Abstract
There have been major developments in deep learning in computer vision since the 2010s. Deep learning has contributed to a wealth of data in medical image processing, and semantic segmentation is a salient technique in this field. This study retrospectively reviews recent studies on the application of deep learning for segmentation tasks in medical imaging and proposes potential directions for future development, including model development, data augmentation processing, and dataset creation. The strengths and deficiencies of studies on models and data augmentation, as well as their application to medical image segmentation, were analyzed. Fully convolutional network developments have led to the creation of the U-Net and its derivatives. Another noteworthy image segmentation model is DeepLab. Regarding data augmentation, due to the low data volume of medical images, most studies focus on means to increase the wealth of medical image data. Generative adversarial networks (GAN) increase data volume via deep learning. Despite the increasing types of medical image datasets, there is still a deficiency of datasets on specific problems, which should be improved moving forward. Considering the wealth of ongoing research on the application of deep learning processing to medical image segmentation, the data volume and practical clinical application problems must be addressed to ensure that the results are properly applied.
1. Introduction
Medical imaging has long functioned as an assistive means for diagnosis and treatment. Advancements in technology have increased the types and qualities of medical images. Lesion detection is one of the primary objectives of medical imaging, as the size and location of lesions are often directly associated with a patient’s diagnosis, treatment, and prognosis. Previously, the size and location of lesions were determined by radiologists through medical image examination. At best, the instruments and software used were only able to enhance the image quality by adjusting the brightness and contrast features to facilitate better observation. Since the development of computer vision algorithms, however, researchers have begun to utilize these algorithms in the field of medical imaging [1].
As the core of deep learning, convolutional neural networks (CNN) (Figure 1) have a considerably long development history [2]. However, due to hardware-related limitations, it was only in the 2010s that breakthroughs in the effectiveness of CNNs were made [3]. Meanwhile, deep learning models that meet specific targets have gradually been proposed, from classification models, to object detection, to object segmentation. Consequently, advancements such as the detection of lung diseases through X-ray [4], the detection of lesion locations [5], and segmentation have been applied in medical imaging. Due to improvements in model performance, deep learning models have exhibited diagnostic capabilities approximating those of clinical physicians, based on the inference of specific datasets. However, in traditional CNN models, there are limitations to segmentation. One of them is about the features extracted from the models. When using a smaller kernel, the features will become more local to the original image. Global information, such as location, may be lost. However, when using a larger kernel, the context of features may decrease [6]. Another limitation is the data availability of biomedical segmentation tasks. Due to privacy in the medical profession, the medical data volume is often small when compared with the data volume in other fields [6]. New models have been developed to solve these problems. Another solution is data augmentation.
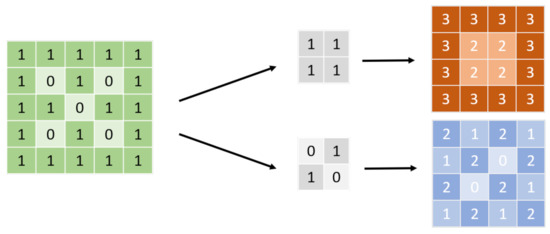
Figure 1.
CNN kernel. Before input, the figure will be transformed into a signal, as shown on the left; 0/1, for example. The arrays in the middle are called the convolutional kernel. The size of the kernel must not be larger than the input. The neural network applies several kernels with different weight compositions to the input to obtain feature maps, which are usually dot products, as shown on the right. The neural network extracts a feature that can make them accomplish the tasks from those kernels.
In this article, we will discuss the models proposed for semantic segmentation and data augmentation techniques, as well as their current applications on clinical data.
2. Fully Convolutional Network (FCN)
One of the most-selected models for segmentation tasks is FCN (Figure 2). The difference between FCNs and traditional convolutional neural networks is that the last layer is not a fully connected layer that enables the model to integrate information. Instead, the final number of output channels is modified through convolutional networks. The main benefit of this approach is that the model has no restrictions from the full connection layer, so the size of the input can be flexible. Table 1 shows the models used for semantic segmentation tasks.
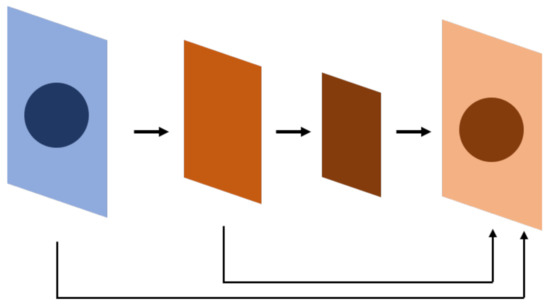
Figure 2.
FCN. The whole neural network is composed of a convolutional neural network, which is different from a conventional neural network, especially on the bottom 1~2 layers. The result is at a pixel level, so the network can afford a segmentation task. FCN uses a convolutional neural network to extract features and uses skip connections to find out the location of the feature at the whole-map level.

Table 1.
Models for segmentation tasks.
The U-Net is an example of an FCN (Figure 3) [6]. The U-Net leverages the extraction features of convolutional networks, in which the upsampling layer and concatenation are used to compare the features of the top layer while simultaneously retaining the features of the bottom layer. The model is thereby able to detect the detailed and general features of objects, thus detecting the location of objects. The U-Net can be referred to as a pixel-level image classification method, in which the segmentation is performed through pixel-wise classification. The U-Net architecture is briefly described as follows:
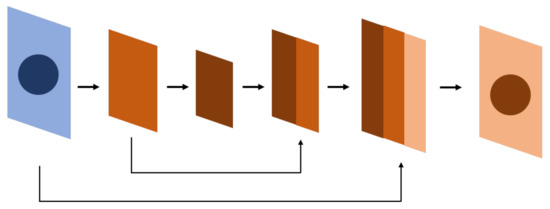
Figure 3.
U-Net, derived from FCN. The result from the FCN experiment shows that the inference will be more precise if the same level feature before downsampling is summed before the feature goes into upsampling. U-Net applies concatenation instead of summation; however, the summation is also used in some research.
- The descending part is also known as the encoding region. It is composed of units called convolutional blocks, which include convolutional and pooling layers and are akin to those of traditional convolutional neural networks and sometimes may include batch normalization layers. Before entering a pooling layer, features are filtered through an activation function that determines whether the extracted features should be transferred to the next layer. The most common activation functions are ReLu and sigmoid, expressed as the following equations:
- 2.
- The ascending part is also known as the decoding region. One of the keys of its architecture is that the size of a feature map is upscaled after passing through the upsampling layer. Another key is that, after upsampling, the feature will be concatenated with the feature retained at the corresponding level in the descending part. The ascending part mainly consists of alternating upsampling and pooling layers. In the upsampling layer, the following methods are usually used to upscale the features:
- Nearest neighbor interpolation: The value assigned is equal to that of the nearest pixel.
- Bilinear interpolation: The value is obtained through bilateral linear interpolation.
- Cubic interpolation: Third-degree polynomials are used to obtain values. Batch normalization, which can still be used, undergoes activation function processing before entering the upsampling layer.
- 3.
- Output layer: Before the result export, the feature reaching the top of the ascending part is processed. The layer is typically a single convolutional layer and usually makes predictions pixel by pixel. The Softmax function can be applied to the layer, as it generates the probability distribution of the classification. If the length and width of an original input image are x and y, respectively, then the output tensor size will be (x,y,c), with c representing the number of classes.
- 4.
- The loss function: Cross-entropy or the dice coefficient is often used. The respective math equations are as follows:
2.1. Modified U-Net Models
2.1.1. Modifying Encoder
The descending part of the U-Net architecture is structurally like the feature extraction process of CNN; hence, the feature extraction part of some CNN classification frameworks, such as VGG or ResNet, is used to replace the convolutional block of descending part. Jakhar et al. [13] applied a simple U-Net structure for segmentation and obtained considerably decent results. Abedalla et al. [8] applied the ResNet model to complete segmentation tasks.
