in Primary Macronodular Adrenal Hyperplasia (PMAH) and the clinical state of variant carriers. J. Endocr. Soc. 2019, 3, 1837–1846. [Google Scholar] [CrossRef] [PubMed]
- Fragoso, M.C.; Alencar, G.A.; Lerario, A.M.; Bourdeau, I.; Almeida, M.Q.; Mendonca, B.B.; Lacroix, A. Genetics of primary macronodular adrenal hyperplasia. J. Endocrinol. 2015, 224, R31–R43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannah-Shmouni, F.; Stratakis, C.A. A gene-based classification of primary adrenocortical hyperplasias. Horm. Metab. Res. 2020, 52, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Chevalier, B.; Vantyghem, M.C.; Espiard, S. Bilateral adrenal hyperplasia: Pathogenesis and treatment. Biomedicines 2021, 9, 1397. [Google Scholar] [CrossRef]
- Yu, L.; Zhang, J.; Guo, X.; Chen, X.; He, Z.; He, Q. ARMC5 mutations in familial and sporadic primary bilateral macronodular adrenal hyperplasia. PLoS ONE 2018, 13, e0191602. [Google Scholar] [CrossRef]
- He, W.T.; Wang, X.; Song, W.; Song, X.D.; Lu, Y.J.; Lv, Y.K.; He, T.; Yu, X.F.; Hu, S.H. A novel nonsense mutation in ARMC5 causes primary bilateral macronodular adrenocortical hyperplasia. BMC Med. Genom. 2021, 14, 126. [Google Scholar] [CrossRef]
- Chasseloup, F.; Bourdeau, I.; Tabarin, A.; Regazzo, D.; Dumontet, C.; Ladurelle, N.; Tosca, L.; Amazit, L.; Proust, A.; Scharfmann, R. Loss of KDM1A in GIP-dependent primary bilateral macronodular adrenal hyperplasia with Cushing’s Syndrome: A multicentre, retrospective, cohort study. Lancet Diabetes Endocrinol. 2021, 9, 813–824. [Google Scholar] [CrossRef]
- Vaczlavik, A.; Bouys, L.; Violon, F.; Giannone, G.; Jouinot, A.; Armignacco, R.; Cavalcante, I.P.; Berthon, A.; Letouzé, E.; Vaduva, P.; et al. KDM1A inactivation causes hereditary food-dependent Cushing syndrome. Genet. Med. 2022, 24, 374–383. [Google Scholar] [CrossRef]
- Cavalcante, I.P.; Berthon, A.; Fragoso, M.C.; Reincke, M.; Stratakis, C.A.; Ragazzon, B.; Bertherat, J. Primary bilateral macronodular adrenal hyperplasia: Definitely a genetic disease. Nat. Rev. Endocrinol. 2022, 18, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Charchar, H.L.S.; Fragoso, M.C.B.V. An overview of the heterogeneous causes of Cushing syndrome resulting from Primary Macronodular Adrenal Hyperplasia (PMAH). J. Endocr. Soc. 2022, 6, bvac041. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freese, N.H.; Norris, D.C.; Loraine, A.E. Integrated genome browser: Visual analytics platform for genomics. Bioinformatics 2016, 32, 2089–2095. [Google Scholar] [CrossRef] [Green Version]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using polyphen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A General framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. Mutationtaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef]
