Lectin Analysis of SARS-CoV-2-Positive Nasopharyngeal Samples Using GLYcoPROFILE® Technology Platform
Abstract
1. Introduction
2. Materials and Methods
2.1. General Remarks
2.2. Antibodies and Reagents
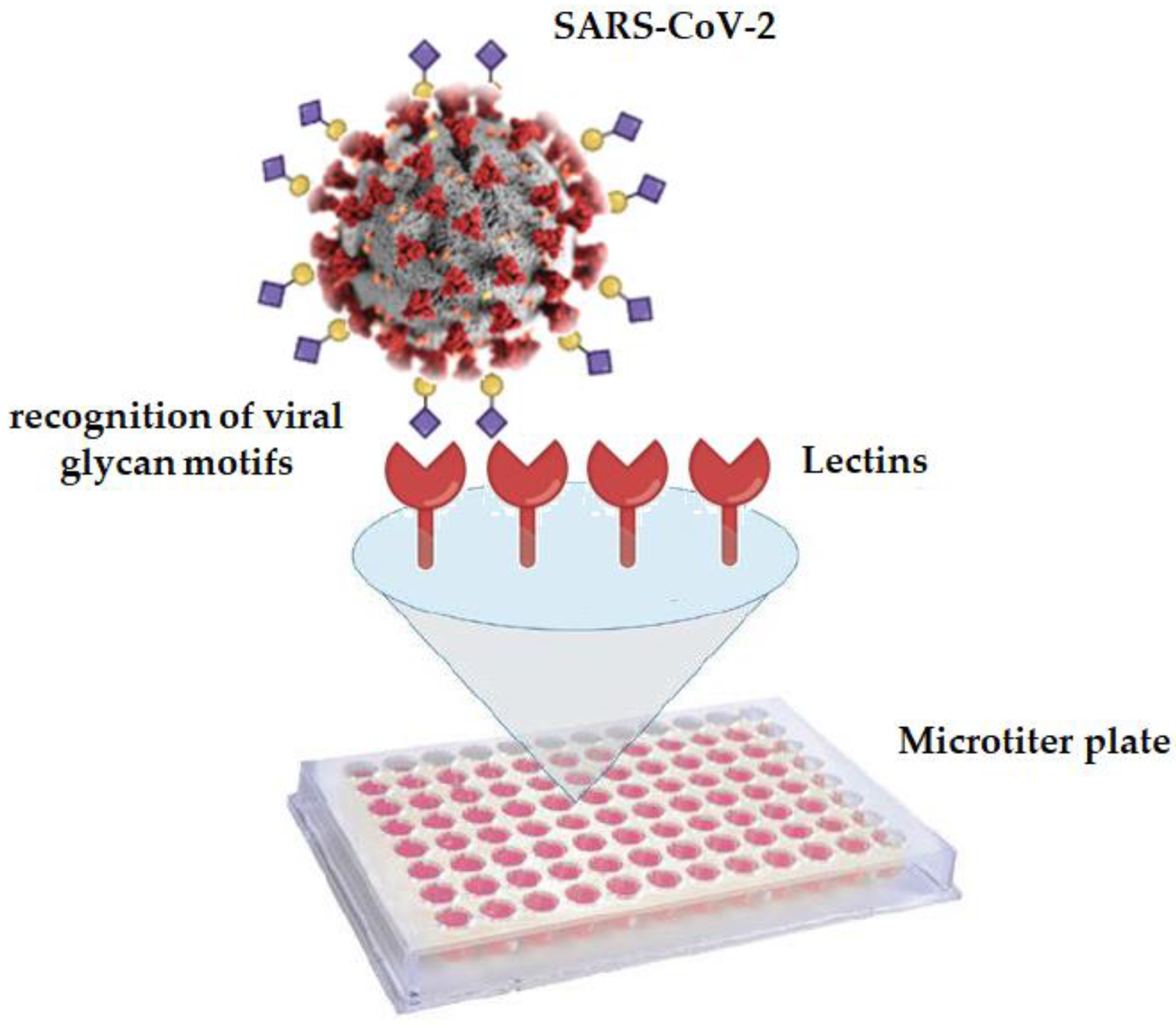
2.3. GLYcoPROFILE® of Nasopharyngeal Samples
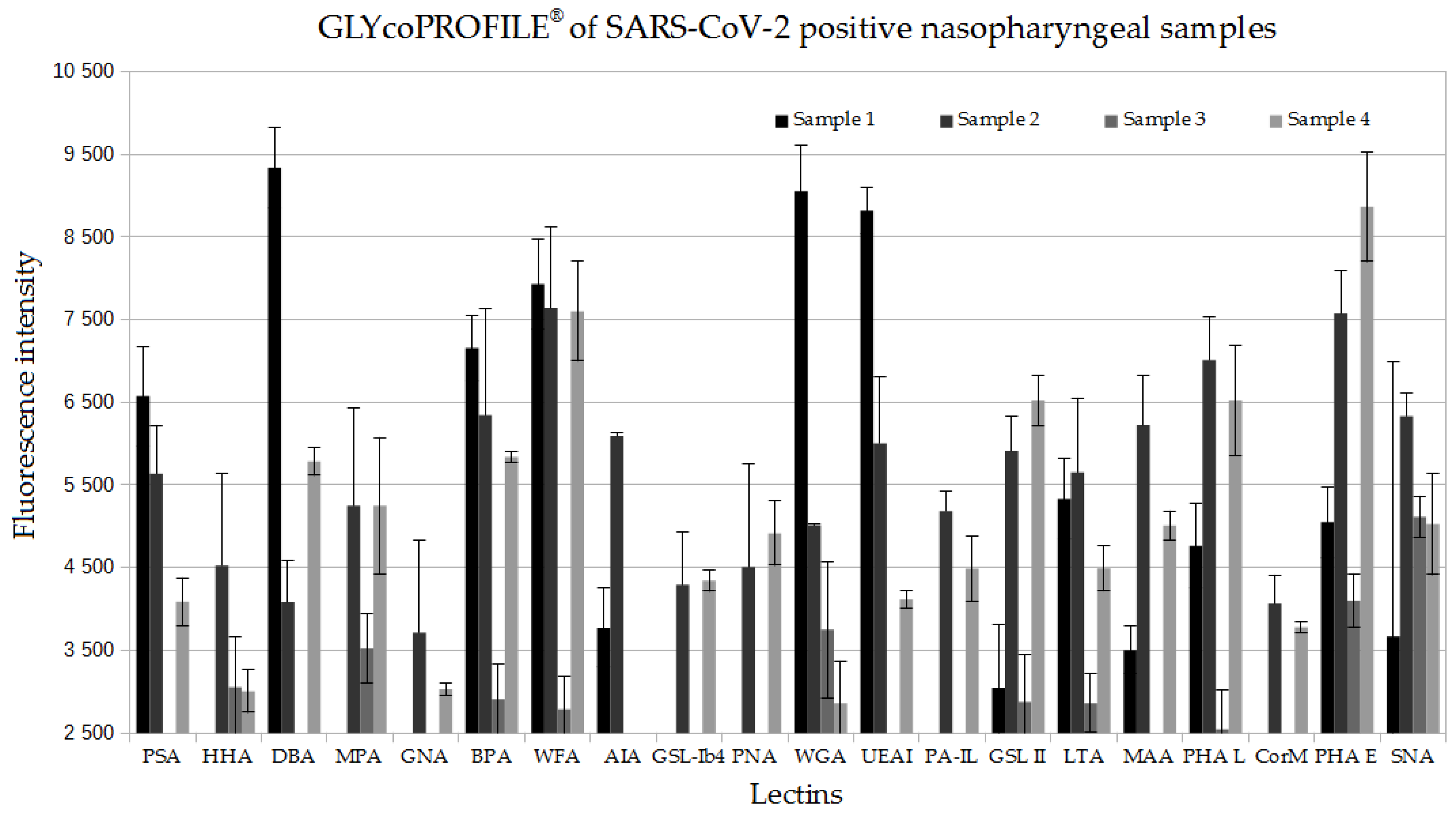
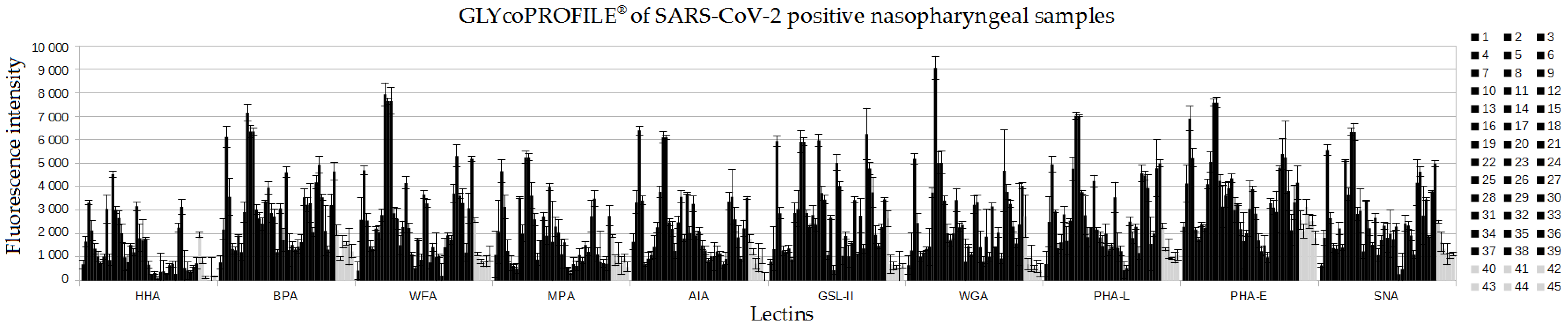
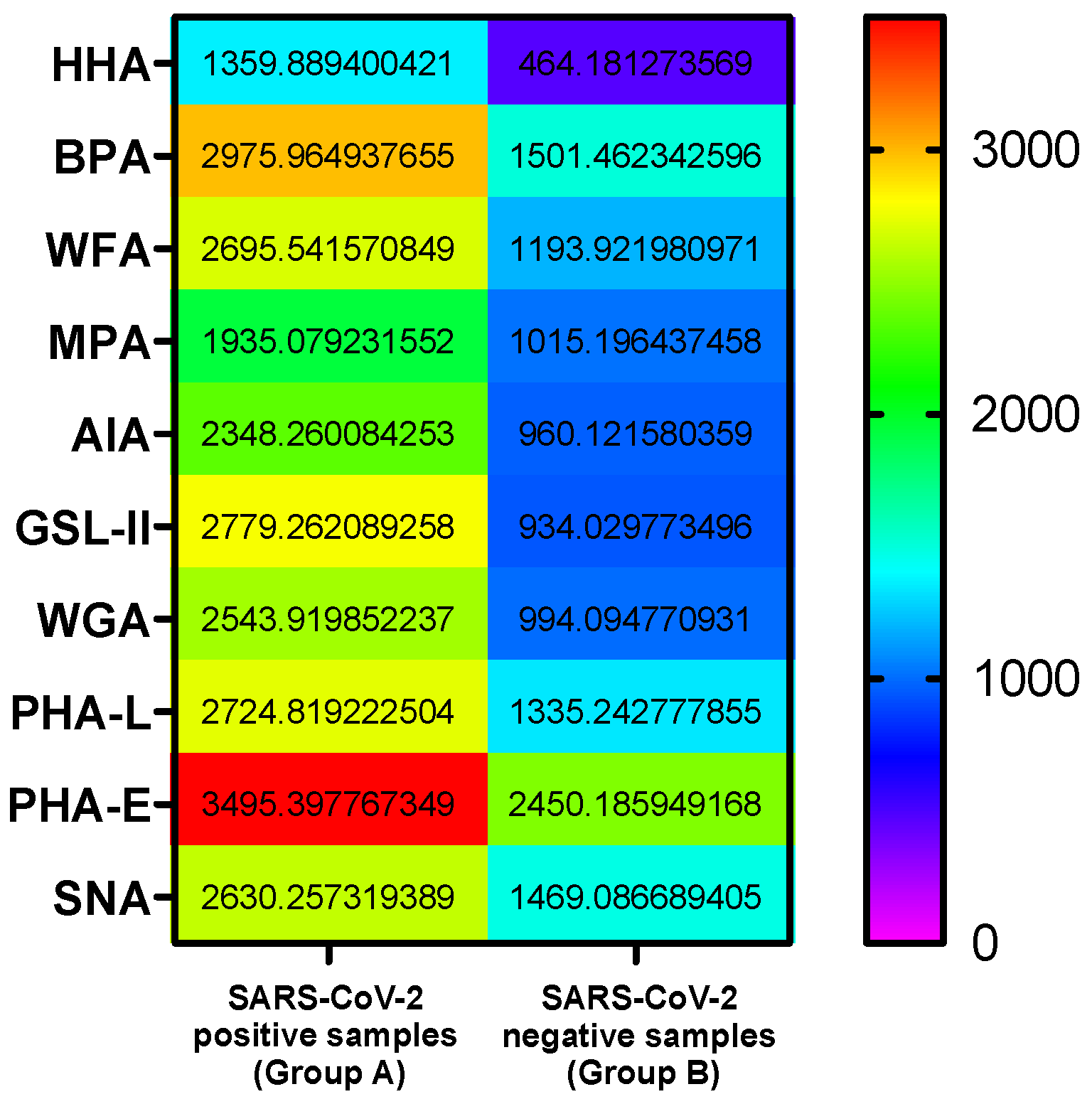
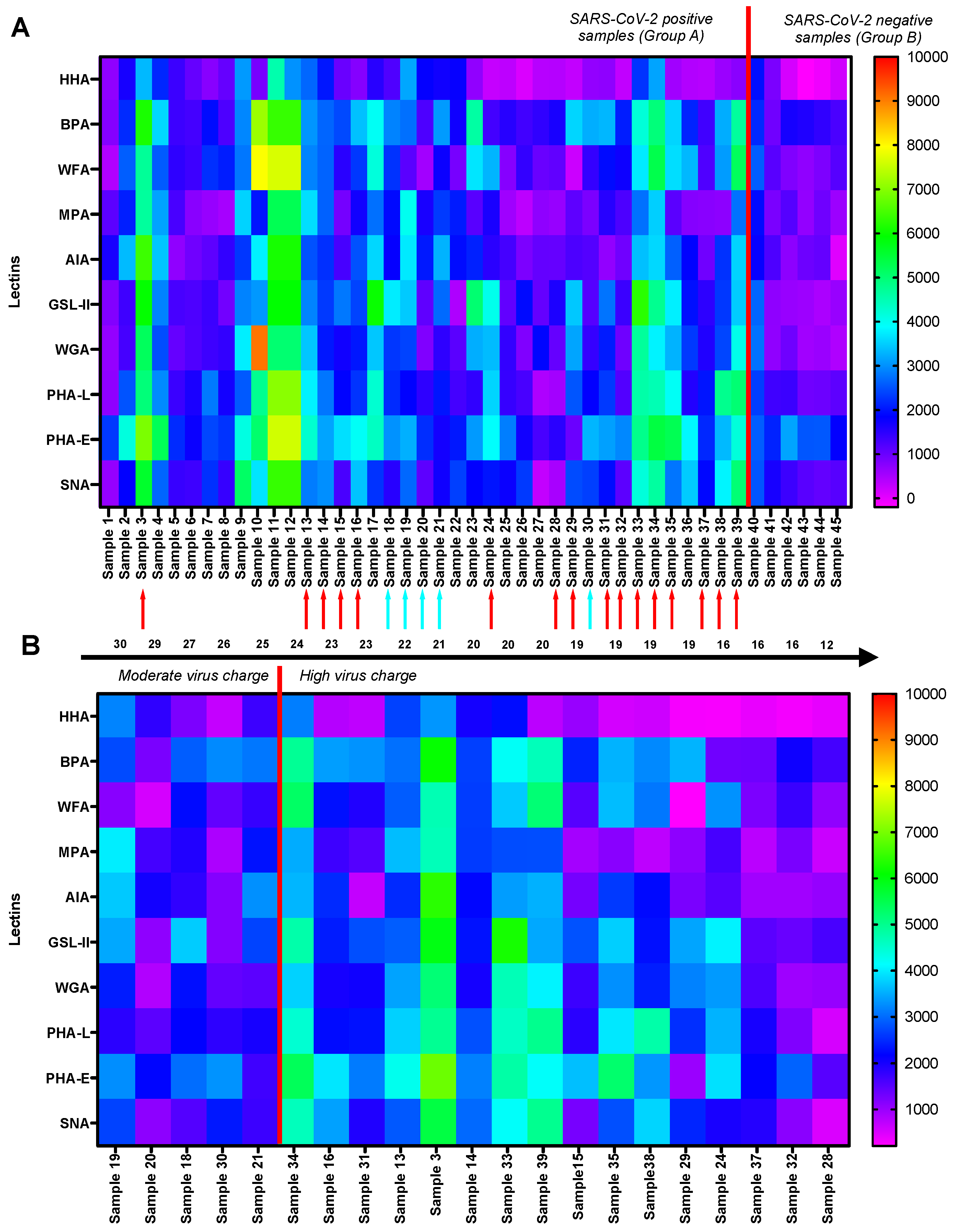
3. Results
3.1. GLYcoPROFILE®—Lectin-Based Glycoprofiling of SARS-CoV-2-Positive Nasopharyngeal Samples
3.2. Interpretation of GLYcoPROFILE®—Discrimination between SARS-CoV-2-Positive and -Negative Crude Nasopharyngeal Samples
4. Discussions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Atzrodt, C.L.; Maknojia, I.; McCarthy, R.D.P.; Oldfield, T.M.; Po, J.; Ta, K.T.L.; Stepp, H.E.; Clements, T.P. A Guide to COVID-19: A global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J. 2020, 287, 3633–3650. [Google Scholar] [CrossRef] [PubMed]