2.1.2. Application of Residual Block
The residual block originates from ResNet (Figure 4a) [14,15]. The core concept is identity mapping. In the past, due to deep learning neural network stacking with convolutional layers, there is gradient loss during backpropagation, and the weights cannot be renewed effectively at the top layer. An identity map has two paths: one extracts features from the CNN layer-by-layer, as traditional CNN layers do, while the other skips those CNN layers and sums the feature extracted from the previous path before entering the next step. The latter path is also known as a skip connection because it does not pass through the stacked CNN layers. Features that pass through a skip connection are retained and extracted in the lower layers. During backpropagation, the skip connection can keep the gradient from vanishing or being lost.
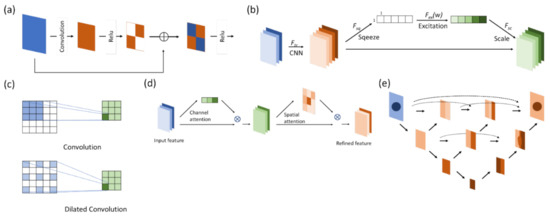
Figure 4.
The blocks or modules used in different neural networks for computer vision deep learning. (a) The residual block applies a skip connection to keep the gradients from vanishing or being lost during backpropagation. (b) The SE module assigns weights to each channel for representation and multiples them to the corresponding feature map to increase the robustness. (c) Dilated convolution expands the reception field to enhance its ability to process objects with blurred borders. (d) The attention module identifies the relationships between regional features so that it detects object borders more effectively. (e) U-Net++ block-fills the space between the encoder and the decoder with convolutional blocks to improve performance.
The encoder–decoder architecture of U-Net is also full of stacked CNN layers in those convolutional blocks. Even though the skip connection already existed in the original U-Net and allowed features to be fully or partially retained before concatenation, some studies have attempted to apply skip connections to other parts of the framework. For example, Isensee et al. [16] modified the original U-Net with skip connections. In the proposed model, there are two paths after reaching the next level layer in the descending part. One path goes through the upsampling, concatenation, and CNN process as the original U-Net did, while the other directly sums up the features of the upper CNN output. In this way, the sensitivity of the U-Net increases.
There was a study that exclusively examined the importance of partial skip connections [17]. In this context, a long skip connection crosses between the encoder–decoder architecture like a concatenated skip connection in the U-Net, while a short skip connection is similar to a residual block in a CNN block. The study found that short skip connections are more stable and efficient for weight updates, as they prevent the weight from hovering around extreme values so that the gradient can descend in a stable manner.
2.1.3. Application of Squeeze-and-Excitation Module
The squeeze-and-excitation (SE) (Figure 4b) module was first proposed by Hu et al. [18] in 2017. The idea of this module is that each channel may add a different contribution in a feature map. Therefore, weights are first submitted by each channel as a representation of their contribution, then they are multiplied by the feature map. In this way, the signals of more contributive channels are enhanced while others are suppressed.
Wang et al. [9] applied SE modules to both spatial and channel feature maps in U-Net by using DenseNet as a backbone. The mean pixel-wise accuracy and Dice similarity coefficient (DSC) were both over 90%.
2.1.4. Application of Dilated Convolution Network
A dilated convolution network (also known as an atrous convolution) (Figure 4c) expands the convolution receptive field while retaining the number of convolution kernels. The mathematic expression is as follows:
where y is the output of the dilated convolution, x is the input, and w is the kernel weight, which can be expressed as follows:
A schematic diagram is shown below. The significance of this process is to increase the receptive field of the CNN kernel to maintain the features of the larger region level as the model goes deeper. Combined with U-Net, this process enables the model to have a more precise prediction at the edge of the object and increases the robustness. A larger scale of dilated CNN kernels is used for learning the location of the edge in an object while smaller dilated CNN models are used for identifying the direction of the edge. When applied to an object with blurred edges, high-rated dilated CNN kernels can determine whether an object is blurry [19] while small-rated dilated CNN kernels can identify how blurred the object is. By doing so, the model can differentiate those similar blurry objects around the target.
Xiao et al. proposed a model that combined U-Net with a dilated CNN called MSDU-Net. In the model, features will be extracted from the input by the dilated CNN before entering U-shaped architecture. As the layer goes deeper, the rate of the dilated CNN (represented by r in Equation (5)) increases. As a result, the receptive field increases while the size of the feature decreases so that it can be the same size as the feature extracted from the upper layer of the encoder. The feature extracted from the upper layer of the encoder and extracted from the corresponding level of the dilated CNN will be concatenated and then transferred to the next level of the encoder [19].
In addition to detecting blurry edges, a dilated CNN kernel is capable of sharpening the edges of an object. Zhou et al. [10] utilized a dilated CNN to attenuate the noise from artifacts in medical imaging. Another study [20] applied dilated CNNs solely to increase the intersection over union (IoU).
2.1.5. Application of the Attention Module
The attention module (Figure 4d) was first used in deep learning models for natural language processing [21]. It figures out the correlation between the query and key to obtain the weight of the value corresponding to the key, and then it sums the weight of the value to get the attention. As a result, the attention module can capture the relationship between texts in a region or paragraph more precisely. The mathematical equation is expressed as follows:
Similar concepts are now used in the computer vision field. Applying attention modules makes the model combine the correlations of regional features, generating better object edge detection results to improve performance.
Yousefi et al. [11] applied the attention module, combined with a dilated CNN, to the U-Net. Attention modules were used within the convolutional block and between the skip connections. The model was tested on a chest CT dataset for esophagus segmentation tasks, and the AUC was 0.76. Cai et al. [12] presented a similar method in which an attention module is placed in the decoder part and is divided into channel attention and spatial attention to allow the model to capture the respective relevance from the channel attention and spatial attention. In a lung and esophageal dataset, the model achieved higher IOU than the U-Net and U-Net++ did. However, the computing speed, GPU memory, and gradient descent may be affected when there are more parameters.
2.1.6. UNet++
The main difference between U-Net++ (Figure 4e) and U-Net in terms of architecture is the design of the skip connection [22,23]. In U-Net++, instead of a long skip connection, the space between the encoder and the decoder is filled with dense convolutional blocks, as shown in Figure 2. The features transmitted between those convolutional blocks are like a decoder–encoder and in a single direction. Meanwhile, the skip connections are connected horizontally between these convolutional blocks, crossing up to three convolutional blocks at most. All these skip connections are horizontal and transmitted from the encoder to the decoder.
The computation of a convolutional block includes convolutions and activation functions. If a convolutional block is not on the far-left side or at the same location as the encoder in a traditional FCN (j > 0), the block will receive additional upsampling information from the convolutional block in the lower layer. With this arrangement, the neural network is akin to a combination of one encoder and multiple decoders. The equation is as follows (Z. Zhou et al. 2018 [23]):
Another difference between the U-Net++ and U-Net is deep supervision, which enables the model to operate in two different modes: accuracy mode and fast mode [23]. In both modes, the network generates an output (x^(0,i)) for each convolutional block in the top layer. In the accuracy mode, all outputs are averaged as the final output. In the fast mode, the model only selects one of the outputs as the final output. The selection is based on the speed of calculation and the size of the network selected by the user. This indicates that the network of the U-Net++ can be different between the training phase and the inference phase. The inference model can be pruned from the trained model according to a user’s needs. Deep supervision combines two loss functions—binary cross-entropy and the Dice coefficient—and is expressed as the following equation:
where Y and represent the actual and predicted probability distribution after flattening, respectively, and N represents the batch size. To sum up, U-Net++ has an encoder–decoder network filled with dense convolutional blocks, while different skip connections cross between the encoder–decoder network and these convolutional blocks so that the gradient flow could be improved; finally, deep supervision is achieved.
U-Net++ has been widely applied in several medical domains. Zhou et al. [22,23] performed a series of experiments using the U-Net++, including histopathological, clinical–medical, and endoscopic imaging. The IOU of the U-Net++ exceeds that of the U-Net by 3% to 5%, which is not affected by using various backbones (e.g., VGG, ResNet, and DenseNet). U-Net++ contains more parameters than U-Net, but it can decrease memory usage to speed up computation by sacrificing a little accuracy through deep supervision, which still makes it a valuable network.
