Combination of Plasma-Based Metabolomics and Machine Learning Algorithm Provides a Novel Diagnostic Strategy for Malignant Mesothelioma
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
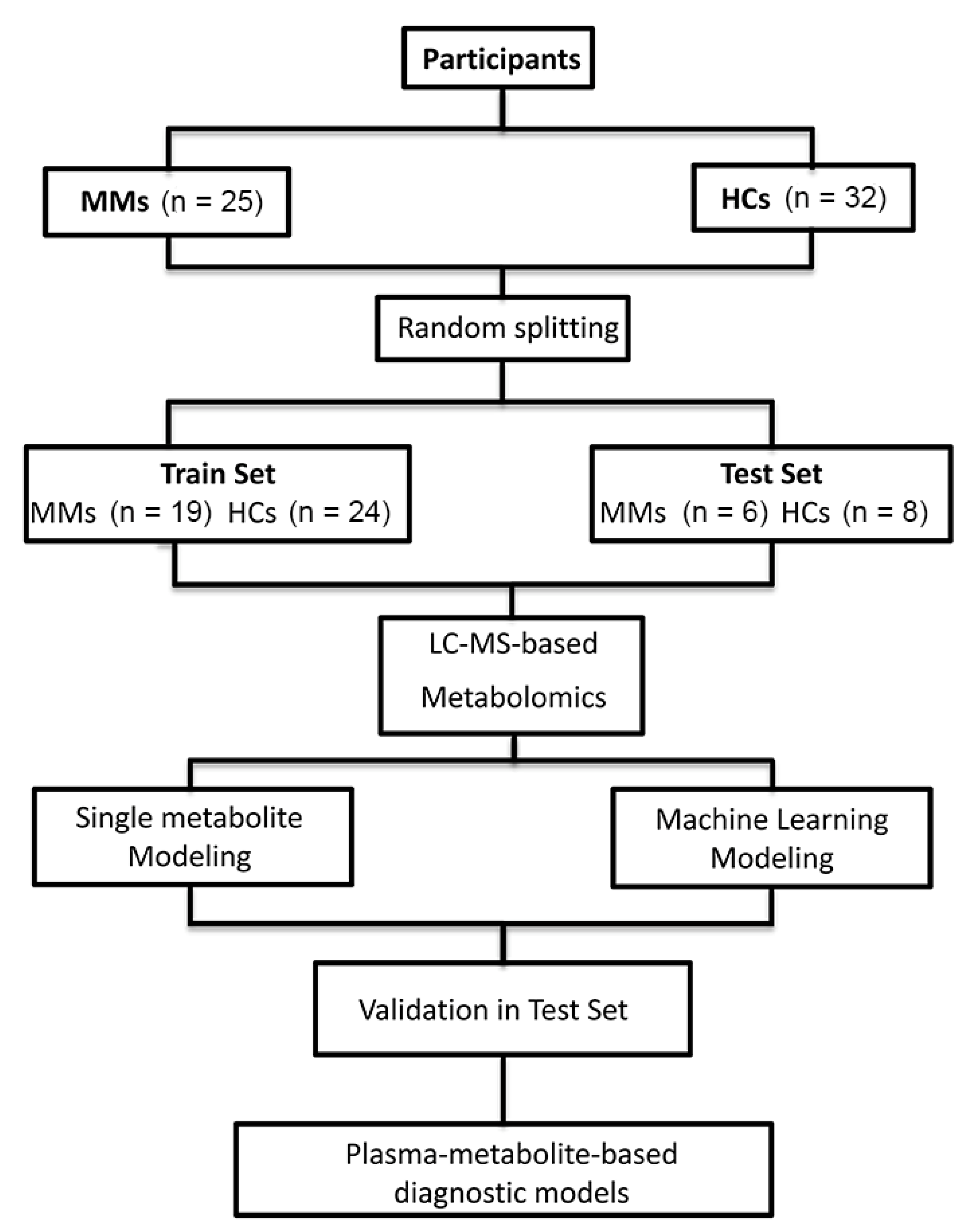
2.2. Study Population and Sample Collection
2.3. Sample Preparation
2.4. LC-MS Analysis
2.5. Metabolomic Data Analysis
2.6. Metabolite-Based Diagnostic Modeling
2.7. Statistical Analyses
3. Results
3.1. Population Characteristics
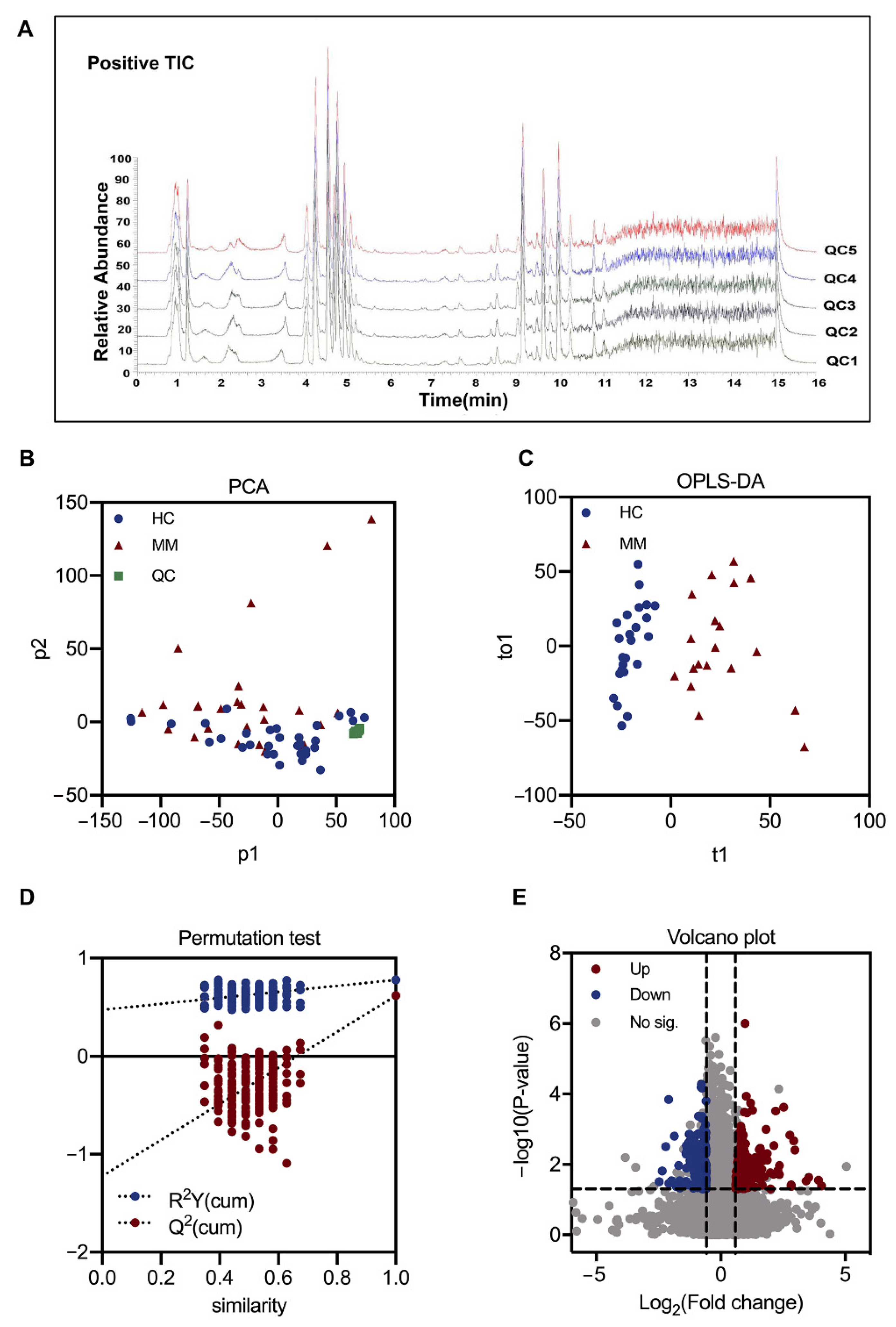
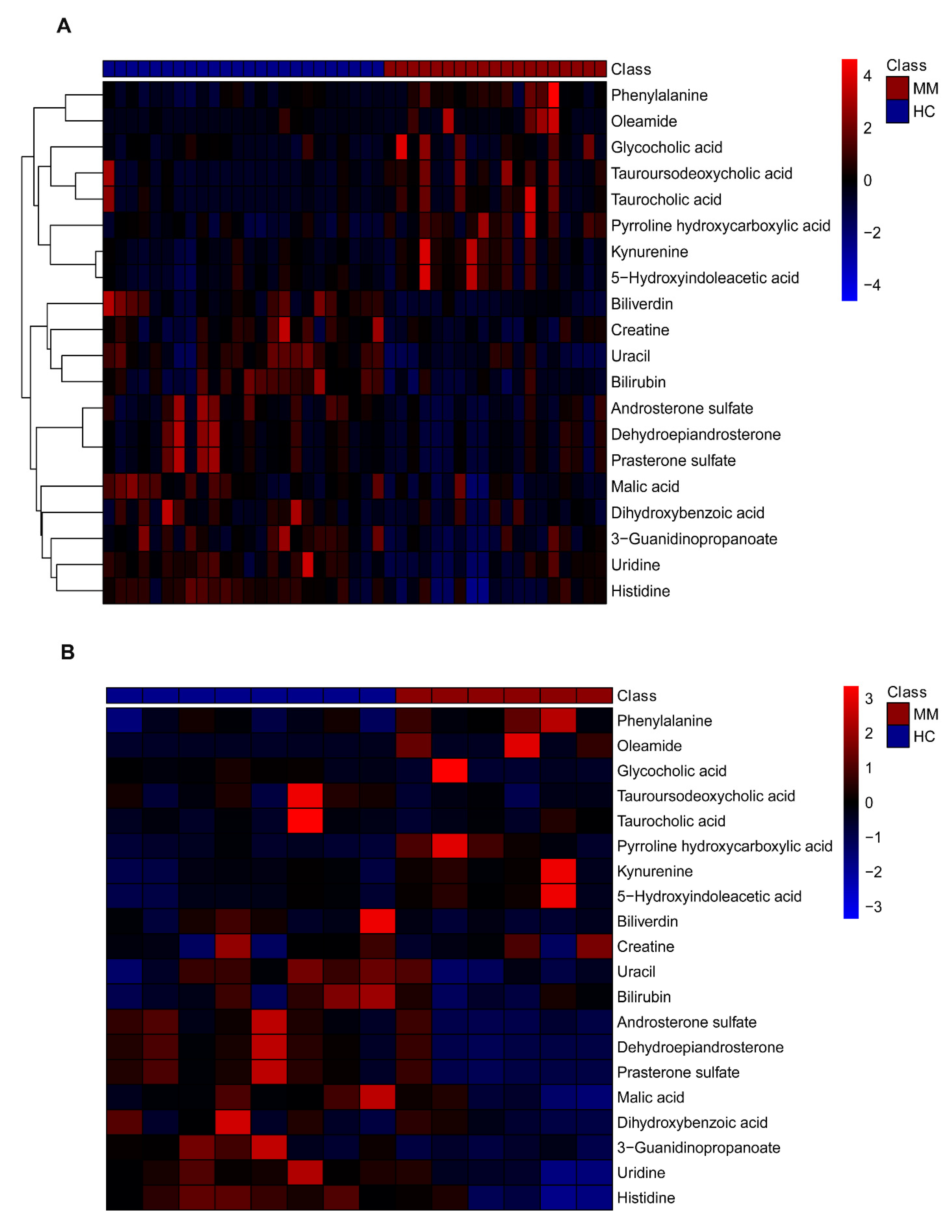
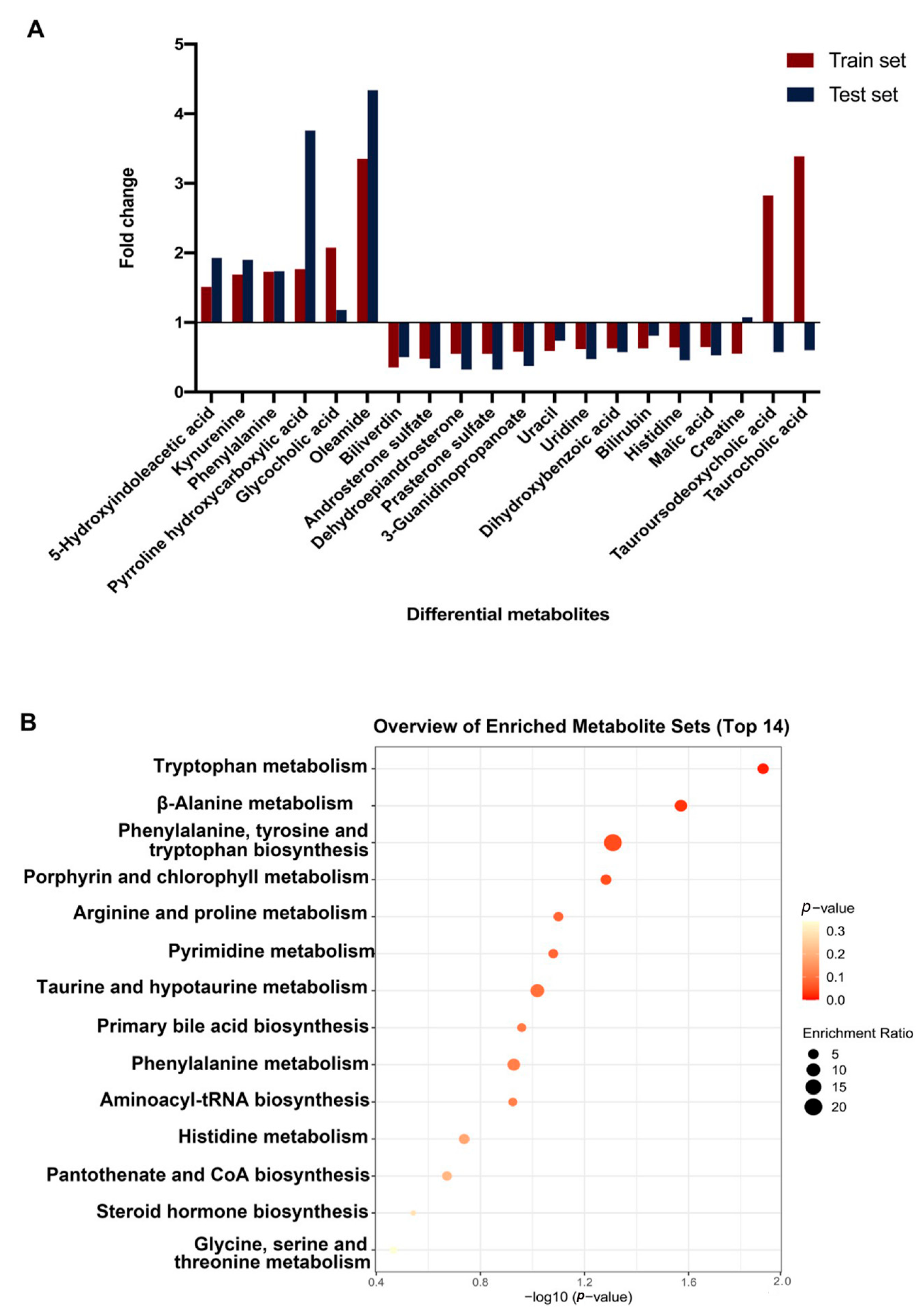
3.2. Plasma Metabolic Shift between MM and HC Groups
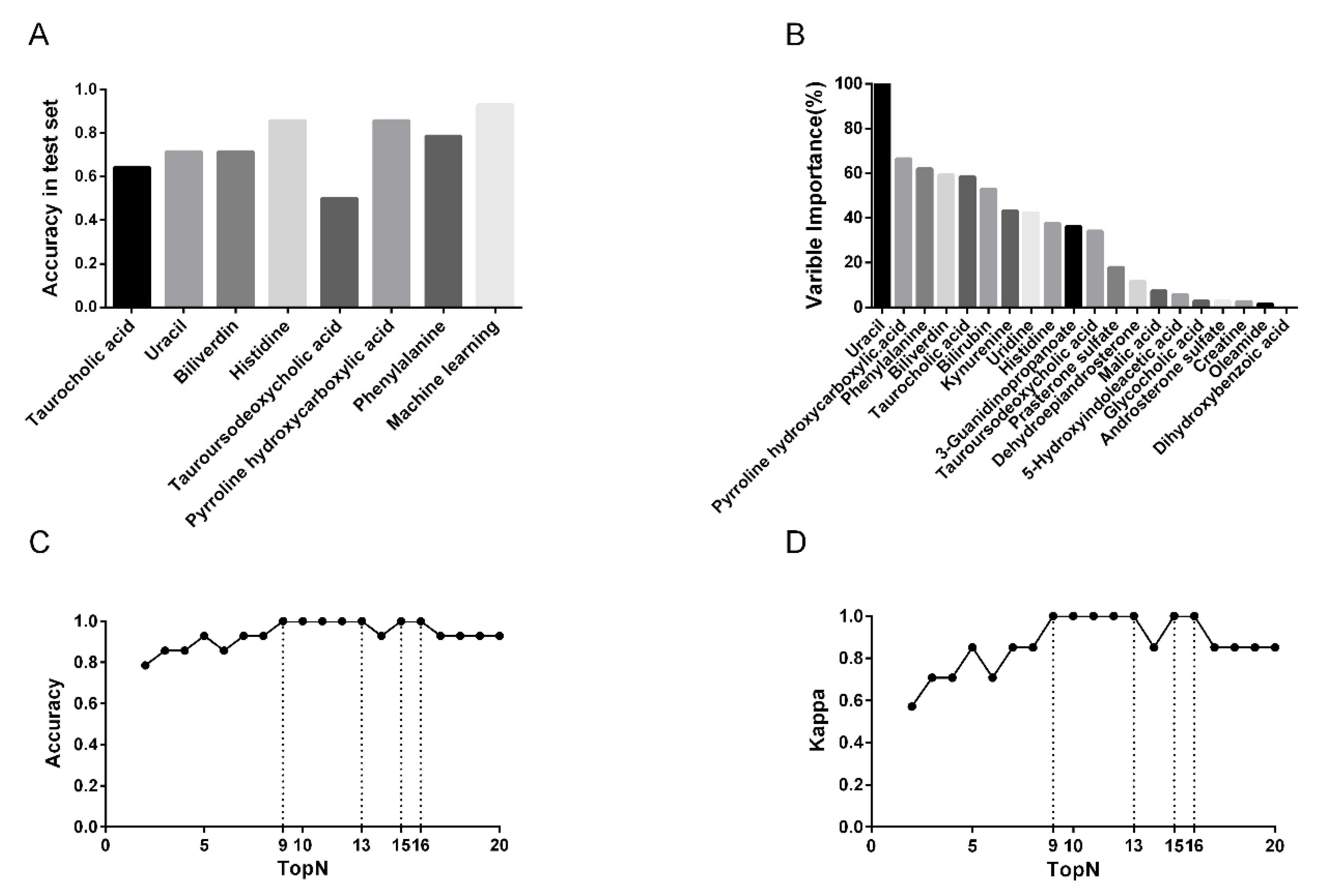
3.3. Predictive Performance of Single-Metabolite-Based Models and Multiple Metabolite-Based Machine Learning Model
3.4. Feature Metabolite Selection for RF Model
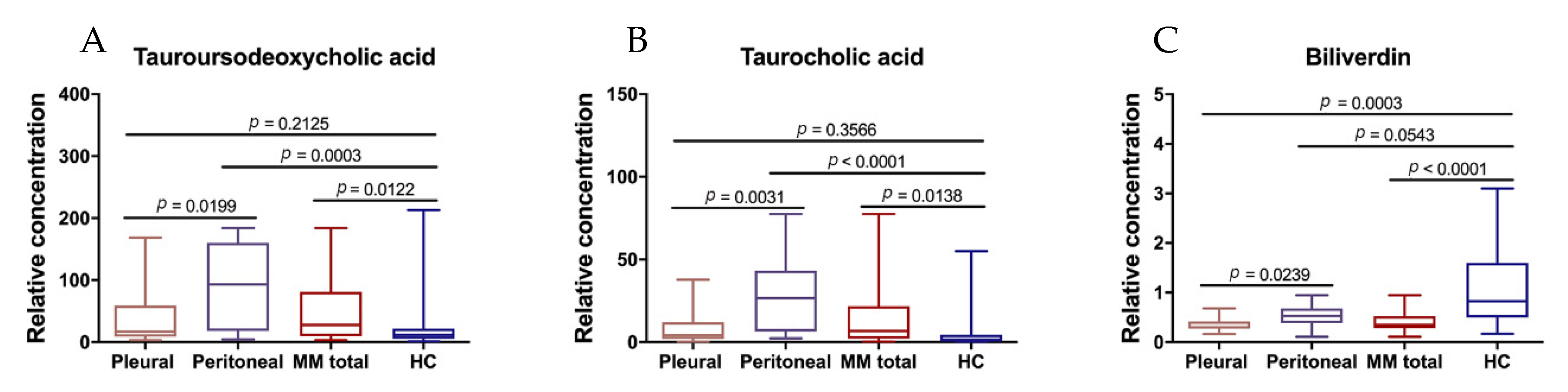
3.5. Differential Metabolites between Peritoneal and Pleural MMs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, B.; van Gerwen, M.; Bonassi, S.; Taioli, E. International Association for the Study of Lung Cancer Mesothelioma Task. Epidemiology of Environmental Exposure and Malignant Mesothelioma. J. Thorac. Oncol. 