- Hamre, D.; Procknow, J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966, 121, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Wevers, B.A.; van der Hoek, L. Recently discovered human coronaviruses. Clin. Lab. Med. 2009, 29, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Pyrc, K.; Berkhout, B.; van der Hoek, L. Identification of new human coronaviruses. Expert. Rev. Anti. Infect. Ther. 2007, 5, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Comish, P.; Kang, R. The hallmarks of COVID-19 disease. PLoS Pathog. 2002, 16, e1008536. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species Severe acute respiratory syndrome related coronavirus: Classifying 2019-nCoV andnaming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Worobey, M.; Levy, J.I.; Serrano, L.M.; Crits-Christoph, A.; Pekar, J.E.; Goldstein, S.A.; Rasmussen, A.L.; Kraemer, M.U.G.; Newman, C.; Koopmans, M.P.G.; et al. The Huanan Seafood Wholesale Market in Wuhan was the early epicenter of the COVID-19 pandemic. Science 2022, 377, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Ochani, R.K.; Asad, A.; Yasmin, F.; Shaikh, S.; Khalid, H.; Batra, S.; Sohail, M.R.; Mahmood, S.F.; Ochani, R.; Arshad, M.H.; et al. COVID-19 pandemic: From origins to outcomes. A comprehensive review of viral pathogenesis, clinical manifestations, diagnostic evaluation, and management. Infez. Med. 2021, 29, 20–36. [Google Scholar] [PubMed]