2.2. Other FCNs
DeepLab
DeepLab (Figure 5a) was first proposed by [24]. The network consists of two parts: atrous convolution and a conditional random field (CRF). They offer solutions to two problems. The first problem is the gradual loss of features through the downsampling process, which can be dealt with using atrous convolution, as mentioned above.
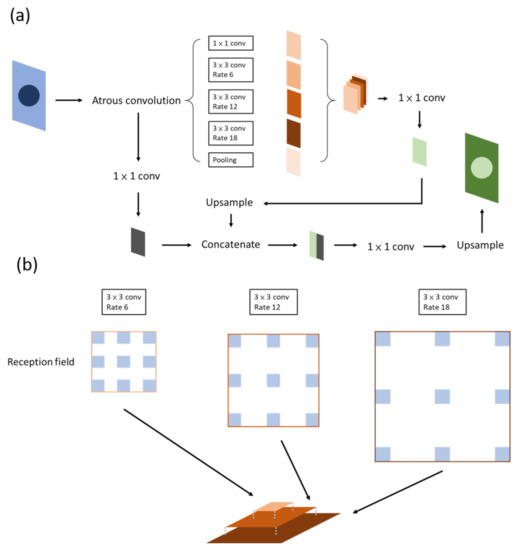
Figure 5.
Deeplab. (a) Version v1; the critical part of the network is the ASPP, or atrous convolution, which is shown in (b).
The other problem is maintaining the spatial stability to detect the centers of objects, which conflicts with the spatial accuracy of the deep convolutional neural network. The CRF combines the local features with classifiers to reinforce the edge detection ability of the model.
It has been claimed that DeepLab possesses three benefits: speed, accuracy, and simple architecture. DeepLab has evolved over time: v1, v2, v3, and, currently, v3+. Atrous spatial pyramid pooling (ASPP) (Figure 5b) is included in v2, in which features are stacked together after being filtered through atrous convolutional layers of different sizes. At present, this method not only allows different levels of features to be retained but the center of an object can be determined effectively as well [25].
In v3, the authors reviewed several common methods of semantic segmentation, such as encoder–decoder, pyramid network, and ASPP in v2. The entire architecture is readjusted first by deepening the convolutional layers, which are similar to ResNet, so as to obtain features near the image level. To ensure that contextual information will not be lost through striding in repeated convolution layers, atrous convolution is used instead when output_stride reaches a certain level. The authors then reviewed ASPP by applying the aforementioned method of increasing the layer number horizontally in the ASPP. A feature map is then concurrently extracted through convolutions with different sizes and rates, concatenated, and then passed through a 1 × 1 convolution before output. This method increases the mIOU to 70–80% [26]. The output_stride is associated with accuracy but takes up a considerable amount of computing resources.
DeepLab also possesses remarkable edge detection capabilities in medical images. Tang et al. performed a two-stage method to accomplish liver segmentation in abdominal CT scans [27]. A faster RCNN was used to detect the location of the liver, and the output was then put into DeepLab for the segmentation task. This approach yielded the lowest volumetric overlap error in the two datasets.
Wang et al. applied DeepLab v3+ to detect gastric cancer in pathology slides [28]. The results were compared with those from a SegNet and a faster RCNN. The sensitivity, specificity, and Dice coefficient (DC) were all over 90%; in addition, the DC from DeepLab v3+ was higher than that of those methods by 12%. Ahmed et al. [29] compared the performance in detecting breast cancers from mammography between DeepLab and mask-RCNN. Following the preprocessing step, in which muscles were filtered, both models achieved an area under the curve (AUC) of more than 0.95 and a mean average precision of 0.8 and 0.75, respectively. Therefore, the authors suggest that both models can be used to assist radiologists in effectively identifying breast cancer lesions through mammography.
3. Data Augmentation
3.1. Traditional Data Processing
Data augmentation (Figure 6) is one of the common methods of reinforcing model performance in deep learning. It allows machines to generate images like the original data, but independent of it—in the model’s view—so as to be used in training. Data augmentation increases the variety of data and reduces the gap between training and validation data to improve model robustness [30]. Rigid and non-rigid transformations are often used in data augmentation. The former includes shifting, rotation, and flipping; the latter includes scaling and shearing.
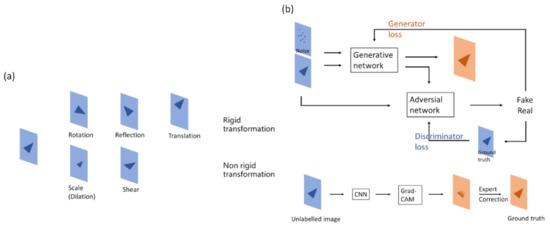
Figure 6.
Method for data augmentation. (a) Making changes to figures, the method can usually be separated as a rigid or non-rigid transformation. Rigid transformation geometrically changes the original image, while non-rigid transformation changes the size or shape with respect to the original image. (b) (Upper) The generative part of the GAN neural network yields figures that have similar properties, which can be used to increase the data volume. (Lower) Grad-CAM shows the heatmap of figures, which can be used for annotations, thus reducing manual workloads.
In medical imaging, rigid transformation is more often used for segmentation tasks than classification tasks. Because of class imbalance, it is better to retain the original image features for model convergence. The pattern of features might be changed in non-rigid transformations, which may lead to poor training results, especially with a small amount of data. In addition, if the left and right sides of an image should be detected by the model, flipping may confuse the model.
Owing to rare data, data augmentation is quite important in medical imaging training tasks. However, there is a paucity of studies that focus on the contribution of data augmentation [31,32,33].
3.2. Generating Data from Models
Another way to augment data is to use deep learning models. A model is trained to generate new data, which are not only transformed from the original data but also the corresponding annotations. Two methods are discussed here: generative adversarial network (GAN) and gradient-weighted class activation mapping (Grad-CAM).
3.2.1. Generating Data from GAN
A GAN consists of a generator network and a discriminator network. The aim of the generator network is to generate an image that is hard for the discriminator to differentiate from the real image, while the aim of the discriminator network is to differentiate the generated image from the real image. Through the adversarial process, the generator is able to create images that look almost real, and the discriminator is also able to detect the subtle difference between the real and generated images [34].
GANs have been applied in medical imaging tasks, including classification [35] and segmentation [36,37,38]. GAN can also be used in data augmentation. Maayan et al. [35] used GAN to generate CT images to improve data volume. The accuracy rose to 0.85, which was higher compared with typical data augmentation methods (0.75–0.8). Veit et al. [38] applied GAN to data augmentation to generate non-contrast CT images based on real-contrast CT images. The performance of the U-Net in segmentation rose from 0.916 to 0.932 (as represented by the Dice score).
There are some limitations to GAN, the biggest of which is convergence. The balance between the generator and the discriminator is difficult to maintain. For example, if the discriminator is overfitted, the generator will have difficulty obtaining its convergence [39]. Therefore, this may affect the quality of the data generated by the model. If the generated images have totally different features or patterns compared with the original data, the purpose of the data augmentation will be lost.
3.2.2. Generating Data from Grad-CAM
It has been a question since deep learning techniques first developed as to how neural networks actually learn. People have tried to figure out the answer by developing methods such as visualizing convolutional networks [40] or based on a gradient point of view [41]. The integrated architecture is an approach called Grad-CAM. A corresponding classification image, also known as a type of heatmap, is derived from the product of computing the weight and the above activation, passing through the ReLu activation of the last convolution layer [42]. This heatmap effectively displays the object to be classified. Therefore, neural networks are perceived to be able to discriminate according to “human thoughts”. From another perspective, Grad-CAM is more similar to judgment rules or the domain knowledge of humans, though the thinking process in the background might not necessarily be similar to that of a human.
The property of the heatmap in Grad-CAM, which matches targets, can be used to help annotate in segmentation. Grad-CAM is used to obtain the approximate location and size of an object, followed by manual modification, which makes annotation more efficient [43].
4. Clinical Datasets and Relevant Studies
4.1. Lung Lesions
Some chest lesions have been studied, including lung lesions such as pneumothorax, lung nodules, pneumonia, cardiac lesions such as ventricle hypertrophy, and bony lesions such as rib fractures. Except for rib fractures, there are obvious targets for segmentation in these lesions, along with open datasets [44,45,46] for constructing a pretrained model so that the training for those tasks is more likely to succeed.