2017, 12, 1031–1045. [Google Scholar] [CrossRef]
- Szolkowska, M.; Blasinska-Przerwa, K.; Knetki-Wroblewska, M.; Rudzinski, P.; Langfort, R. Malignant pleural mesothelioma: Main topics of American Society of Clinical Oncology clinical practice guidelines for diagnosis and treatment. J. Thorac. Dis. 2018, 10 (Suppl. 17), S1966–S1970. [Google Scholar] [CrossRef]
- Carbone, M.; Adusumilli, P.S.; Alexander, H.R., Jr.; Baas, P.; Bardelli, F.; Bononi, A.; Bueno, R.; Felley-Bosco, E.; Galateau-Salle, F.; Jablons, D.; et al. Mesothelioma: Scientific clues for prevention, diagnosis, and therapy. CA Cancer J. Clin. 2019, 69, 402–429. [Google Scholar] [CrossRef]
- Zhao, J.; Zuo, T.; Zheng, R.; Zhang, S.; Zeng, H.; Xia, C.; Yang, Z.; Chen, W. Epidemiology and trend analysis on malignant mesothelioma in China. Chin. J. Cancer Res. 2017, 29, 361–368. [Google Scholar] [CrossRef]
- De Lara, P.T.; Cecconi, V.; Hiltbrunner, S.; Yagita, H.; Friess, M.; Bode, B.; Opitz, I.; Vrugt, B.; Weder, W.; Stolzmann, P.; et al. Gemcitabine Synergizes with Immune Checkpoint Inhibitors and Overcomes Resistance in a Preclinical Model and Mesothelioma Patients. Clin. Cancer Res. 2018, 24, 6345–6354. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.A.; Aerts, J.G.; Popat, S.; Fennell, D.A. Novel insights into mesothelioma biology and implications for therapy. Nat. Rev. Cancer 2017, 17, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Calabrò, L.; Morra, A.; Cornelissen, R.; Aerts, J.; Maio, M. Immune checkpoint blockade therapy of mesothelioma: A clinical and radiological challenge. Cancer Immunol. Immunother. 2018, 67, 1317–1324. [Google Scholar] [CrossRef]
- Wang, C.; Peng, J.; Kuang, Y.; Zhang, J.; Dai, L. Metabolomic analysis based on 1H-nuclear magnetic resonance spectroscopy metabolic profiles in tuberculous, malignant and transudative pleural effusion. Mol. Med. Rep. 2017, 16, 1147–1156. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Torresano, L.; Nuevo-Tapioles, C.; Santacatterina, F.; Cuezva, J.M. Metabolic reprogramming and disease progression in cancer patients. Biochim. Biophys. Acta BBA—Mol. Basis Dis. 2020, 1866, 165721. [Google Scholar] [CrossRef]