- Khan, M.; Adil, S.F.; Alkhathlan, H.Z.; Tahir, M.N.; Saif, S.; Khan, M.; Khan, S.T. COVID-19: A global challenge with old history, epidemiology and progress so far. Molecules 2020, 26, 39. [Google Scholar] [CrossRef] [PubMed]
- Piret, J.; Boivin, G. Pandemics throughout history. Front. Microbiol. 2021, 11, 631736. [Google Scholar] [CrossRef] [PubMed]
- da Silva, L.C.N.; Mendonça, J.S.P.; de Oliveira, W.F.; Batista, K.L.R.; Zagmignan, A.; Viana, I.F.T.; Correia, M.T.D.S. Exploring lectin-glycan interactions to combat COVID-19: Lessons acquired from other enveloped viruses. Glycobiology 2021, 31, 358–371. [Google Scholar] [CrossRef] [PubMed]
- Sabzian-Molaei, F.; NasiriKhalili, M.A.; Sabzian-Molaei, M.; Shahsavarani, H.; Pour, A.F.; MolaeiRad, A.; Hadi, A. Urtica dioica Agglutinin: A plant protein candidate for inhibition of SARS-CoV-2 receptor-binding domain for control of COVID-19 Infection. PLoS ONE 2022, 17, e0268156. [Google Scholar] [CrossRef] [PubMed]