Table 2 shows research on lung lesion segmentation. Singadkar et al. [47] applied an FCN-based neural network, combined with residual blocks in the decoder section and long skip connections between the encoder and decoder, for lung nodule segmentation in CT scan images. They successfully reached an average Dice score of 0.95 and a Jaccard index of 0.887. Abedalla et al. [8] utilized multiple U-Net models with different backbones in each network for training and used a method similar to ensemble learning, in which four models are first summated according to fixed weights and then subjected to a threshold in order to accomplish segmentation via a pneumothorax during in inference phase. The weights and thresholds are manually adjusted. The network achieved a DSC of 0.86 in the 2019 Pneumothorax Challenge dataset.

Table 2.
Research on lung lesion segmentation.
4.2. Brain Lesions
Brain lesion detection includes brain tumors, strokes, traumatic brain injuries, and brain metastases. BraTS [56] (Figure 7a) is a brain tumor dataset with labels not only the location and size but also the cell type of tumors, primarily low-grade and high-grade gliomas. Magnetic resonance imaging (MRI) scans are divided into pretreatment and posttreatment images. In addition, each patient is scanned via instruments with varying magnetic field intensities (1.5 and 3 T) and protocols (2D and 3D). There are four major types of MR images: T1, T1c, T2, and FLAIR. The tumor edge is difficult to identify in segmentation tasks because of infiltrations, particularly those of high-grade gliomas, and the variety of degrees of contrast enhancement across different MRI scans.
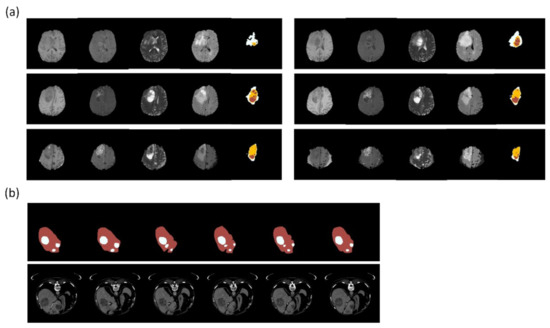
Figure 7.
Clinical dataset for training segmentation model. (a) BraTS is a dataset for glioblastoma multiforme. The figures show part of a case series with different MRI weights (from the left: T1, T1ce, T2, FLAIR) and annotations (rightmost: white—perifocal edema; yellow—tumor; red—necrosis). (b) LiTSis a dataset about liver and liver tumor segmentation. The figures show part of a case series with annotations (upper: red—liver; white—tumor) and CT images (lower). Reference: (a) from BRATS (Menze et al. [25,56,57]); (b) from LiTS (Bilic et al. [58]). The figures were illustrated by using Python 3.6 from the datasets.
Table 3 shows research on segmenting brain lesions. Isensee et al. [16] attempted to modify the structure of the U-Net architecture by using batch normalization and short skip connections such as s residual block in ResNet instead of a traditional convolutional block. Finally, they summated the outputs of each layer in the ascending part before entering the output part. The Dice coefficient was superior to that of the traditional U-Net architecture. In summary, most of the leading models in the BraTS dataset over the years have been based on U-Net architecture. Some of them have been modified from convolutional blocks, while others have been adjusted at the ascending part.
The intensity of stroke lesions in CT images can change over time after examination, especially infarction strokes. [59] In addition to CT, MRI datasets such as ISLES [60] have been established in recent years. Models trained with those datasets not only determine the location and region of stroke lesions but also facilitate physicians to determine the severity of brain damage and may predict the prognosis and potential of recovery. Zhang et al. [61] developed a multi-plane neural network structure to segment stroke lesions from diffusion-weighted magnetic resonance images. In contrast with the direct usage of 3D neural networks, they applied three neural networks that correspond to images on three different planes—axial, coronal, and sagittal—then integrated them into a multi-angle neural network, which is called a multi-plane fusion network. This neural network offers both segmentation and detection functions and can retain the original information from the input. Based on images from three different planes, the edges of lesions can be identified more accurately. The authors achieved a Dice coefficient of 62.2% and a sensitivity of 71.7% in the ISLES dataset.

Table 3.
Research on brain lesion segmentation.
Table 3.
Research on brain lesion segmentation.
| Authors (Year) | Method | Medical Image | Performance | Notes |
|---|---|---|---|---|
| Brain tumor | ||||
| Havaei et al. [62] (2016) | Deep CNN | Magnetic resonance images | DC 1: 0.88 | Cascade architecture using pre-output concatenation |
| Pereira et al. [63] (2016) | CNN-based | Magnetic resonance images | DC: 0.88 | Patch extraction from an image before entering the CNN |
| Isensee et al. [16] (2018) | 3D U-Net | Magnetic resonance images | DC: 0.85 | Modified from U-Net; summation for multi-level features |
| Xu et al. [33] (2020) | U-Net | Magnetic resonance images | DC: 0.87 | Attention-U-Net |
| McKinley et al. [64] (2018) | deepSCAN | Magnetic resonance images | Mean DC ET 2: 0.7 WT 3: 0.86 TC 4: 0.71 | Bottleneck CNN design; dilated convolution |
| Stroke | ||||
| Wang et al.[65] (2016) | Deep Lesion Symmetry ConvNet | Magnetic resonance images | Mean DSC 5: 0.63 | Combined unilateral (local) and bilateral (global) voxel descriptor |
| Monteiro et al. [66] (2020) | DeepMedic | Computed tomography | Differs according to size | Three parallel 3D CNNs for different resolutions |
| Zhang et al. [61] (2020) | U-Net | Magnetic resonance images | DSC: 0.62 IoU 6: 0.45 | FPN for extraction first |
1 Dice coefficient, 2 enhanced tumor, 3 whole tumor, 4 tumor core, 5 Dice similarity coefficient, 6 intersection over union.
4.3. Abdomen
Abdominal Organ Segmentation
The solid organs in the abdomen such as the liver, kidneys, spleen, and pancreas, as well as lower abdomen organs such as the prostate, have more prominent edges and distinct intensity values compared with the background, which is usually fat or peritoneum. Thus, they are obvious targets for segmentation. Convincing results could be achieved with traditional computer vision techniques [67,68]. Regarding the urinary bladder, due to its prominent edges, despite being a hollow organ, segmentation tasks could still be accomplished with trained models (particularly in the case of a distended bladder) [69]. There is a wealth of data focusing on abdominal organ segmentation [70,71,72]. In recent years, the application of deep learning for segmentation tasks has been considerably robust [73,74].
5. Abdominal Lesion Segmentation
Relevant studies on abdominal tumor segmentation mostly focus on the liver, kidneys, prostate, and urinary bladder. Manjunath et al. [75] trained a U-Net model with ResNet as the backbone to segment the liver and liver tumors from CT images and achieved a DSC of 96.35 and an accuracy of 99.71% in the liver tumor segmentation (LiTS) dataset [58] (Figure 7b). Vorontsov et al. [76] developed an FCN network trained from LiTS and a local dataset and made inferences to segment metastatic liver lesions from colorectal cancers in another set of CT scan images from several local hospitals. Despite experiencing limitations such as small data volume and the wide spread of patient origins, the model achieved a positive predictive value of over 80% for lesions larger than 1 cm. Liang et al. [77] applied a square-window CNN to segment pancreatic cancers. The model was trained from the multi-phased MRIs of 27 patients, with extraction and augmentation from 245,000 normal and 230,000 abnormal patch images. The model was tested on the images of 13 patients, and it achieved a DSC of 0.73, thus solidifying it as an assistive means for radiation oncologists to ascertain the gross tumor volume. Pellicer-Valero et al. [78] purposed a Retina U-Net trained from two MRI datasets to analyze the prostate and tumors and to predict their Gleason grade group. Chen et al. [79] applied the 3D AlexNet for prostate cancer segmentation and achieved an accuracy of 0.92 and a DSC of 0.977.
