Improving the Detection Sensitivity of a New Rapid Diagnostic Technology for Severe Acute Respiratory Syndrome Coronavirus 2 Using a Trace Amount of Saliva
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Primers and Probes
2.3. RT-PCR
2.4. Detection of SARS-CoV-2 RNA in Mouthwash Obtained from Patients with COVID-19
3. Results
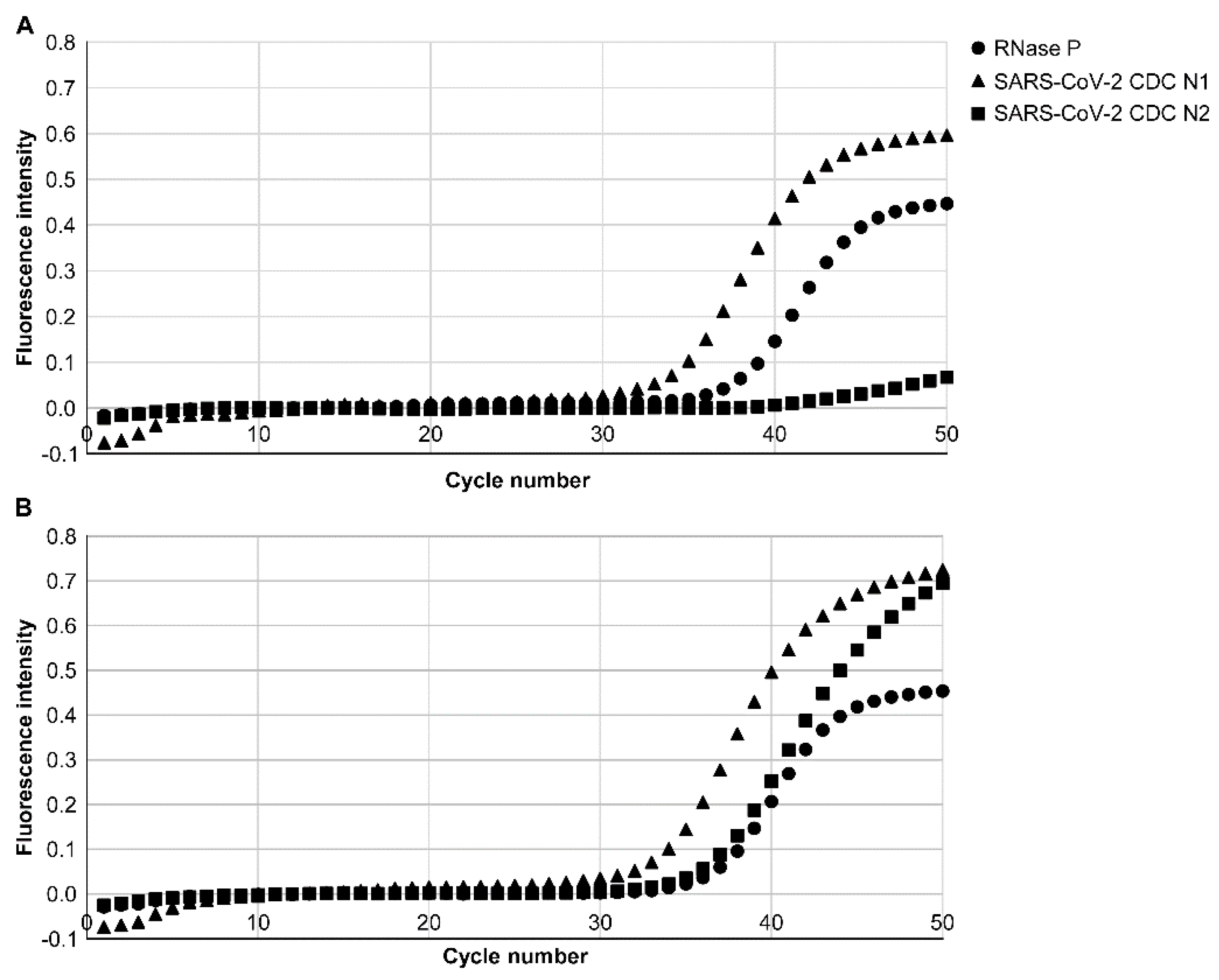
3.1. Analytical Limits of Detection (LoD)
3.2. Detection of CDC N1 and N2 of SARS-CoV-2 Viral RNA from Patients with COVID-19
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The disease and tools for detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef]
- Evans, R.W. Diagnostic Testing for SARS-CoV-2; WHO: Geneva, Switzerland, 2020; Available online: http://www.who.int (accessed on 20 November 2020).