- Landemarre, L.; Duverger, E. Lectin glycoprofiling of recombinant therapeutic Interleukin-7. In Glycosylation Engineering of Biopharmaceuticals: Methods and Protocols, Methods in Molecular Biology; Beck, A., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 221–226. [Google Scholar]
- Assailly, C.; Bridot, C.; Saumonneau, A.; Lottin, P.; Roubinet, B.; Krammer, E.-M.; François, F.; Vena, F.; Landemarre, L.; Dorta, D.A.; et al. Polyvalent Transition-State Analogues of Sialyl Substrates Strongly Inhibit Bacterial Sialidases. Chem. Eur. J. 2021, 27, 3142–3150. [Google Scholar] [CrossRef] [PubMed]
- Shajahan, A.; Supekar, N.T.; Gleinich, A.S.; Azadi, P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 2020, 30, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Brissonnet, Y.; Assailly, C.; Saumonneau, A.; Bouckaert, J.; Maillasson, M.; Petitot, C.; Roubinet, B.; Didak, B.; Landemarre, L.; Bridot, C.; et al. Multivalent thiosialosides and their synergistic interaction with pathogenic sialidases. Chem. Eur. J. 2019, 25, 2358–2365. [Google Scholar] [CrossRef] [PubMed]
- Cauwel, M.; Sivignon, A.; Bridot, C.; Nongbe, M.C.; Deniaud, D.; Roubinet, B.; Landemarre, L.; Felpin, F.; Bouckaert, J.; Barnich, N.; et al. Heptylmannose-functionalized cellulose for the binding and specific detection of pathogenic E. coli. Chem. Commun. 2019, 55, 10158–10161. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, Z.; Li, M.; Wang, X. Comparative review of SARS-CoV-2, SARS-CoV, MERS-CoV, and influenza arespiratory viruses. Front. Immunol. 2020, 11, 552909. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Zhao, Y.; Dong, J.; Liang, S.; Guo, M.; Liu, X.; Wang, X.; Huang, Z.; Sun, X.; Zhang, Z.; et al. Coinfection with influenza A virus enhances SARS-CoV-2 infectivity. Cell Res. 2021, 31, 395–403. [Google Scholar] [CrossRef] [PubMed]
| Lectin Abbreviation | Common Name | Glycan Specificity |
|---|---|---|
| PSA | Pisum sativum agglutinin | Manα6(Manα3)Man |
| HHA | Hippeastrum hybrid agglutinin | terminal and internal Man |
| DBA | Dolichos biflorus agglutinin | αGalNAc; terminal GalNAc; GalNAcα3GalNAc |
| MPA | Maclurapomifera agglutinin | Galβ3GalNAc (T antigen), Galα6Glc (melibiose) |
| GNA | Galanthus nivalis agglutinin | Manα6(Manα3)Man |