In 2016, Cha et al. [80] conducted a pilot study to evaluate whether deep-learning neural networks can segment urinary bladder tumors in CT images and further assess the treatment outcomes. The results showed that the assessment of tumor size changes from the model was not inferior to those derived from humans according to the World Health Organization (WHO) and the Response Evaluation Criteria in Solid Tumors (RECIST) guidelines [81]. The authors subsequently conducted a study that combines radiomic feature analysis [82], whereby lesions are segmented by the deep learning model to determine changes in tumor size in contrast-enhanced CTs before and after treatment. Conclusively, the method offers higher accuracy for determining tumor volume, but it is not necessarily superior to human judgment for determining treatment efficacy. Nonetheless, the model can be used as an effective tool for radiologists in diagnosis. Table 4 summarizes the research on the segmentation of abdomens mentioned above.
Physicians choose different tools for examination according to the location of tumors and, therefore, obtain quite different images from traditional image scopes. For example, endoscopy is used for gastrointestinal tumors. Unlike CT or MRI images presenting the lesion from a general anatomy point of view, endoscopic images only present the lesion in the regional field via the scope passing through the lumen, such as when using endoscopic ultrasound (EUS) to diagnose pancreatic tumors. The endoscopic images are generated in real-time, so they are like streams instead of slices of images. Physicians can still get snapshots that they are interested in, but, in general, it has no regular interval between the snapshot images as CT or MRI images do. For this reason, it is better to perform tumor segmentation in real-time, as with the stream, where the results are displayed to the physician. Li et al. [83] trained a U-Net to segment EUS-observed GIST tumors, which are derived from the gastric stroma. The authors made some modifications to the model to address the problems of shadowing and size differences, which often appear in EUS imaging. The model achieved a DSC of over 0.9 in the test data. Tonozuka et al. [84] used a U-Net to segment pancreatic tumors in EUS and achieved a median IoU of 0.77. The results were affected by unclear tumor margins but not by respiratory movements.

Table 4.
Research on abdomen segmentation.
Table 4.
Research on abdomen segmentation.
| Authors (Year) | Method | Medical Image | Performance | Notes |
|---|---|---|---|---|
| Abdominal organ | Differs between organs | |||
| Landman et al. [85] (2018) | FCN | Magnetic resonance images | DSC 1: 0.56–0.93 | |
| Gibson et al. [86] (2018) | Dense V-Networks | Computed tomography | DC 2: 0.76–0.9 | High-resolution activation maps; batch-wise spatial dropout |
| Kim et al. [73] (2020) | 3D U-Net Atlas-based | Computed tomography | DC: 0.60–0.96 DC: 0.15–0.81 | Multi-organs were tested; the U-Net result could be comparable to that of an interobserver |
| Kart et al. [74] (2021) | nn-UNet | Magnetic resonance images | DC: 0.82–0.9 | |
| Abdominal tumor | ||||
| Abdel-Massieh et al. [87] (2010) | CV 3 method | Computed tomography | Overlap error: 0.22 | Gaussian blurring; isodata threshold |
| Abd-Elaziz et al. [88] (2014) | CV method | Computed tomography | Error rate: 0.002–0.012 | Regional pixel growing and morphological processing |
| Manjunath et al. [75] (2021) | ResUNet | Computed tomography | DSC: 0.96 | Replacing convolutional blocks with residual blocks |
| Vorontsov et al. [76] (2019) | FCN | Computed tomography | DSC per lesion: (automated) <10 mm: 0.14 10–20mm: 0.53 >20 mm: 0.68 | Two-step segmentation: the first is FCN for livers, and the second is FCN for lesions in livers |
| Liang et al. [77] (2020) | Square-window based CNN | Magnetic resonance images | DSC: 0.73 on the test set | |
| Pellicer-Valero et al. [78] (2021) | Retina U-Net | Magnetic resonance images | DSC: (prostate) 0.915 | Two 3D CNNs: the first one takes a T2-weighted MRI as the input, and the second one takes an MRI and the output from the first one as inputs |
| Chen et al. [79] (2020) | 3D AlexNet | Magnetic resonance images | DSC: 0.97 | |
| Li et al. [83] (2020) | MRBS-U-Net | Endoscopic ultrasound | DSC: 0.92 |
1 Dice similarity coefficient, 2 Dice coefficient, 3 computer vision.
6. Discussion
There have been major advancements in the application of deep learning for medical imaging segmentation since the development of the U-Net architecture. Meanwhile, FCN-related studies and techniques are making the model more robust. We think there are two major points to make sure the model is successfully trained:
(1) Data. Sufficient data are necessary for model convergence. In addition, well-defined task objectives lead to clear data labeling, while domain knowledge is critical to maintaining label consistency. To prevent class imbalance, labeled data should be sufficient and should not be skewed toward a particular class [89]. However, class imbalance is a common problem in semantic segmentation. U-Net may overcome this problem [6]. While researchers have investigated other models or approaches to deal with data problems, the best way is still to ensure the data structure [90,91,92], which requires the assistance of data scientists or software to evaluate the distribution of the class of the label. The data quality should also be involved. To achieve this, the production of data should be consistent, and unnecessary interferences and irrelevant information should be removed so that the model will not learn from the information that has nothing to do with what the model should learn during training.
(2) Model type. Due to the nature of segmentation tasks, full convolution networks are currently the solution. Edge detection of the object may be a key point for future developments. In addition to model architecture, loss function is also important. The literature has shown that, in addition to model structure modifications, adjustments of loss function have also been used to raise the performance of the model [22,23].
On the other hand, from the perspective of clinical medicine, there are still some obstacles to research:
(1) Poor data volume. Patients are a relatively small population compared with the whole human population. In addition, data security and privacy policies lead to insufficient data influence, making it challenging to establish an effective dataset.
(2) Inconsistency in data quality between medical institutes. Due to differences between examination procedures, instruments, and image processing methods, the images generated by those institutes may vary in terms of contrast, brightness, or resolution. These issues may occur between and even within medical institutes.
(3) The low reliability of machine-assisted diagnostic tools is perceived by clinical workers. The results of previous research suggest that making image-based diagnoses using deep learning models is not inferior to experienced physicians or experts in certain circumstances. Moreover, the model may even effectively compensate for less-experienced clinicians [93]. Nevertheless, there are different procedures and protocols in each medical department, which results in two issues: Whether a model can be seamlessly integrated into an environment, and whether the system can be acceptable with an inserted model. The solutions to these issues hinge on the collaboration between medical professionals, information professionals, and researchers. The inclusion of multidisciplinary professionals is crucial for expediting the process.
One of the directions for future developments in semantic segmentation is federated learning, which was initially an approach proposed by Google for smartphone keyboard input prediction [94]. Numerous medical departments can train the model on their own dataset to contribute weights to the final model. The data do not need to pass through a platform, so the patient information is secure, and the model is able to obtain diverse and large data volumes as well. However, there may be some problems. One of them is the variety of data volumes. Medical institutes that can provide larger data volumes would make greater contributions to the model. If the gap in data volumes between institutes is too large, such that most of the data are provided from one or only a few institutes, the model will have a similar or even worse performance than those only trained on their own dataset, which will lower their willingness to participate. Another problem is cost-effectiveness. The weights are contributed by each medical institute, and how the gradient is backpropagated significantly affects the performance. By using general averaging methods, hospitals possessing higher data volumes may contribute more weights, which results in a model that is biased toward hospitals, affecting the model’s generalization. By using other methods, it may benefit only those providing fewer data instead. Since those institutes owning larger data volumes will be more likely to make larger investments in maintaining the dataset, the cost-effectiveness problem will become a key concern. The differences between the imaging qualities of each medical institute will also raise concerns about the performance of generalization [95]. Thus, there are still some obstacles needed to be overcome in federated learning.
7. Conclusions
In this review, we reviewed several FCN models for their applications in medical image segmentation. There have been an increasing number of advancements since the development of U-Net: some frameworks deal with blurred objects, and other frameworks are good at detecting object burdens.
Despite the lower accessibility of medical data, they are still applicable to train models with critical data augmentation. We summarized two main skills for data augmentation: figure transformation and the GAN network. Although Grad-CAM may not generate figures from original images, it can assist in labeling by generating heatmaps of images.