- Quang, D.; Chen, Y.; Xie, X. Dann: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 2015, 31, 761–763. [Google Scholar] [CrossRef]
- Davydov, E.V.; Goode, D.L.; Sirota, M.; Cooper, G.M.; Sidow, A.; Batzoglou, S. Identifying a high fraction of the human genome to be under selective constraint using Gerp++. PLoS Comput. Biol. 2010, 6, e1001025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nykamp, K.; Anderson, M.; Powers, M.; Garcia, J.; Herrera, B.; Ho, Y.Y.; Kobayashi, Y.; Patil, N.; Thusberg, J.; Westbrook, M.; et al. Sherloc: A comprehensive refinement of the ACMG-AMP variant classification criteria. Genet. Med. 2017, 19, 1105–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rad, I.; Honardoost, M. Structural modification in hepatitis c virus envelope protein; potential viral strategy against interferon therapy. Int. J. Pept. Res. Ther. 2020, 26, 171–179. [Google Scholar] [CrossRef]
- Shahsavari Alavijeh, M.; Rad, I.; Hatamie, S. Magnetic nanocomposite’s mechanism of action during the hyperthermia treatment of the breast cancer. Appl. Nanosci. 2021, 11, 2739–2746. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the expasy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Holland, O.B.; Brown, H.; Kuhnert, L.; Fairchild, C.; Risk, M.; Gomez-Sanchez, C.E. Further evaluation of saline infusion for the diagnosis of primary aldosteronism. Hypertension 1984, 6, 717–723. [Google Scholar] [CrossRef] [Green Version]
- Stowasser, M.; Ahmed, A.H.; Cowley, D.; Wolley, M.; Guo, Z.; McWhinney, B.C.; Ungerer, J.P.; Gordon, R.D. Comparison of seated with recumbent saline suppression testing for the diagnosis of primary aldosteronism. J. Clin. Endocrinol. Metab. 2018, 103, 4113–4124. [Google Scholar] [CrossRef] [Green Version]
- Espiard, S.; Drougat, L.; Libé, R.; Assié, G.; Perlemoine, K.; Guignat, L.; Barrande, G.; Brucker-Davis, F.; Doullay, F.; Lopez, S.; et al. ARMC5 mutations in a large cohort of primary macronodular adrenal hyperplasia: Clinical and functional consequences. J. Clin. Endocrinol. Metab. 2015, 100, E926–E935. [Google Scholar] [CrossRef] [Green Version]
- Kar, G.; Gursoy, A.; Keskin, O. Human cancer protein-protein interaction network: A structural perspective. PLoS Comput. Biol. 2009, 5, e1000601. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, H.; Wanyan, Y.; Liu, K.; Lv, T.; Li, M.; Chen, Y. Effect of hydrophobicity on the anticancer activity of fatty-acyl-conjugated CM4 in breast cancer cells. ACS Omega 2020, 5, 21513–21523. [Google Scholar] [CrossRef] [PubMed]
- Ellard, S.; Baple, E.L.; Callaway, A.; Berry, I.; Forrester, N.; Turnbull, C.; Owens, M.; Eccles, D.M.; Abbs, S.; Scott, R.; et al. ACGS best practice guidelines for variant classification in rare disease 2020. Assoc. Clin. Genom. Sci. 2020, 1–33. Available online: https://www.acgs.uk.com/media/11631/uk-practice-guidelines-for-variant-classification-v4-01-2020.pdf (accessed on 20 November 2022).