- Ravi, N.; Cortade, D.L.; Ng, E.; Wang, S.X. Diagnostics for SARS-CoV-2 detection: A comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020, 165, 112454. [Google Scholar] [CrossRef]
- Tang, Y.W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. Laboratory diagnosis of COVID-19: Current issues and challenges. J. Clin. Microbiol. 2020, 58, e00512-20. [Google Scholar] [CrossRef]
- Vashist, S.K. In vitro diagnostic assays for COVID-19: Recent advances and emerging trends. Diagnostics 2020, 10, 202. [Google Scholar] [CrossRef]
- Adachi, D.; Johnson, G.; Draker, R.; Ayers, M.; Mazzulli, T.; Talbot, P.J.; Tellier, R. Comprehensive detection and identification of human coronaviruses, including the SARS-sssociated coronavirus, with a single RT-PCR assay. J. Virol. Methods 2004, 122, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Porte, L.; Legarraga, P.; Vollrath, V.; Aguilera, X.; Munita, J.M.; Araos, R.; Pizarro, G.; Vial, P.; Iruretagoyena, M.; Dittrich, S.; et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int. J. Infect. Dis. 2020, 99, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Joung, C.J.; Ladha, A.; Saito, M.; Segel, M.; Bruneau, R.; Huang, M.W.; Kim, N.-G.; Yu, X.; Li, J.; Walker, B.D.; et al. Point-of-Care Testing for COVID-19 Using Sherlock Diagnostics. medRxiv 2020. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7273289/ (accessed on 10 December 2020).
- Gibani, M.M.; Toumazou, C.; Sohbati, M.; Sahoo, R.; Karvela, M.; Hon, T.K.; De Mateo, S.; Burdett, A.; Leung, K.Y.F.; Barnett, J.; et al. Assessing a novel, lab-free, point-of-care test for SARS-CoV-2 (CovidNudge): A diagnostic accuracy study. Lancet Microbe 2020, 1, e300–e307. [Google Scholar] [CrossRef]
- Cheng, M.P.; Papenburg, J.; Desjardins, M.; Kanjilal, S.; Quach, C.; Libman, M.; Dittrich, S.; Yansouni, C.P. Diagnostic testing for severe acute pespiratory syndrome-related coronavirus 2: A narrative review. Ann. Intern. Med. 2020, 172, 726–734. [Google Scholar] [CrossRef]
- Sheridan, C. Fast, Portable tests come online to curb coronavirus pandemic. Nat. Biotechnol. 2020, 38, 515–518. [Google Scholar] [CrossRef]
- Ji, T.; Liu, Z.; Wang, G.; Guo, X.; Akbar Khan, S.; Lai, C.; Chen, H.; Huang, S.; Xia, S.; Chen, B.; et al. Detection of COVID-19: A review of the current literature and future perspectives. Biosens. Bioelectron. 2020, 166, 112455. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Wang, T.; Zhang, B.; Luo, Y.; Mao, L.; Wang, F.; Wu, S.; Sun, Z. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin. Transl. Immunol. 2020, 9, e01136. [Google Scholar] [CrossRef] [PubMed]
- Kontou, P.I.; Braliou, G.G.; Dimou, N.L.; Nikolopoulos, G.; Bagos, P.G. Antibody tests in detecting SARS-CoV-2 infection: A meta-analysis. Diagnostics 2020, 10, 319. [Google Scholar] [CrossRef] [PubMed]