- Khan, I.; Nam, M.; Kwon, M.; Seo, S.-S.; Jung, S.; Han, J.S.; Hwang, G.-S.; Kim, M.K. LC/MS-Based Polar Metabolite Profiling Identified Unique Biomarker Signatures for Cervical Cancer and Cervical Intraepithelial Neoplasia Using Global and Targeted Metabolomics. Cancers 2019, 11, 511. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Gao, D.; Wang, Y.; Jin, F.; Wu, Q.; Liu, H. Application of metabolomics to investigate the antitumor mechanism of flavopiridol in MCF-7 breast cancer cells. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1025, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wang, L.; Hu, X.; Chen, S.; Tao, Y.; Su, H.; Yang, J.; Xu, W.; Vedarethinam, V.; Wu, S.; et al. Machine learning of serum metabolic patterns encodes early-stage lung adenocarcinoma. Nat. Commun. 2020, 11, 3556. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef]
- Chen, Z.; Dai, Y.; Huang, X.; Chen, K.; Gao, Y.; Li, N.; Wang, D.; Chen, A.; Yang, Q.; Hong, Y.; et al. Combined Metabolomic Analysis of Plasma and Tissue Reveals a Prognostic Risk Score System and Metabolic Dysregulation in Esophageal Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 1545. [Google Scholar] [CrossRef]
- Yang, Z.; Song, Z.; Chen, Z.; Guo, Z.; Jin, H.; Ding, C.; Hong, Y.; Cai, Z. Metabolic and lipidomic characterization of malignant pleural effusion in human lung cancer. J. Pharm. Biomed. Anal. 2020, 180, 113069. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gaudino, G.; Pass, H.; Carbone, M.; Yang, H. Diagnostic and prognostic biomarkers for malignant mesothelioma: An update. Transl. Lung Cancer Res. 2017, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Creeden, J.F.; Gordon, D.M.; Stec, D.E.; Hinds, T.D. Bilirubin as a metabolic hormone: The physiological relevance of low levels. Am. J. Physiol. Endocrinol. Metab. 2021, 320, E191–E207. [Google Scholar] [CrossRef]
- Rao, P.; Suzuki, R.; Mizobuchi, S.; Yamaguchi, T.; Sasaguri, S. Bilirubin exhibits a novel anti-cancer effect on human adenocarcinoma. Biochem. Biophys. Res. Commun. 2006, 342, 1279–1283. [Google Scholar] [CrossRef] [PubMed]
