A Novel Saliva RT-LAMP Workflow for Rapid Identification of COVID-19 Cases and Restraining Viral Spread
Abstract
:1. Introduction
2. Methods
2.1. Subjects
2.2. Saliva Collection
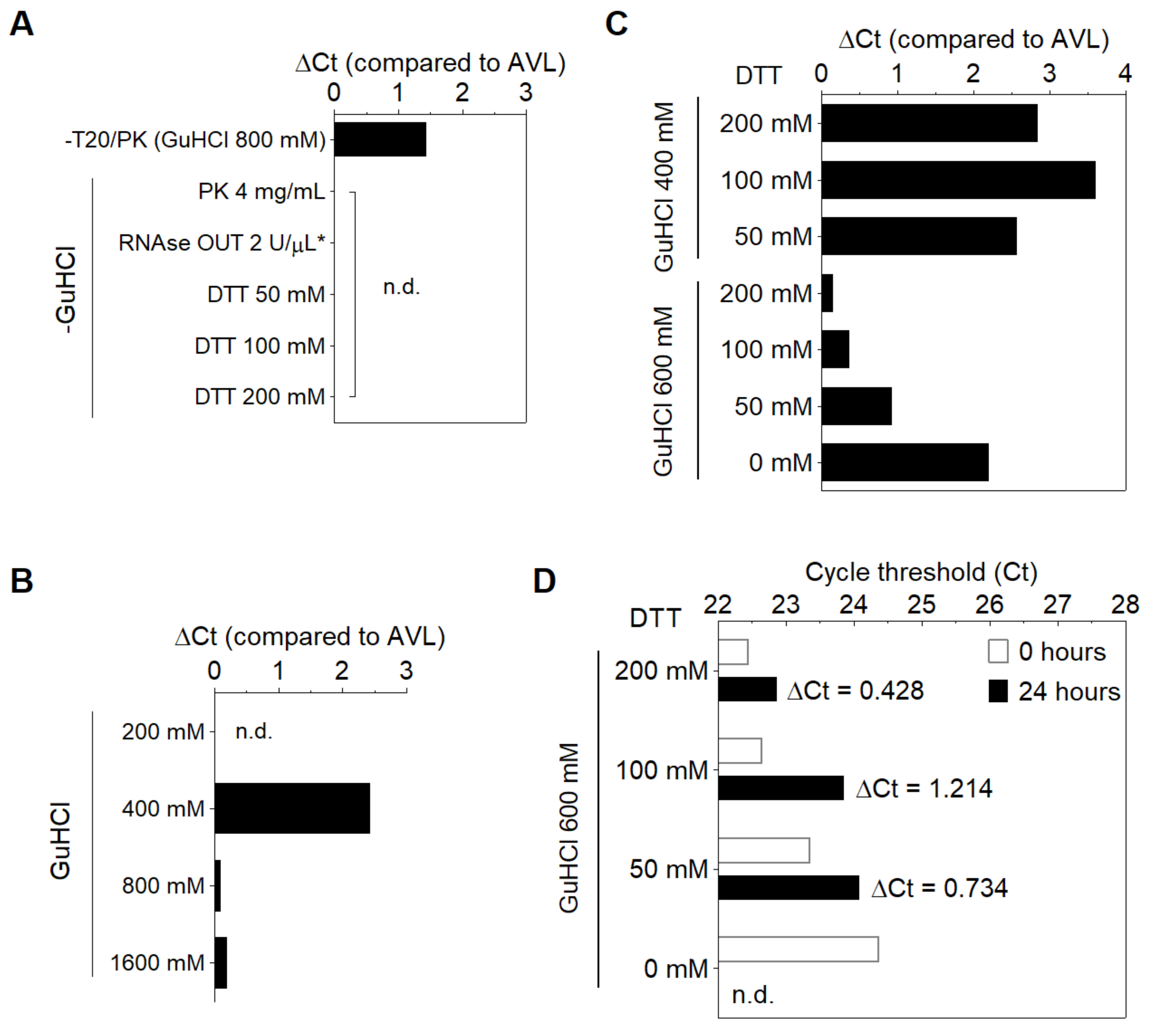
2.3. DGS Preparation
2.4. RT-LAMP and rtRT-LAMP Reactions
2.5. RNA Isolation and RT-PCR Reactions on Saliva Samples
2.6. Simulation of Saliva Positive Controls with SARS-CoV-2 RNA
2.7. Statistical Analyses
3. Results
3.1. Stabilization of SARS-CoV-2 RNA in Saliva
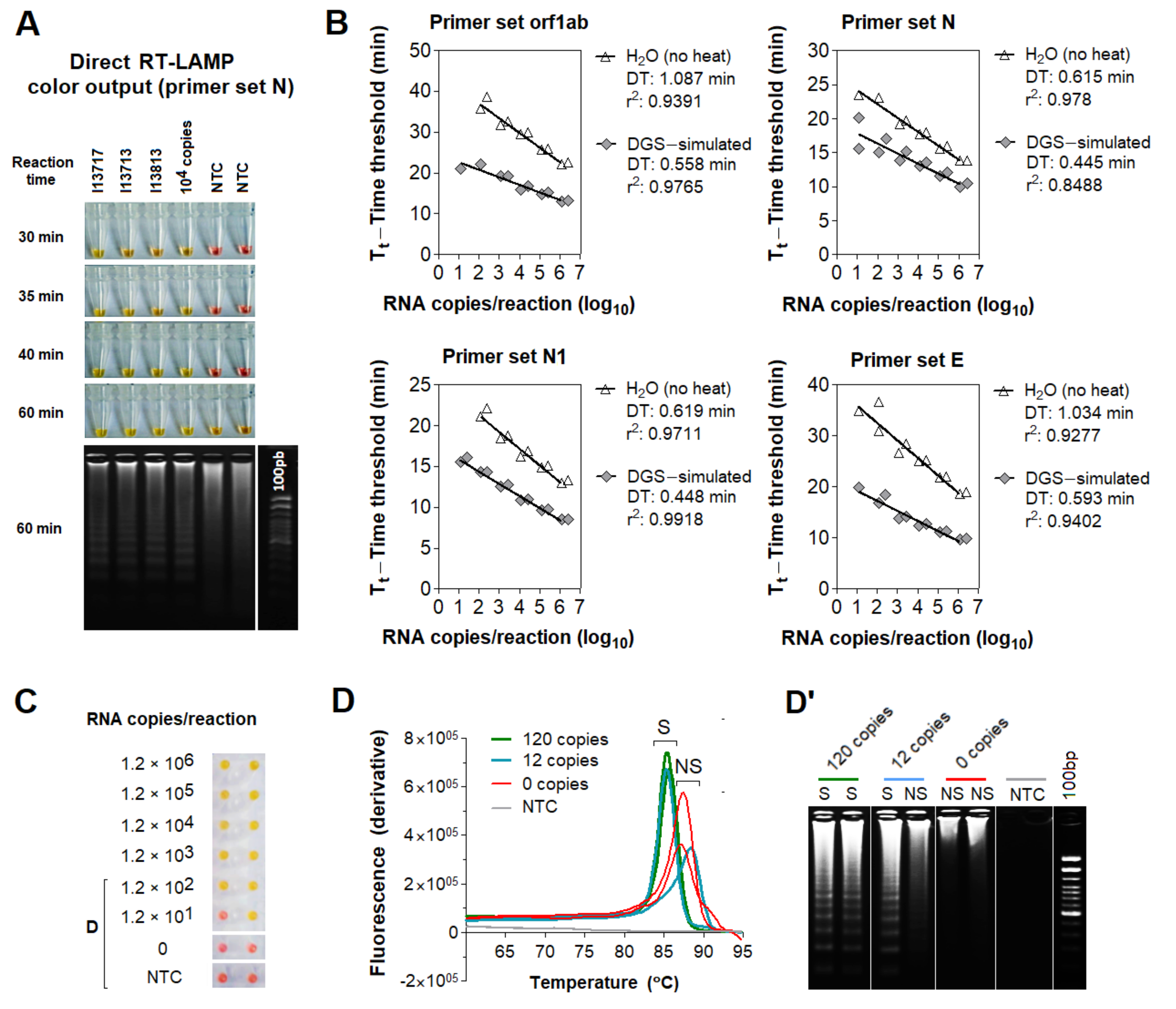
3.2. Compatibility of DGS with Direct RT-LAMP in Saliva
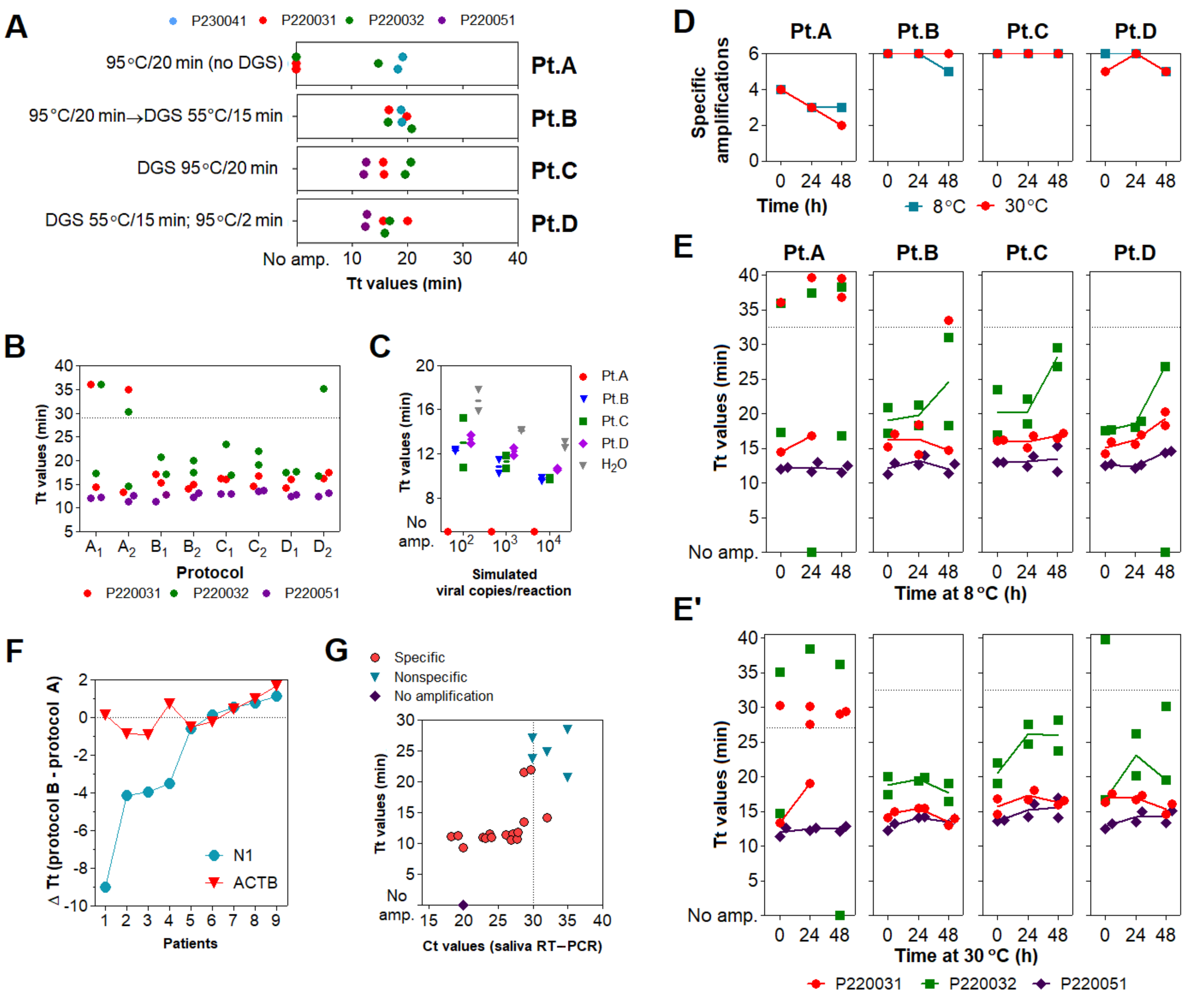
3.3. Optimization of the DGS Workflow
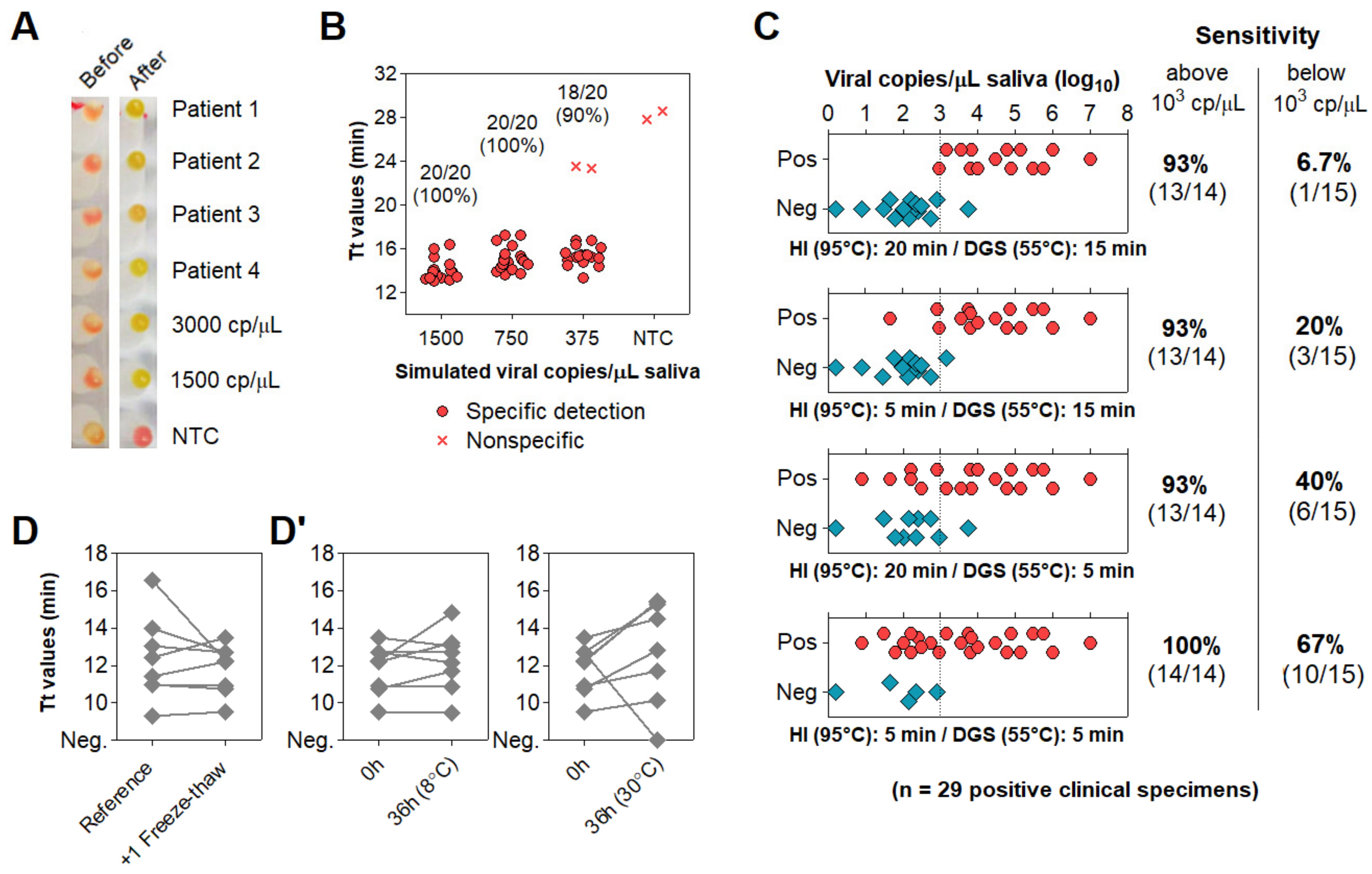
3.4. Optimization of Color Discrimination and Sample Processing Time
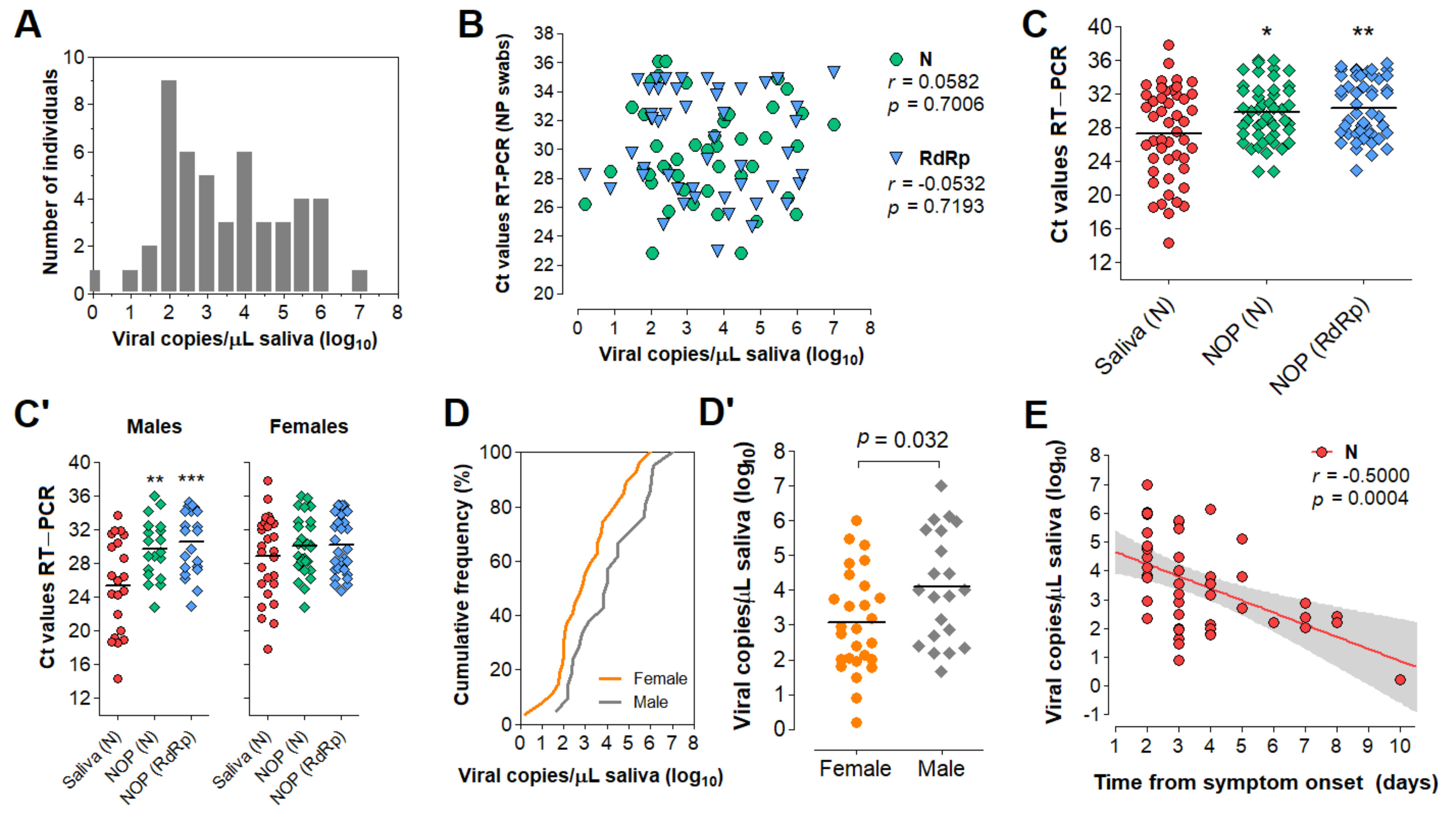
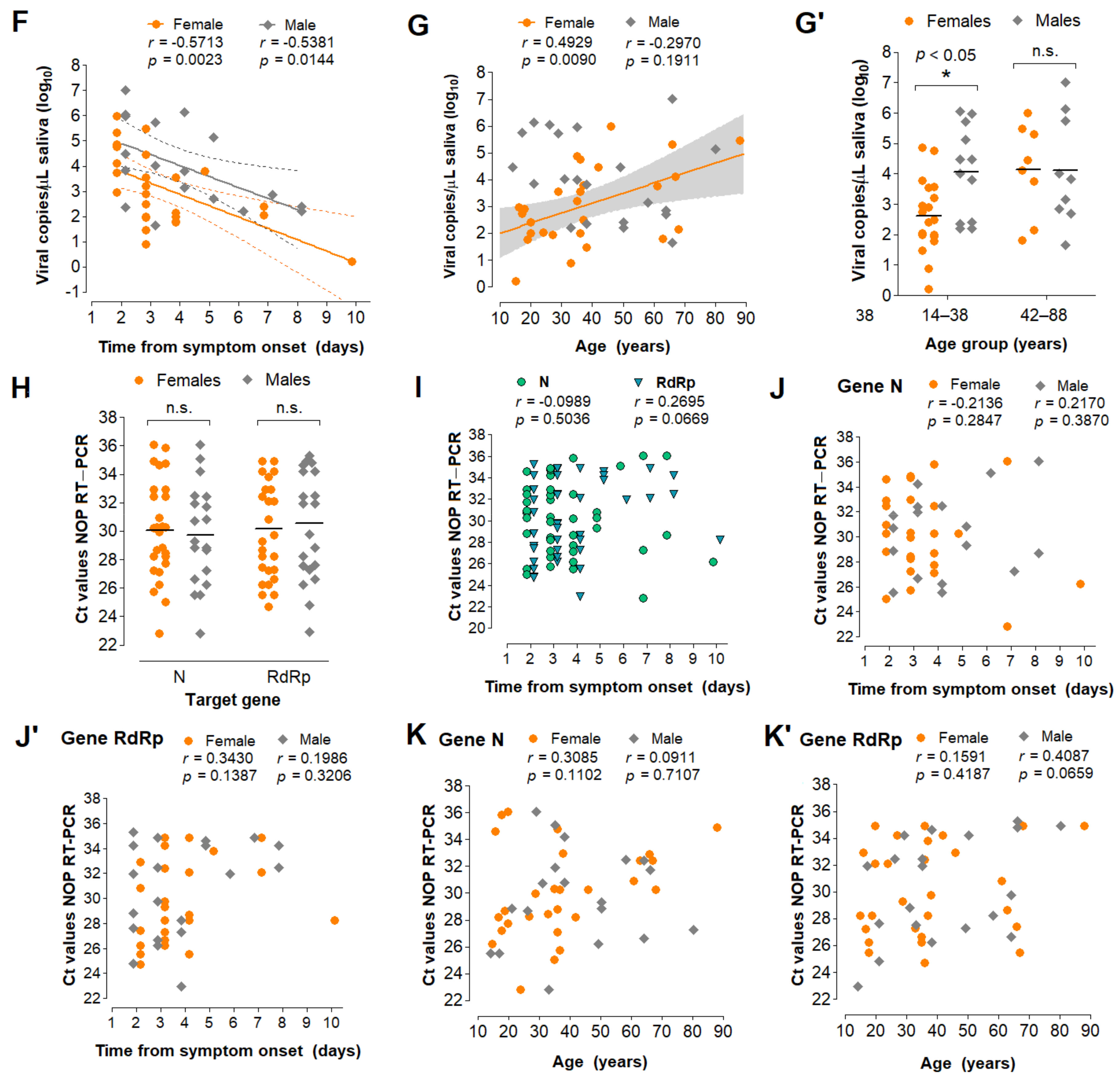
3.5. Characterization of the Diagnostic Properties of Saliva Compared to NOP Swabs
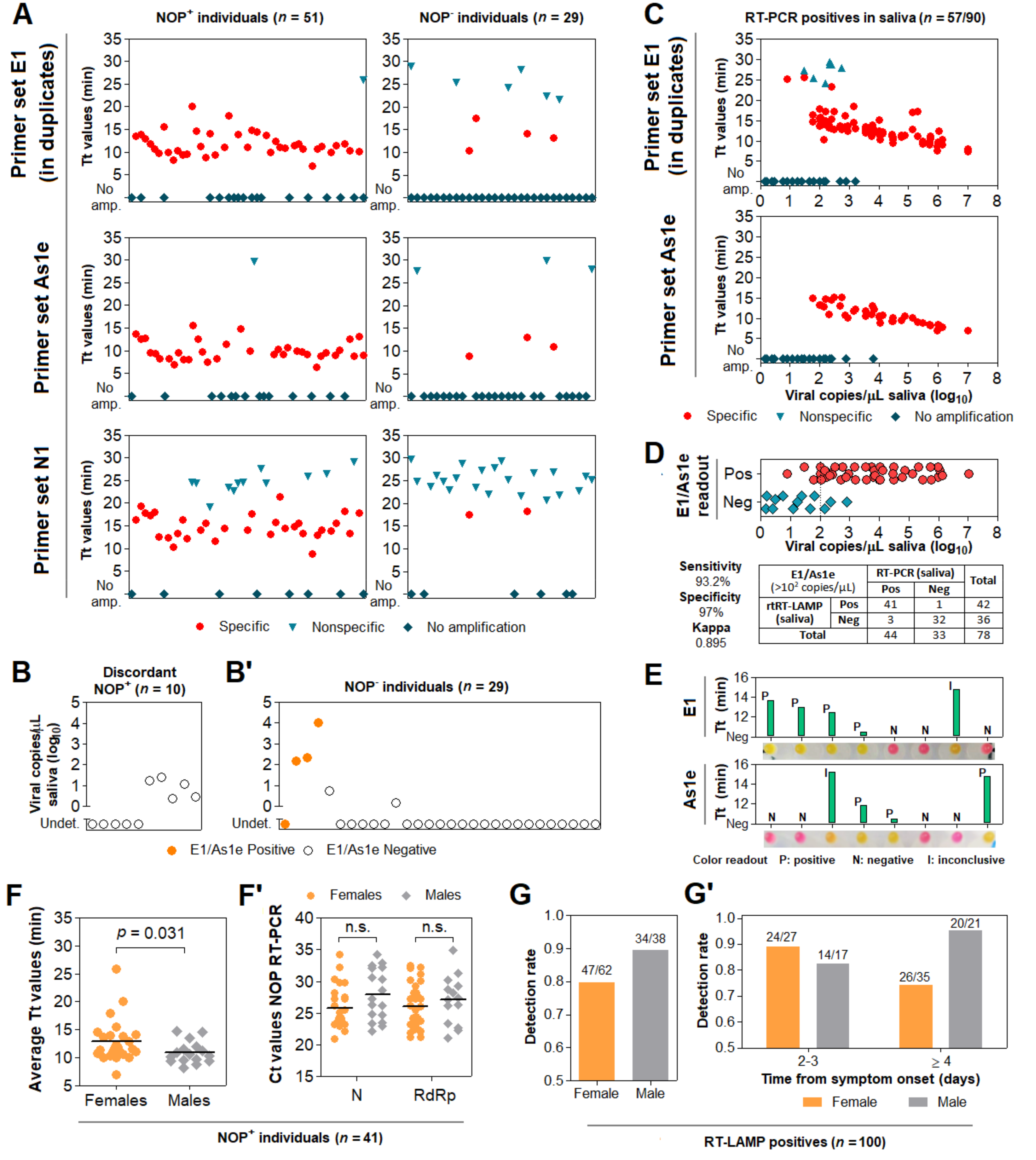
3.6. Direct RT-LAMP in 80 Saliva Samples from Symptomatic Individuals
3.7. Clinical Performance of the Direct RT-LAMP Workflow
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, N.; Pérez, P.; Kato, T.; Mikami, Y.; Okuda, K.; Gilmore, R.C.; Conde, C.D.; Gasmi, B.; Stein, S.; Beach, M.; et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 2021, 27, 892–903. [Google Scholar] [CrossRef]
- Fakheran, O.; Dehghannejad, M.; Khademi, A. Saliva as a diagnostic specimen for detection of SARS-CoV-2 in suspected patients: A scoping review. Infect. Dis. Poverty 2020, 9, 100. [Google Scholar] [CrossRef]