| BPA | Bauhinia purpurea agglutinin | Galβ3GalNAc (T-antigen), GalNAc, Gal |
| WFA | Wisteria floribunda agglutinin | GalNAcα6Gal, GalNAcα3GalNAc, GalNAc |
| AIA | Artocarpus intergrifolia agglutinin | Galβ3GalNAc |
| GSL-Ib4 | Griffoniasimplicifolia isoB4 | αGal |
| PNA | Arachis hypogaea agglutinin | Galβ3GalNAc |
| WGA | Triticum vulgare agglutinin | (GlcNAcβ4)n, Neu5Ac; poly(N-acetyllactosamine) |
| UEA-I | Ulex europaeus | GlcNAcβ4GlcNAc oligomers; Fucα2Galβ4GlcNAc |
| PA-IL | Pseudomonas aeruginosa lectin A (Lec A) | terminal αGal |
| GSL II | Griffonia simplicifolia lectin | α or βGlcNAc; αgalactosylated tri/tetra antennary glycans; core 3 O-glycans |
| LTA | Lotus tetragonolobus agglutinin | Fuc, LeX but not LeA, 2-FucαGalβ4Glc |
| MAA | Maackiaamurensis agglutinin | Neu5Acα2, 3Gal4GalNAc |
| PHA-L | Phaseolus vulgaris agglutinin | Complex glycans |
| PHA-E | Phaseolus vulgaris agglutinin | Complex glycans |
| CorM | Coregonus lavaretusmarenae | Rha |
| SNA | Sambucus nigra agglutinin | Neu5Acα2,6Gal/GalNAc |
| Lectins | p-Value |
|---|---|
| GSL-II | 0.002 |
| WGA | 0.005 |
| BPA | 0.007 |
| PHA-L | 0.007 |
| WFA | 0.024 |