- Khoei, N.S.; Jenab, M.; Murphy, N.; Banbury, B.L.; Carreras-Torres, R.; Viallon, V.; Kühn, T.; Bueno-De-Mesquita, B.; Aleksandrova, K.; Cross, A.J.; et al. Circulating bilirubin levels and risk of colorectal cancer: Serological and Mendelian randomization analyses. BMC Med. 2020, 18, 229. [Google Scholar] [CrossRef]
- Horsfall, L.J.; Burgess, S.; Hall, I.; Nazareth, I. Genetically raised serum bilirubin levels and lung cancer: A cohort study and Mendelian randomisation using UK Biobank. Thorax 2020, 75, 955–964. [Google Scholar] [CrossRef]
- Synakiewicz, A.; Stanislawska-Sachadyn, A.; Sawicka-Zukowska, M.; Galezowska, G.; Ratajczyk, J.; Owczarzak, A.; Skuza, M.; Wolska, L.; Stachowicz-Stencel, T. Correction to: Plasma free amino acid profiling as metabolomic diagnostic and prognostic biomarker in paediatric cancer patients: A follow-up study. Amino Acids 2021, 53, 139–141. [Google Scholar] [CrossRef]
- Najumudeen, A.K.; Ceteci, F.; Fey, S.K.; Hamm, G.; Steven, R.T.; Hall, H.; Nikula, C.J.; Dexter, A.; Murta, T.; Race, A.M.; et al. The amino acid transporter SLC7A5 is required for efficient growth of KRAS-mutant colorectal cancer. Nat. Genet. 2021, 53, 16–26. [Google Scholar] [CrossRef]
- Platten, M.; Nollen, E.A.A.; Röhrig, U.F.; Fallarino, F.; Opitz, C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019, 18, 379–401. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-L.; Zhang, A.-H.; Miao, J.-H.; Sun, H.; Yan, G.-L.; Wu, F.-F.; Wang, X.-J. Targeting regulation of tryptophan metabolism for colorectal cancer therapy: A systematic review. RSC Adv. 2019, 9, 3072–3080. [Google Scholar] [CrossRef]
- Zhang, J.; Pavlova, N.N.; Thompson, C.B. Cancer cell metabolism: The essential role of the nonessential amino acid, glutamine. EMBO J. 2017, 36, 1302–1315. [Google Scholar] [CrossRef] [PubMed]
- Qamar, A.; Mysore, K.S.; Senthil-Kumar, M. Role of proline and pyrroline-5-carboxylate metabolism in plant defense against invading pathogens. Front. Plant Sci. 2015, 6, 503. [Google Scholar] [CrossRef]