- Leal, F.E.; Mendes-Correa, M.C.; Buss, L.F.; Costa, S.F.; Bizario, J.C.S.; de Souza, S.R.P.; Thomaz, O.; Tozetto-Mendoza, T.R.; Villas-Boas, L.S.; de Oliveira-da Silva, L.C.; et al. Clinical features and natural history of the first 2073 suspected COVID-19 cases in the Corona São Caetano primary care programme: A prospective cohort study. BMJ Open 2021, 11, e042745. [Google Scholar] [PubMed]
- Rabe, B.A.; Cepko, C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl. Acad. Sci. USA 2020, 117, 24450–24458. [Google Scholar] [CrossRef] [PubMed]
- Alves, P.A.; de Ellen, G.O.; Franco-Luiz, A.P.M.; Almeida, L.T.; Gonçalves, A.B.; Borges, I.A.; Rocha, F.d.S.; Rocha, R.P.; Bezerra, M.F.; Miranda, P.; et al. Clinical validation of colorimetric RT-LAMP, a fast, highly sensitive and specific COVID-19 molecular diagnostic tool that is robust to detect SARS-CoV-2 variants of concern. medRxiv 2021. [Google Scholar] [CrossRef]
- Lamb, L.E.; Bartolone, S.N.; Ward, E.; Chancellor, M.B. Rapid detection of novel coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS ONE 2020, 15, e0234682. [Google Scholar] [CrossRef]
- Lalli, M.A.; Langmade, J.S.; Chen, X.; Fronick, C.C.; Sawyer, C.S.; Burcea, L.C.; Wilkinson, M.N.; Fulton, R.S.; Heinz, M.; Buchser, W.J.; et al. Rapid and Extraction-Free Detection of SARS-CoV-2 from Saliva by Colorimetric Reverse-Transcription Loop-Mediated Isothermal Amplification. Clin. Chem. 2021, 67, 415–424. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [Green Version]
- Auerswald, H.; Yann, S.; Dul, S.; In, S.; Dussart, P.; Martin, N.J.; Karlsson, E.A.; Garcia-Rivera, J.A. Assessment of inactivation procedures for SARS-CoV-2. J. Gen. Virol. 2021, 102, 001539. [Google Scholar] [CrossRef]
- Ranoa, D.R.E.; Holland, R.L.; Alnaji, F.G.; Green, K.J.; Wang, L.; Brooke, C.B.; Burke, M.D.; Fan, T.M.; Hergenrother, P.J. Saliva-Based Molecular Testing for SARS-CoV-2 that Bypasses RNA Extraction. BioRxiv 2020. [Google Scholar] [CrossRef]