Furthermore, we also reviewed studies related to model performance in clinical datasets. Generally, the proposed models showed good performance in testing datasets, and even in some clinical images collected by researchers. However, model performance remains a challenge after deployment. To overcome the gap between the dataset performance and clinical practice, more local data should be collected to train new models or to perform transfer learning with pre-trained models. In addition, it is necessary to constantly improve the model through cooperation with data scientists, data analysts, and clinical practitioners.
Author Contributions
Conceptualization, D.-W.L., S.-Y.H., and R.-J.H.; methodology, W.-L.H.; writing—original draft preparation, S.-Y.H. and R.-J.H.; writing—review and editing, W.-L.H. and D.-W.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Buddhist Tzu Chi Medical Foundation, Grant number TCRD-110-15, IMAR-110-01-08, TCRD111-67, TCRD111-75 and the APC was funded by Buddhist Tzu Chi Medical Foundation.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Figure 7a: BRATS. This is an open dataset; the website is as follows:http://braintumorsegmentation.org/ (accessed on 8 October 2022). Figure 7b: LiTS. This is an open dataset; the website is as follows: https://www.kaggle.com/datasets/andrewmvd/liver-tumor-segmentation (accessed on 3 October 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cootes, T.F.; Taylor, C.J. Statistical Models of Appearance for Medical Image Analysis and Computer Vision. Med. Imaging 2001, 4322, 236–249. [Google Scholar] [CrossRef]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-Based Learning Applied to Document Recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Hwang, S.; Kim, H.-E.; Jeong, J.; Kim, H.-J. A novel approach for tuberculosis screening based on deep convolutional neural networks. Med. Imaging 2016, 9785, 750–757. [Google Scholar] [CrossRef]
- Dou, Q.; Chen, H.; Yu, L.; Qin, J.; Heng, P.A. Multilevel Contextual 3-D CNNs for False Positive Reduction in Pulmonary Nodule Detection. IEEE Trans. Biomed. Eng. 2017, 64, 1558–1567. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef]
- Ciresan, D.; Giusti, A.; Gambardella, L.; Schmidhuber, J. Deep Neural Networks Segment Neuronal Membranes in Electron Microscopy Images. Adv. Neural Inf. Process. Syst. 2012, 25. [Google Scholar]
- Abedalla, A.; Abdullah, M.; Al-Ayyoub, M.; Benkhelifa, E. Chest X-ray pneumothorax segmentation using U-Net with EfficientNet and ResNet architectures. PeerJ Comput. Sci. 2021, 7, e607. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, Q.; Luo, G.; Liu, Z.; Huang, J.; Zhou, Y.; Zhou, Y.; Xu, W.; Cheng, J.-Z. Automated segmentation and diagnosis of pneumothorax on chest X-rays with fully convolutional multi-scale ScSE-DenseNet: A retrospective study. BMC Med. Informatics Decis. Mak. 2020, 20, 317. [Google Scholar] [CrossRef]
- Zhou, H.; Zhu, Y.; Wang, Q.; Xu, J.; Li, G.; Chen, D.; Dong, Y.; Zhang, H. Multi-Scale Dilated Convolution Neural Network for Image Artifact Correction of Limited-Angle Tomography. IEEE Access 2020, 8, 1567–1576. [Google Scholar] [CrossRef]
- Yousefi, S.; Sokooti, H.; Elmahdy, M.S.; Lips, I.M.; Shalmani, M.T.M.; Zinkstok, R.T.; Dankers, F.J.W.M.; Staring, M. Esophageal Tumor Segmentation in CT Images Using a Dilated Dense Attention Unet (DDAUnet). IEEE Access 2021, 9, 99235–99248. [Google Scholar] [CrossRef]
- Cai, Y.; Wang, Y. MA-Unet: An improved version of Unet based on multi-scale and attention mechanism for medical image segmentation. In Proceedings of the Third International Conference on Electronics and Communication; Network and Computer Technology (ECNCT 2021), Harbin, China, 3–5 December 2021. [Google Scholar] [CrossRef]
- Jakhar, K.; Kaur, A.; Gupta, M. Pneumothorax Segmentation: Deep Learning Image Segmentation to Predict Pneumothorax. arXiv 2019. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In European Conference on Computer Vision; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2016; Volume 9908, pp. 630–645. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Isensee, F.; Kickingereder, P.; Wick, W.; Bendszus, M.; Maier-Hein, K.H. Brain tumor segmentation and radiomics survival prediction: Contribution to the BRATS 2017 challenge. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 10670, pp. 287–297. [Google Scholar] [CrossRef]
- Drozdzal, M.; Vorontsov, E.; Chartrand, G.; Kadoury, S.; Pal, C. The Importance of Skip Connections in Biomedical Image Segmentation. In Deep Learning and Data Labeling for Medical Applications; Springer: Cham, Switzerland, 2016; Volume 10008, pp. 179–187. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Xiao, X.; Yang, F.; Sadovnik, A. Msdu-net: A multi-scale dilated u-net for blur detection. Sensors 2021, 21, 1873. [Google Scholar] [CrossRef] [PubMed]
- Piao, S.; Liu, J. Accuracy Improvement of Unet Based on Dilated Convolution. J. Phys. Conf. Ser. 2019, 1345, 052066. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans. Med. Imaging 2020, 39, 1856–1867. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2018; Volume 11045, pp. 3–11. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S.; et al. The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification. arXiv 2021, arXiv:2107.02314. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv 2017. [Google Scholar] [CrossRef]
- Tang, W.; Zou, D.; Yang, S.; Shi, J. DSL: Automatic Liver Segmentation with Faster R-CNN and Deeplab. In Artificial Neural Networks and Machine Learning; Springer International Publishing: Cham Switzerland, 2018; Volume 11140. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X. Medical image recognition and segmentation of pathological slices of gastric cancer based on Deeplab v3+ neural network. Comput. Methods Programs Biomed. 2021, 207, 106210. [Google Scholar] [CrossRef]
- Ahmed, L.; Iqbal, M.M.; Aldabbas, H.; Khalid, S.; Saleem, Y.; Saeed, S. Images data practices for Semantic Segmentation of Breast Cancer using Deep Neural Network. J. Ambient Intell. Humaniz. Comput. 2020. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Eaton-Rosen, Z.; Bragman, F.; Ourselin, S.; Cardoso, M.J. Improving Data Augmentation for Medical Image Segmentation. In Proceedings of the 1st Conference on Medical Imaging with Deep Learning, Amsterdam, The Netherlands, 4–6 July 2018. [Google Scholar]
- Abdollahi, B.; Tomita, N.; Hassanpour, S. Data Augmentation in Training Deep Learning Models for Medical Image Analysis. In Deep Learners and Deep Learner Descriptors for Medical Applications; Springer: Cham, Switzerland, 2020; Volume 186, pp. 167–180. [Google Scholar] [CrossRef]
- Xu, J.; Li, M.; Zhu, Z. Automatic data augmentation for 3d medical image segmentation. In Medical Image Computing and Computer-Assisted Intervention 2020; Lecture Notes in Computer Science; Springer Science and Business Media Deutschland GmbH: Berlin/Heidelberg, Germany, 2020; Volume 12261, pp. 378–387. [Google Scholar] [CrossRef]
- Creswell, A.; White, T.; Dumoulin, V.; Arulkumaran, K.; Sengupta, B.; Bharath, A.A. Generative adversarial networks: An overview. IEEE Signal Process. Mag. 2018, 35, 53–65. [Google Scholar] [CrossRef]
- Frid-Adar, M.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. Synthetic data augmentation using GAN for improved liver lesion classification. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 289–293. [Google Scholar] [CrossRef]
- Zhao, C.; Han, J.; Jia, Y.; Gou, F. Lung nodule detection via 3D U-net and contextual convolutional neural network. In Proceedings of the 2018 International Conference on Networking and Network Applications (NaNA), Xi’an, China, 12–15 October 2018; pp. 356–361. [Google Scholar] [CrossRef]
- Rezaei, M.; Yang, H.; Harmuth, K.; Meinel, C. Conditional generative adversarial refinement networks for unbalanced medical image semantic segmentation. In Proceedings of the 2019 IEEE Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 7–11 January 2019; pp. 1836–1845. [Google Scholar] [CrossRef]
- Sandfort, V.; Yan, K.; Pickhardt, P.J.; Summers, R.M. Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Sci. Rep. 2019, 9, 16884. [Google Scholar] [CrossRef]
- Barnett, S.A. Convergence Problems with Generative Adversarial Networks (GANs). arXiv 2018. [Google Scholar] [CrossRef]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. In Proceedings of the 2nd International Conference on Learning Representations, ICLR 2014-Workshop Track Proceedings, Banff, AB, Canada, 14–16 April 2014; pp. 1–8. [Google Scholar]
- Selvaraju, R.R.; Chattopadhyay, P.; Elhoseiny, M.; Sharma, T.; Batra, D.; Parikh, D.; Lee, S. Choose Your Neuron: Incorporating Domain Knowledge through Neuron-Importance. In Computer Vision–ECCV 2018; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11217, pp. 540–556. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef]
- Shi, X.; Khademi, S.; Li, Y.; van Gemert, J. Zoom-Cam: Generating fine-grained pixel annotations from image labels. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 10289–10296. [Google Scholar] [CrossRef]
- LUNA16 Dataset|Papers With Code. Available online: https://paperswithcode.com/dataset/luna16 (accessed on 26 December 2021).