- Alencar, G.A.; Lerario, A.M.; Nishi, M.Y.; Mariani, B.M.; Almeida, M.Q.; Tremblay, J.; Hamet, P.; Bourdeau, I.; Zerbini, M.C.; Pereira, M.A.; et al. ARMC5 mutations are a frequent cause of primary macronodular adrenal hyperplasia. J. Clin. Endocrinol. Metab. 2014, 99, E1501–E1509. [Google Scholar] [CrossRef] [Green Version]
- Berthon, A.; Faucz, F.R.; Espiard, S.; Drougat, L.; Bertherat, J.; Stratakis, C.A. Age-dependent effects of ARMC5 haploinsufficiency on adrenocortical function. Hum. Mol. Genet. 2017, 26, 3495–3507. [Google Scholar] [CrossRef] [Green Version]
- Swain, J.M.; Grant, C.S.; Schlinkert, R.T.; Thompson, G.B.; van Heerden, J.A.; Lloyd, R.V.; Young, W.F. Corticotropin-independent macronodular adrenal hyperplasia: A clinicopathologic correlation. Arch. Surg. 1998, 133, 541–545; discussion 45–46. [Google Scholar] [CrossRef] [Green Version]
- Ohashi, A.; Yamada, Y.; Sakaguchi, K.; Inoue, T.; Kubo, M.; Fushimi, H. A natural history of Adrenocorticotropin-Independent Bilateral Adrenal Macronodular Hyperplasia (AIMAH) from preclinical to clinically overt Cushing’s Syndrome. Endocr. J. 2001, 48, 677–683. [Google Scholar] [CrossRef] [Green Version]
- Lacroix, A. ACTH-independent macronodular adrenal hyperplasia. Best Pract. Res. Clin. Endocrinol. Metab. 2009, 23, 245–259. [Google Scholar] [CrossRef]
- Albiger, N.M.; Regazzo, D.; Rubin, B.; Ferrara, A.M.; Rizzati, S.; Taschin, E.; Ceccato, F.; Arnaldi, G.; Giraldi, F.P.; Stigliano, A.; et al. A multicenter experience on the prevalence of ARMC5 mutations in patients with primary bilateral macronodular adrenal hyperplasia: From genetic characterization to clinical phenotype. Endocrine 2017, 55, 959–968. [Google Scholar] [CrossRef]
- Kirschner, M.A.; Powell, R.D., Jr.; Lipsett, M.B. Cushing’s Syndrome: Nodular cortical hyperplasia of adrenal glands with clinical and pathological features suggesting adrenocortical tumor. J. Clin. Endocrinol. Metab. 1964, 24, 947–955. [Google Scholar] [CrossRef]
- Cavalcante, I.P.; Nishi, M.; Zerbini, M.C.N.; Almeida, M.Q.; Brondani, V.B.; Botelho, M.; Tanno, F.Y.; Srougi, V.; Chambo, J.L.; Mendonca, B.B.; et al. The role of ARMC5 in human cell cultures from nodules of Primary Macronodular Adrenocortical Hyperplasia (PMAH). Mol. Cell. Endocrinol. 2018, 460, 36–46. [Google Scholar] [CrossRef]
- Assié, G.; Libé, R.; Espiard, S.; Rizk-Rabin, M.; Guimier, A.; Luscap, W.; Barreau, O.; Lefèvre, L.; Sibony, M.; Guignat, L.; et al. ARMC5 mutations in macronodular adrenal hyperplasia with Cushing’s Syndrome. N. Engl. J. Med. 2013, 369, 2105–2114. [Google Scholar] [CrossRef] [PubMed]
- Stratakis, C.A.; Berthon, A. Molecular mechanisms of ARMC5 mutations in adrenal pathophysiology. Curr. Opin. Endocr. Metab. Res 2019, 8, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cui, L.; Gao, J.P.; Yan, W.H.; Jin, N.; Chen, K.; Zang, L.; Du, J.; Wang, X.L.; Guo, Q.H.; et al. Whole-genome sequencing revealed armadillo repeat containing 5 (ARMC5) mutation in a Chinese family with ACTH-independent macronodular adrenal hyperplasia. Endocr. J. 2018, 65, 269–279. [Google Scholar] [CrossRef] [Green Version]
- Davidson, C.J.; Zeringer, E.; Champion, K.J.; Gauthier, M.P.; Wang, F.; Boonyaratanakornkit, J.; Jones, J.R.; Schreiber, E. Improving the limit of detection for sanger sequencing: A comparison of methodologies for kras variant detection. Biotechniques 2012, 53, 182–188. [Google Scholar] [CrossRef]