- Nixon, G.J.; Svenstrup, H.F.; Donald, C.E.; Carder, C.; Stephenson, J.M.; Morris-Jones, S.; Huggett, J.F.; Foy, C.A. A novel approach for evaluating the performance of real time quantitative loop-mediated isothermal amplification-based methods. Biomol. Detect. Quantif. 2014, 2, 4–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamb, L.E.; Bartolone, S.N.; Ward, E.; Chancellor, M.B. Rapid Detection of Novel Coronavirus (COVID-19) by Reverse Transcription-Loop-Mediated Isothermal Amplification. medRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, Y.; Ren, G.; Buss, J.; Barry, A.J.; Patton, G.C.; Tanner, N.A. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. BioTechniques 2020, 69, 178–185. [Google Scholar] [CrossRef]
- Dudley, D.M.; Newman, C.M.; Weiler, A.M.; Ramuta, M.D.; Shortreed, C.G.; Heffron, A.S.; Accola, M.A.; Rehrauer, W.M.; Friedrich, T.C.; O’Connor, D.H. Optimizing direct RT-LAMP to detect transmissible SARS-CoV-2 from primary nasopharyngeal swab samples. PLoS ONE 2020, 15, e0244882. [Google Scholar] [CrossRef]
- Mallmann, L.; Schallenberger, K.; Demolliner, M.; Antunes Eisen, A.K.; Hermann, B.S.; Heldt, F.H.; Hansen, A.W.; Spilki, F.R.; Fleck, J.D. Pre-treatment of the clinical sample with Proteinase K allows detection of SARS-CoV-2 in the absence of RNA extraction. BioRxiv 2020. [Google Scholar] [CrossRef]
- Ladha, A.; Joung, J.; Abudayyeh, O.; Gootenberg, J.; Zhang, F. A 5-min RNA preparation method for COVID-19 detection with RT-qPCR. medRxiv 2020. [Google Scholar] [CrossRef]
- Kellner, M.J.; Ross, J.J.; Schnabl, J.; Dekens, M.P.S.; Heinen, R.; Tanner, N.A.; Fritsche-Polanz, R.; Traugott, M.; Seitz, T.; Zoufaly, A.; et al. Scalable, rapid and highly sensitive isothermal detection of SARS-CoV-2 for laboratory and home testing. BioRxiv 2020. [Google Scholar] [CrossRef]