- Pedrosa, J.; Aresta, G.; Ferreira, C.; Rodrigues, M.; Leitão, P.; Carvalho, A.S.; Rebelo, J.; Negrão, E.; Ramos, I.; Cunha, A.; et al. LNDb: A Lung Nodule Database on Computed Tomography. arXiv 2019. [Google Scholar] [CrossRef]
- SIIM-ACR Pneumothorax Segmentation|Kaggle. Available online: https://www.kaggle.com/c/siim-acr-pneumothorax-segmentation (accessed on 1 January 2022).
- Singadkar, G.; Mahajan, A.; Thakur, M.; Talbar, S. Deep Deconvolutional Residual Network Based Automatic Lung Nodule Segmentation. J. Digit. Imaging 2020, 33, 678–684. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, M.; Liu, Z.; Liu, Z.; Gu, D.; Zang, Y.; Dong, D.; Gevaert, O.; Tian, J. Central focused convolutional neural networks: Developing a data-driven model for lung nodule segmentation. Med. Image Anal. 2017, 40, 172–183. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, M.; Gevaert, O.; Tang, Z.; Dong, D.; Liu, Z.; Jie, T. A multi-view deep convolutional neural networks for lung nodule segmentation. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Korea, 11–15 July 2017; pp. 1752–1755. [Google Scholar] [CrossRef]
- Maqsood, M.; Yasmin, S.; Mehmood, I.; Bukhari, M.; Kim, M. An Efficient DA-Net Architecture for Lung Nodule Segmentation. Mathematics 2021, 9, 1457. [Google Scholar] [CrossRef]
- Meraj, T.; Rauf, H.T.; Zahoor, S.; Hassan, A.; Lali, M.I.; Ali, L.; Bukhari, S.A.C.; Shoaib, U. Lung nodules detection using semantic segmentation and classification with optimal features. Neural Comput. Appl. 2021, 33, 10737–10750. [Google Scholar] [CrossRef]
- Usman, M.; Lee, B.-D.; Byon, S.S.; Kim, S.-H.; Lee, B.-i.; Sho, I.G. Volumetric lung nodule segmentation using adaptive ROI with multi-view residual learning. Sci. Rep. 2020, 10, 12839. [Google Scholar] [CrossRef] [PubMed]
- Keetha, N.V.; Babu P, S.A.; Annavarapu, C.S.R. U-Det: A Modified U-Net architecture with bidirectional feature network for lung nodule segmentation. arXiv 2020. [Google Scholar] [CrossRef]
- Ozdemir, O.; Russell, R.L.; Berlin, A.A. A 3D Probabilistic Deep Learning System for Detection and Diagnosis of Lung Cancer Using Low-Dose CT Scans. arXiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Hesemian, M.H.; Jia, W.; He, X.; Kennedy, P.J. Atrous Convolution for Binary Semantic Segmentation of Lung Nodule. ICASSP 2019-2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1015–1019. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Nat. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef]
- Bilic, P.; Christ, P.F.; Vorontsov, E.; Chlebus, G.; Chen, H.; Dou, Q.; Fu, C.-W.; Han, X.; Heng, P.-A.; Hesser, J.; et al. The Liver Tumor Segmentation Benchmark (LiTS). arXiv 2019. [Google Scholar] [CrossRef]
- Norton, G.A.; Kishore, P.R.; Lin, J. CT contrast enhancement in cerebral infarction. AJR Am. J. Roentgenol. 1978, 131, 881–885. [Google Scholar] [CrossRef]
- Liew, S.-L.; Anglin, J.M.; Banks, N.W.; Sondag, M.; Ito, K.L.; Kim, H.; Chan, J.; Ito, J.; Jung, C.; Khoshab, N.; et al. A large, source dataset of stroke anatomical brain images and manual lesion segmentations. Sci. Data 2018, 5, 180011. [Google Scholar] [CrossRef]
- Zhang, L.; Song, R.; Wang, Y.; Zhu, C.; Liu, J.; Yang, J.; Liu, L. Ischemic Stroke Lesion Segmentation Using Multi-Plane Information Fusion. IEEE Access 2020, 8, 45715–45725. [Google Scholar] [CrossRef]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.-M.; Larochelle, H. Brain Tumor Segmentation with Deep Neural Networks. arXiv 2015. [Google Scholar] [CrossRef]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- McKinley, R.; Jungo, A.; Wiest, R.; Reyes, M. Pooling-Free Fully Convolutional Networks with Dense Skip Connections for Semantic Segmentation, with Application to Brain Tumor Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2017; Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2017; Volume 10670. [Google Scholar] [CrossRef]
- Wang, Y.; Katsaggelos, A.K.; Wang, X.; Parrish, T.B. A deep symmetry convnet for stroke lesion segmentation. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016; pp. 111–115. [Google Scholar] [CrossRef]
- Monteiro, M.; Newcombe, V.F.J.; Mathieu, F.; Adatia, K.; Kamnitsas, K.; Ferrante, E.; Das, T.; Whitehouse, D.; Rueckert, D.; Menon, D.K.; et al. Multiclass semantic segmentation and quantification of traumatic brain injury lesions on head CT using deep learning: An algorithm development and multicentre validation study. Lancet Digit. Health 2020, 2, e314–e322. [Google Scholar] [CrossRef]
- Okada, T.; Linguraru, M.G.; Hori, M.; Suzuki, Y.; Summers, R.M.; Tomiyama, N.; Sato, Y. Multi-Organ Segmentation in Abdominal CT Images. In Proceedings of the 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Diego, CA, USA, 28 August–1 September 2012; pp. 3986–3989. [Google Scholar] [CrossRef]
- Jawarneh, M.; Rajeswari, M.; Ramachandram, D.; Lutfi, I. Segmentation of abdominal volume dataset slices guided by single annotated image. In Proceedings of the 2009 2nd International Conference on Biomedical Engineering and Informatics, Tianjin, China, 17–19 October 2009; pp. 4–8. [Google Scholar] [CrossRef]
- Cha, K.H.; Hadjiiski, L.; Samala, R.K.; Chan, H.P.; Caoili, E.M.; Cohan, R.H. Urinary bladder segmentation in CT urography using deep-learning convolutional neural network and level sets. Med. Phys. 2016, 43, 1882–1896. [Google Scholar] [CrossRef] [PubMed]
- MICCAI 2015 Multi-Atlas Abdomen Labeling Challenge. Available online: https://paperswithcode.com/dataset/miccai-2015-multi-atlas-abdomen-labeling (accessed on 4 November 2022).