- Quinn, G.P.; Pal, T.; Murphy, D.; Vadaparampil, S.T.; Kumar, A. High-risk consumers’ perceptions of preimplantation genetic diagnosis for hereditary cancers: A systematic review and meta-analysis. Genet. Med. 2012, 14, 191–200. [Google Scholar] [CrossRef] [Green Version]
- Rechitsky, S.; Verlinsky, O.; Chistokhina, A.; Sharapova, T.; Ozen, S.; Masciangelo, C.; Kuliev, A.; Verlinsky, Y. Preimplantation genetic diagnosis for cancer predisposition. Reprod. Biomed. Online 2002, 5, 148–155. [Google Scholar] [CrossRef]
- Vriesen, N.; Carmany, E.P.; Natoli, J.L. Clinical outcomes of preimplantation genetic testing for hereditary cancer syndromes: A systematic review. Prenat. Diagn. 2022, 42, 201–211. [Google Scholar] [CrossRef]



| Analytical Characteristic | Proband III-6 |
|---|---|
| Total number of reads | 121,128,400 |
| Average read length (bp) | 150 |
| Target region (Mbp) | 36 |
| % Bases QV > 30 | 98.16 |
| % Initial mappable reads | 99 |
| % Minimum coverage of target regions (For depth 1×, 5×, 10×, 25×, 50× and 100×) | 97.7, 97.3, 96.9, 94.7, 85.4 and 52.7 |
| % Of duplicate reads (pre-alignment) | 25 |
| % Of duplicate reads (post-alignment) | 6 |
| % On target reads (post-alignment) | 55 66,811,296 (reads) |
| Patient | Proband III-6 |
|---|---|
| Variant Definition | |
| -Gene name | ARMC5 (NM_001105247.2) |
| -Varian name | c.2105C>A |
| -Protein change | p. Ala702Glu |
| -Chromosome position (GRCh37) | Chr16: 31477507 |
| -Zygosity | Heterozygote |
| In-silico predictive tools | |
| -CADD (Phred score) | 25 (deleterious) |
| -DANN | 0.9948 (deleterious) |
| -GERP | 5 |
| -Mutation taster | Disease-causing |
| -Polyphen | Probably-damaging |
| Population databases | |
| -1000 GP | - |
| -ExAC | - |
| -ESP | - |
| -GnomAD | - |
| -Iranome | - |
| Related phenotypes (OMIM number) | ACTH-independent macronodular adrenal hyperplasia 2/AIMAH2 (OMIM: 615954) |
| Variant classification (Evidence based on ACMG guideline) (Evidence based on Sherloc) | Likely pathogenic (PM2, PP3, PP4-strong, and PP1) (PM2, PP3, PP4, PP1, and LAB-assay points) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eghbali, M.; Cheraghi, S.; Samanian, S.; Rad, I.; Meghdadi, J.; Akbari, H.; Honardoost, M. A Novel ARMC5 Germline Variant in Primary Macronodular Adrenal Hyperplasia Using Whole-Exome Sequencing. Diagnostics 2022, 12, 3028. https://doi.org/10.3390/diagnostics12123028
Eghbali M, Cheraghi S, Samanian S, Rad I, Meghdadi J, Akbari H, Honardoost M. A Novel ARMC5 Germline Variant in Primary Macronodular Adrenal Hyperplasia Using Whole-Exome Sequencing. Diagnostics. 2022; 12(12):3028. https://doi.org/10.3390/diagnostics12123028
Chicago/Turabian StyleEghbali, Maryam, Sara Cheraghi, Sara Samanian, Iman Rad, Jafar Meghdadi, Hamideh Akbari, and Maryam Honardoost. 2022. "A Novel ARMC5 Germline Variant in Primary Macronodular Adrenal Hyperplasia Using Whole-Exome Sequencing" Diagnostics 12, no. 12: 3028. https://doi.org/10.3390/diagnostics12123028
APA StyleEghbali, M., Cheraghi, S., Samanian, S., Rad, I., Meghdadi, J., Akbari, H., & Honardoost, M. (2022). A Novel ARMC5 Germline Variant in Primary Macronodular Adrenal Hyperplasia Using Whole-Exome Sequencing. Diagnostics, 12(12), 3028. https://doi.org/10.3390/diagnostics12123028