- Ott, I.M.; Strine, M.S.; Watkins, A.E.; Boot, M.; Kalinich, C.C.; Harden, C.A.; Vogels, C.B.F.; Casanovas-Massana, A.; Moore, A.J.; Muenker, M.C.; et al. Simply saliva: Stability of SARS-CoV-2 detection negates the need for expensive collection devices. medRxiv 2020. [Google Scholar] [CrossRef]
- Welch, S.R.; Davies, K.A.; Buczkowski, H.; Hettiarachchi, N.; Green, N.; Arnold, U.; Jones, M.; Hannah, M.J.; Evans, R.; Burton, C.; et al. Analysis of Inactivation of SARS-CoV-2 by Specimen Transport Media, Nucleic Acid Extraction Reagents, Detergents, and Fixatives. J. Clin. Microbiol. 2020, 58, e01713-20. [Google Scholar] [CrossRef]
- Feng, W.; Newbigging, A.M.; Le, C.; Pang, B.; Peng, H.; Cao, Y.; Wu, J.; Abbas, G.; Song, J.; Wang, D.-B.; et al. Molecular Diagnosis of COVID-19: Challenges and Research Needs. Anal. Chem. 2020, 92, 10196–10209. [Google Scholar] [CrossRef]
- Khan, M.A.; Wolf, D.P.; Litt, M. Effect of mucolytic agents on the rheological properties of tracheal mucos. Biochim. Biophys. Acta Gen. Subj. 1976, 444, 369–373. [Google Scholar] [CrossRef]
- Batejat, C.; Grassin, Q.; Manuguerra, J.-C.; Leclercq, I. Heat inactivation of the Severe Acute Respiratory Syndrome Coronavirus 2. J. Biosaf. Biosecur. 2020, 3, 1–3. [Google Scholar]
- Burton, J.; Love, H.; Richards, K.; Burton, C.; Summers, S.; Pitman, J.; Easterbrook, L.; Davies, K.; Spencer, P.; Killip, M.; et al. The effect of heat-treatment on SARS-CoV-2 viability and detection. J. Virol. Methods 2021, 290, 114087. [Google Scholar] [CrossRef] [PubMed]
- Ben-Assa, N.; Naddaf, R.; Gefen, T.; Capucha, T.; Hajjo, H.; Mandelbaum, N.; Elbaum, L.; Rogov, P.; King, D.A.; Kaplan, S.; et al. Direct on-the-spot detection of SARS-CoV-2 in patients. Exp. Biol. Med. 2020, 245, 1187–1193. [Google Scholar] [CrossRef]