| SNA | 0.033 |
| AIA | 0.048 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seničar, M.; Roubinet, B.; Daniellou, R.; Prazuck, T.; Landemarre, L. Lectin Analysis of SARS-CoV-2-Positive Nasopharyngeal Samples Using GLYcoPROFILE® Technology Platform. Diagnostics 2022, 12, 2860. https://doi.org/10.3390/diagnostics12112860
Seničar M, Roubinet B, Daniellou R, Prazuck T, Landemarre L. Lectin Analysis of SARS-CoV-2-Positive Nasopharyngeal Samples Using GLYcoPROFILE® Technology Platform. Diagnostics. 2022; 12(11):2860. https://doi.org/10.3390/diagnostics12112860
Chicago/Turabian StyleSeničar, Mateja, Benoît Roubinet, Richard Daniellou, Thierry Prazuck, and Ludovic Landemarre. 2022. "Lectin Analysis of SARS-CoV-2-Positive Nasopharyngeal Samples Using GLYcoPROFILE® Technology Platform" Diagnostics 12, no. 11: 2860. https://doi.org/10.3390/diagnostics12112860
APA StyleSeničar, M., Roubinet, B., Daniellou, R., Prazuck, T., & Landemarre, L. (2022). Lectin Analysis of SARS-CoV-2-Positive Nasopharyngeal Samples Using GLYcoPROFILE® Technology Platform. Diagnostics, 12(11), 2860. https://doi.org/10.3390/diagnostics12112860