- Infantino, M.; Grossi, V.; Lari, B.; Bambi, R.; Perri, A.; Manneschi, M.; Terenzi, G.; Liotti, I.; Ciotta, G.; Taddei, C.; et al. Diagnostic accuracy of an automated chemiluminescent immunoassay for anti-SARS-CoV-2 IgM and IgG antibodies: An Italian experience. J. Med. Virol. 2020, 92, 1671–1675. [Google Scholar] [CrossRef] [PubMed]
- Chuan, J.; Gong, B.; Shuai, P.; Zhou, Y.; Zhang, Y.; Jiang, Z.; Zhang, D.; Liu, X.; Ma, S. Detection of serum IgM and IgG for COVID-19 diagnosis. Sci. China 2020, 63, 777–780. [Google Scholar]
- Wen, T.; Huang, C.; Shi, F.J.; Zeng, X.Y.; Lu, T.; Ding, S.N.; Jiao, Y.J. Development of a lateral flow immunoassay strip for rapid detection of IgG antibody against SARS-CoV-2 virus. Analyst 2020, 145, 5345–5352. [Google Scholar] [CrossRef]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and clinical application of a papid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020, 92, 1518–1524. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Adriano, A.; Berhane, S.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; Beese, S.; et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020, 8, CD013705. Available online: https://www.cochranelibrary.com/cdsr/doi/10.1002/14651858.CD013705/full (accessed on 10 December 2020).
- Tokuyama-Toda, R.; Muraoka, M.; Terada-Ito, C.; Ide, S.; Horiuchi, T.; Amemiya, T.; Fukuoka, A.; Hamada, Y.; Sejima, S.; Satomura, K. Feasibility of rapid diagnostic technology for SARS-CoV-2 virus using a trace amount of saliva. Diagnostics 2021, 11, 2024. [Google Scholar] [CrossRef]
- Jung, Y.J.; Park, G.S.; Moon, J.H.; Ku, K.; Beak, S.H.; Kim, S.; Park, E.C.; Park, D.; Lee, J.H.; Byeon, C.W.; et al. Comparative analysis of primer-probe sets for the laboratory confirmation of SARS-CoV-2. bioRxiv 2020, 11, 2513–2523. [Google Scholar] [CrossRef]
- Shirato, K.; Nao, N.; Kawase, M.; Kageyama, T. An ultra-rapid real-time RT-PCR method using PCR1100 for detecting human orthopneumovirus. Jpn. J. Infect. Dis. 2020, 73, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Vogels, C.B.F.; Brito, A.F.; Wyllie, A.L.; Fauver, J.R.; Ott, I.M.; Kalinich, C.C.; Petrone, M.E.; Casanovas-Massana, A.; Catherine Muenker, M.; Moore, A.J.; et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat. Microbiol. 2020, 5, 1299–1305. [Google Scholar] [CrossRef] [PubMed]
- Freire-Paspuel, B.; Garcia-Bereguiain, M.A. Analytical sensitivity and clinical performance of a triplex RT-qPCR assay using CDC N1, N2, and RP targets for SARS-CoV-2 diagnosis. Int. J. Infect. Dis. 2021, 102, 14–16. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Hamilton, K.A.; Lobos, A.; Hughes, B.; Staley, C.; Sadowsky, M.J.; Harwood, V.J. Evaluation of process limit of detection and quantification variation of SARS-CoV-2 RT-qPCR and RT-dPCR assays for wastewater surveillance. Water Res. 2022, 213, 118–132. [Google Scholar] [CrossRef] [PubMed]