- Asprino, P.; Bettoni, F.; Camargo, A.; Coelho, D.; Coppini, G.; Correa, I.; Freitas, E.; Inoue, L.; Kitajima, J.P.; Kuroki, M.; et al. A Scalable Saliva-based, Extraction-free RT-LAMP Protocol for SARS-Cov-2 Diagnosis. medRxiv 2020. [Google Scholar] [CrossRef]
- Flynn, M.J.; Snitser, O.; Yelin, I.; Flynn, J.; Green, S.; Szwarcwort, M.; Kishony, R.; Elowitz, M.B. A simple direct RT-LAMP SARS-CoV-2 saliva diagnostic. medRxiv 2020. [Google Scholar] [CrossRef]
- Yoshikawa, R.; Abe, H.; Igasaki, Y.; Negishi, S.; Goto, H.; Yasuda, J. Development and evaluation of a rapid and simple diagnostic assay for COVID-19 based on loop-mediated isothermal amplification. PLoS Negl. Trop. Dis. 2020, 14, e0008855. [Google Scholar] [CrossRef]
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.; Kirrmaier, D. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12, eabc7075. [Google Scholar] [CrossRef]
- Wei, S.; Kohl, E.; Djandji, A.; Morgan, S.; Whittier, S.; Mansukhani, M. Direct diagnostic testing of SARS-CoV-2 without the need for prior RNA extraction. Sci. Rep. 2021, 11, 2402. [Google Scholar] [CrossRef]
- Schellenberg, J.J.; Ormond, M.; Keynan, Y. Extraction-free RT-LAMP to detect SARS-CoV-2 is less sensitive but highly specific compared to standard RT-PCR in 101 samples. J. Clin. Virol. 2021, 136, 104764. [Google Scholar] [CrossRef]
- Fowler, V.L.; Armson, B.; Gonzales, J.L.; Wise, E.L.; Howson, E.L.A.; Vincent-Mistiaen, Z. A highly effective reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 infection. J. Infect. 2021, 82, 117–125. [Google Scholar] [CrossRef]