- Rister, B.; Yi, D.; Shivakumar, K.; Nobashi, T.; Rubin, D.L. Ct-ORG, a new dataset for multiple organ segmentation in computed tomography. Sci. Data 2020, 7, 381. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhang, Y.; Gu, S.; Zhu, C.; Ge, C.; Zhang, Y.; An, X.; Wang, C.; Wang, Q.; Liu, X.; et al. AbdomenCT-1K: Is Abdominal Organ Segmentation a Solved Problem. IEEE Trans. Pattern Anal. Mach. Intell. 2021. [Google Scholar] [CrossRef]
- Kim, H.; Jung, J.; Kim, J.; Cho, B.; Kwak, J.; Jang, J.Y.; Lee, S.-W.; Lee, J.-G.; Yoon, S.M. Abdominal multi-organ auto-segmentation using 3D-patch-based deep convolutional neural network. Sci. Rep. 2020, 10, 6204. [Google Scholar] [CrossRef]
- Kart, T.; Fischer, M.; Küstner, T.; Hepp, T.; Bamberg, F.; Winzeck, S.; Glocker, B.; Rueckert, D.; Gatidis, S. Deep Learning-Based Automated Abdominal Organ Segmentation in the UK Biobank and German National Cohort Magnetic Resonance Imaging Studies. Investig. Radiol. 2021, 56, 401–408. [Google Scholar] [CrossRef]
- Manjunath, R.V.; Kwadiki, K. Automatic liver and tumour segmentation from CT images using Deep learning algorithm. Results Control Optim. 2022, 6, 100087. [Google Scholar] [CrossRef]
- Vorontsov, E.; Cerny, M.; Régnier, P.; Di Jorio, L.; Pal, C.J.; Lapointe, R.; Vandenbroucke-Menu, F.; Turcotte, S.; Kadoury, S.; Tang, A. Deep Learning for Automated Segmentation of Liver Lesions at CT in Patients with Colorectal Cancer Liver Metastases. Radiol. Artif. Intell. 2019, 1, 180014. [Google Scholar] [CrossRef]
- Liang, Y.; Schott, D.; Zhang, Y.; Wang, Z.; Nasief, H.; Paulson, E.; Hall, W.; Knechtges, P.; Erickson, B.; Li, X.A. Auto-segmentation of pancreatic tumor in multi-parametric MRI using deep convolutional neural networks. Radiother. Oncol. 2020, 145, 193–200. [Google Scholar] [CrossRef]
- Pellicer-Valero, O.J.; Jiménez, J.L.M.; Gonzalez-Perez, V.; Ramón-Borja, J.L.C.; García, I.M.; Benito, M.B.; Gómez, P.P.; Rubio-Briones, J.; Rupérez, M.J.; Martín-Guerrero, J.D. Deep Learning for fully automatic detection, segmentation, and Gleason Grade estimation of prostate cancer in multiparametric Magnetic Resonance Images. Sci. Rep. 2022, 12, 2975. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wan, Z.; Zhang, J.; Li, W.; Chen, Y.; Li, Y.; Duan, Y. Medical image segmentation and reconstruction of prostate tumor based on 3D AlexNet. Methods Programs Biomed. 2021, 200, 105878. [Google Scholar] [CrossRef] [PubMed]
- Cha, K.H.; Hadjiiski, L.M.; Samala, R.; Chan, H.-P.; Cohan, R.H.; Caoili, E.M.; Paramagul, C.; Alva, A.; Weizer, A.Z. Bladder Cancer Segmentation in CT for Treatment Response Assessment: Application of Deep-Learning Convolution Neural Network—A Pilot Study. Tomography 2016, 2, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Tirkes, T.; Hollar, M.A.; Tann, M.; Kohli, M.D.; Akisik, F.; Sandrasegaran, K. Response criteria in oncologic imaging: Review of traditional and new criteria. RadioGraphics 2013, 33, 1323–1341. [Google Scholar] [CrossRef] [PubMed]
- Cha, K.H.; Hadjiiski, L.; Chan, H.-P.; Weizer, A.Z.; Alva, A.; Cohan, R.H.; Caoili, E.M.; Paramagul, C.; Samala, R.K. Bladder Cancer Treatment Response Assessment in CT using Radiomics with Deep-Learning. Sci. Rep. 2017, 7, 8738. [Google Scholar] [CrossRef]
- Li, X.; Guo, Y.; Jiang, F.; Xu, L.; Shen, F.; Jin, Z.; Wang, Y. Multi-Task Refined Boundary-Supervision U-Net (MRBSU-Net) for Gastrointestinal Stromal Tumor Segmentation in Endoscopic Ultrasound (EUS) Images. IEEE Access 2020, 8, 5805–5816. [Google Scholar] [CrossRef]
- Tonozuka, R.; Itoi, T.; Nagata, N.; Kojima, H.; Sofuni, A.; Tsuchiya, T.; Ishii, K.; Tanaka, R.; Nagakawa, Y.; Mukai, S. Deep learning analysis for the detection of pancreatic cancer on endosonographic images: A pilot study. J. Hepatobiliary Pancreat. Sci. 2021, 28, 95–104. [Google Scholar] [CrossRef]
- Bobo, M.F.; Bao, S.; Huo, Y.; Yao, Y.; Virostko, J.; Plassard, A.J.; Lyu, I.; Assad, A.; Abramson, R.G.; Hilmes, M.A.; et al. Fully Convolutional Neural Networks Improve Abdominal Organ Segmentation. Proc. SPIE Int. Soc. Opt. Eng. 2018, 10574, 105742V. [Google Scholar] [CrossRef]
- Gibson, E.; Giganti, F.; Hu, Y.; Bonmati, E.; Bandula, S.; Gurusamy, K.; Davidson, B.; Pereira, S.P.; Clarkson, M.J.; Barratt, D.C. Automatic Multi-Organ Segmentation on Abdominal CT With Dense V-Networks. IEEE Trans. Med. Imaging 2018, 37, 1822–1834. [Google Scholar] [CrossRef]
- Abdel-massieh, N.H.; Hadhoud, M.M.; Amin, K.M. Fully automatic liver tumor segmentation from abdominal CT scans. In Proceedings of the 2010 International Conference on Computer Engineering & Systems, Cairo, Egypt, 30 November–2 December 2010; pp. 197–202. [Google Scholar] [CrossRef]
- Abd-Elaziz, O.F.; Sayed, M.S.; Abdullah, M.I. Liver tumors segmentation from abdominal CT images using region growing and morphological processing. In Proceedings of the 2014 International Conference on Engineering and Technology (ICET), Cairo, Egypt, 19–20 April 2014; pp. 1–6. [Google Scholar] [CrossRef]
- Johnson, J.M.; Khoshgoftaar, T.M. Survey on deep learning with class imbalance. J. Big Data 2019, 6, 27. [Google Scholar] [CrossRef]
- Rezaei, M.; Yang, H.; Meinel, C. Recurrent generative adversarial network for learning imbalanced medical image semantic segmentation. Multimed. Tools Appl. 2020, 79, 15329–15348. [Google Scholar] [CrossRef]
- Estabrooks, A.; Jo, T.; Japkowicz, N. A multiple resampling method for learning from imbalanced data sets. Comput. Intell. 2004, 20, 18–36. [Google Scholar] [CrossRef]
- Li, Z.; Kamnitsas, K.; Glocker, B. Overfitting of Neural Nets Under Class Imbalance: Analysis and Improvements for Segmentation. In Medical Image Computing and Computer-Assisted Intervention 2019; Lecture Notes in Computer Science; Springer Science and Business Media Deutschland GmbH: Berlin/Heidelberg, Germany, 2019; Volume 11766, pp. 402–410. [Google Scholar] [CrossRef]
- Yi, P.H.; Kim, T.K.; Yu, A.C.; Bennett, B.; Eng, J.; Lin, C.T. Can AI outperform a junior resident? Comparison of deep neural network to first-year radiology residents for identification of pneumothorax. Emerg. Radiol. 2020, 27, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Andrew, G.; Eichner, H.; Sun, H.; Li, W.; Kong, N.; Ramage, D.; Beaufays, F. Applied Federated Learning: Improving Google Keyboard Query Suggestions. arXiv 2018. [Google Scholar] [CrossRef]
- Ng, D.; Lan, X.; Yao, M.M.S.; Chan, W.P.; Feng, M. Federated learning: A collaborative effort to achieve better medical imaging models for individual sites that have small labelled datasets. Quant. Imaging Med. Surg. 2021, 11, 852–857. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).