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Haider, N.; Abbasi, A.F.; Jaferi, U.; Prakash, S.; Balendra, V. The emerging SARS-CoV-2 variants of concern. Ther. Adv. Infect. Dis. 2021, 8, 1–10. [Google Scholar] [CrossRef]
- Fiolet, T.; Kherabi, Y.; MacDonald, C.-J.; Ghosn, J.; Peiffer-Smadja, N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: A narrative review. Clin. Microbiol. Infect. 2022, 28, 202–221. [Google Scholar] [CrossRef]
- Boehm, E.; Kronig, I.; Neher, R.A.; Eckerle, I.; Vetter, P.; Kaiser, L. Novel SARS-CoV-2 variants: The pandemics within the pandemic. Clin. Microbiol. Infect. 2021, 27, 1109–1117. [Google Scholar] [CrossRef]

| Patient | Age/Sex | Ver. 1 Ct Value | Ver. 2 Ct Value | ||||
|---|---|---|---|---|---|---|---|
| RNase P | CDC N1 | CDC N2 | RNase P | CDC N1 | CDC N2 | ||
| 1 | 90/M | 35.3 | 39.0 | (-) | 33.3 | 37.4 | 43.2 |
| 2 | 56/F | 37.8 | 37.0 | (-) | 34.1 | 33.9 | 46.3 |
| 3 | 50/M | 31.1 | 45.5 | (-) | 29.7 | 39.9 | 46.1 |
| 4 | 25/F | 36.0 | 33.0 | (-) | 35.0 | 32.0 | 34.0 |
| 5 | 70/M | 33.5 | 30.0 | (-) | 31.3 | 28.7 | 38.3 |
| 6 | 90/M | 32.0 | 35.2 | (-) | 30.3 | 31.4 | 35.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tokuyama-Toda, R.; Terada-Ito, C.; Muraoka, M.; Horiuchi, T.; Amemiya, T.; Fukuoka, A.; Hamada, Y.; Takebe, Y.; Ogawa, T.; Fujii, S.; et al. Improving the Detection Sensitivity of a New Rapid Diagnostic Technology for Severe Acute Respiratory Syndrome Coronavirus 2 Using a Trace Amount of Saliva. Diagnostics 2022, 12, 2568. https://doi.org/10.3390/diagnostics12112568
Tokuyama-Toda R, Terada-Ito C, Muraoka M, Horiuchi T, Amemiya T, Fukuoka A, Hamada Y, Takebe Y, Ogawa T, Fujii S, et al. Improving the Detection Sensitivity of a New Rapid Diagnostic Technology for Severe Acute Respiratory Syndrome Coronavirus 2 Using a Trace Amount of Saliva. Diagnostics. 2022; 12(11):2568. https://doi.org/10.3390/diagnostics12112568
Chicago/Turabian StyleTokuyama-Toda, Reiko, Chika Terada-Ito, Masaaki Muraoka, Toshikatsu Horiuchi, Tsuyoshi Amemiya, Airi Fukuoka, Yoshiki Hamada, Yusuke Takebe, Takashi Ogawa, Seiko Fujii, and et al. 2022. "Improving the Detection Sensitivity of a New Rapid Diagnostic Technology for Severe Acute Respiratory Syndrome Coronavirus 2 Using a Trace Amount of Saliva" Diagnostics 12, no. 11: 2568. https://doi.org/10.3390/diagnostics12112568
APA StyleTokuyama-Toda, R., Terada-Ito, C., Muraoka, M., Horiuchi, T., Amemiya, T., Fukuoka, A., Hamada, Y., Takebe, Y., Ogawa, T., Fujii, S., Kikuta, T., Sejima, S., & Satomura, K. (2022). Improving the Detection Sensitivity of a New Rapid Diagnostic Technology for Severe Acute Respiratory Syndrome Coronavirus 2 Using a Trace Amount of Saliva. Diagnostics, 12(11), 2568. https://doi.org/10.3390/diagnostics12112568