- Anahtar, M.N.; McGrath, G.E.G.; Rabe, B.A.; Tanner, N.A.; White, B.A.; Lennerz, J.K.M. Clinical Assessment and Validation of a Rapid and Sensitive SARS-CoV-2 Test Using Reverse Transcription Loop-Mediated Isothermal Amplification Without the Need for RNA Extraction. In Open Forum Infectious Diseases; Oxford University Press: Cary, NC, USA, 2021; Volume 8. [Google Scholar]
- Marks, M.; Millat-Martinez, P.; Ouchi, D.; Roberts, C.H.; Alemany, A.; Corbacho-Monné, M.; Ubals, M.; Tobias, A.; Tebé, C.; Ballana, E.; et al. Transmission of COVID-19 in 282 clusters in Catalonia, Spain: A cohort study. Lancet Infect. Dis. 2021, 21, 629–636. [Google Scholar] [CrossRef]
- Larremore, D.B.; Wilder, B.; Lester, E.; Shehata, S.; Burke, J.M.; Hay, J.A.; Tambe, M.; Mina, M.J.; Parker, R. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci. Adv. 2021, 7, eabd5393. [Google Scholar] [CrossRef]
- Peckham, H.; de Gruijter, N.M.; Raine, C.; Radziszewska, A.; Ciurtin, C.; Wedderburn, L.R.; Rosser, E.C.; Webb, K.; Deakin, C.T. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020, 11, 6317. [Google Scholar] [CrossRef]
- Ho, F.K.; Petermann-Rocha, F.; Gray, S.R.; Jani, B.D.; Katikireddi, S.V.; Niedzwiedz, C.L.; Foster, H.; Hastie, C.E.; Mackay, D.F.; Gill, J.M.R.; et al. Is older age associated with COVID-19 mortality in the absence of other risk factors? General population cohort study of 470,034 participants. PLoS ONE 2020, 15, e0241824. [Google Scholar] [CrossRef]
- Chua, G.T.; Wong, J.S.C.; To, K.K.W.; Lam, I.C.S.; Yau, F.Y.S.; Chan, W.H.; Ho, P.P.K.; Duque, J.S.R.; Yip, C.C.Y.; Ng, A.C.K.; et al. Saliva viral load better correlates with clinical and immunological profiles in children with coronavirus disease 2019. Emerg. Microbes Infect. 2021, 10, 235–241. [Google Scholar] [CrossRef]
- Silva, J.; Lucas, C.; Sundaram, M.; Israelow, B.; Wong, P.; Klein, J. Saliva viral load is a dynamic unifying correlate of COVID-19 severity and mortality. medRxiv 2021. [Google Scholar] [CrossRef]
- Takahashi, T.; Ellingson, M.K.; Wong, P.; Israelow, B.; Lucas, C.; Klein, J.; Silva, J.; Mao, T.; Oh, J.E.; Tokuyama, M.; et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature 2020, 588, 315–320. [Google Scholar] [CrossRef]
- Castelli, E.C.; de Castro, M.V.; Naslavsky, M.S.; Scliar, M.O.; Silva, N.S.B.; Andrade, H.S. Immunogenetics of resistance to SARS-CoV-2 infection in discordant couples. medRxiv 2021. [Google Scholar] [CrossRef]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.; Wang, W.; Wang, Y.; Litvinova, M.; Luo, K.; Ren, L. Infectivity, susceptibility, and risk factors associated with SARS-CoV-2 transmission under intensive contact tracing in Hunan, China. Nat. Commun. 2021, 12, 1533. [Google Scholar] [CrossRef] [PubMed]
| Composition | Ct | |
|---|---|---|
| AVL | Guanidinium thiocyanate (50–70%) | 23.15 |
| PK1 | PK (2 mg/mL) | ND |
| PK2 | PK (0.4 mg/mL) | ND |
| PK3 | PK (0.4 mg/mL) + T20 (10%) | ND |
| PK4 | PK (0.4 mg/mL) + T20 (10%) + Tris-HCl pH 8.0 (30 mM) | ND |
| PK5 | PK (0.4 mg/mL) + Tris-HCl pH 8.0 (30 mM) | ND |
| PK6 | PK (0.4 mg/mL) + T20 (10%) + GuHCl (800 mM) + Tris-HCl pH 8.0 (30 mM) | 23.9 |
| Component | DGS | DGS:Saliva (1:1) | Carryover into RT-LAMP (1.25 µL Input) |
|---|---|---|---|
| GuHCl | 600 mM | 300 mM | 30 mM |
| DTT | 200 mM | 100 mM | 10 mM |
| Tris-HCl pH 8.0 | 30 mM | 15 mM | 1.5 mM |
| Saliva | - | 100 µL | 0.625 µL |
| Total volume | 100 µL | 200 µL | 12.5 µL |
| Component | DGS | DGS:Saliva (2:1) | Carryover into RT-LAMP (1 µL Input) |
|---|---|---|---|
| GuHCl | 600 mM | 400 mM | 32 mM |
| DTT | 200 mM | 133.3 mM | 10.66 mM |
| Tris-HCl pH 8.0 | 30 mM | 20 mM | 1.6 mM |
| Saliva | - | 100 µL | 0.33 µL |
| Total volume | 200 µL | 300 µL | 12.5 µL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kobayashi, G.S.; Brito, L.A.; Moreira, D.d.P.; Suzuki, A.M.; Hsia, G.S.P.; Pimentel, L.F.; de Paiva, A.P.B.; Dias, C.R.; Lourenço, N.C.V.; Oliveira, B.A.; et al. A Novel Saliva RT-LAMP Workflow for Rapid Identification of COVID-19 Cases and Restraining Viral Spread. Diagnostics 2021, 11, 1400. https://doi.org/10.3390/diagnostics11081400
Kobayashi GS, Brito LA, Moreira DdP, Suzuki AM, Hsia GSP, Pimentel LF, de Paiva APB, Dias CR, Lourenço NCV, Oliveira BA, et al. A Novel Saliva RT-LAMP Workflow for Rapid Identification of COVID-19 Cases and Restraining Viral Spread. Diagnostics. 2021; 11(8):1400. https://doi.org/10.3390/diagnostics11081400
Chicago/Turabian StyleKobayashi, Gerson Shigeru, Luciano Abreu Brito, Danielle de Paula Moreira, Angela May Suzuki, Gabriella Shih Ping Hsia, Lylyan Fragoso Pimentel, Ana Paula Barreto de Paiva, Carolina Regoli Dias, Naila Cristina Vilaça Lourenço, Beatriz Araujo Oliveira, and et al. 2021. "A Novel Saliva RT-LAMP Workflow for Rapid Identification of COVID-19 Cases and Restraining Viral Spread" Diagnostics 11, no. 8: 1400. https://doi.org/10.3390/diagnostics11081400
APA StyleKobayashi, G. S., Brito, L. A., Moreira, D. d. P., Suzuki, A. M., Hsia, G. S. P., Pimentel, L. F., de Paiva, A. P. B., Dias, C. R., Lourenço, N. C. V., Oliveira, B. A., Manuli, E. R., Corral, M. A., Cavaçana, N., Mitne-Neto, M., Sales, M. M., Dell’ Aquila, L. P., Filho, A. R., Parrillo, E. F., Mendes-Corrêa, M. C., ... Passos-Bueno, M. R. (2021). A Novel Saliva RT-LAMP Workflow for Rapid Identification of COVID-19 Cases and Restraining Viral Spread. Diagnostics, 11(8), 1400. https://doi.org/10.3390/diagnostics11081400