- Schoemaker, M.H.; De La Rosa, L.C.; Buist-Homan, M.; Vrenken, T.E.; Havinga, R.; Poelstra, K.; Haisma, H.; Jansen, P.L.M.; Moshage, H. Tauroursodeoxycholic acid protects rat hepatocytes from bile acid-induced apoptosis via activation of survival pathways. Hepatology 2004, 39, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, C.; Bernstein, H.; Garewal, H.; Dinning, P.; Jabi, R.; Sampliner, R.E.; McCuskey, M.K.; Panda, M.; Roe, D.J.; L’Heureux, L.; et al. A bile acid-induced apoptosis assay for colon cancer risk and associated quality control studies. Cancer Res. 1999, 59, 2353–2357. [Google Scholar]
- Ridlon, J.M.; Wolf, P.G.; Gaskins, H.R. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes 2016, 7, 201–215. [Google Scholar] [CrossRef]

| Feature | HC (n = 32) | MM (n = 25) | p-Value a |
|---|---|---|---|
| Gender | 0.83 | ||
| Male | 17 (53.1%) | 14 (56.0%) | |
| Female | 15 (46.9%) | 11 (44.0%) | |
| Age | 0.99 | ||
| Mean ± SD | 55.8 ± 7.4 | 55.7 ± 10.9 | |
| Site | NA | ||
| Pleural mesothelioma | NA | 17 (68.0%) | |
| Peritoneal mesothelioma | NA | 8 (32.0%) | |
| Asbestos exposure | NA | ||
| Yes | NA | 11 (44.0%) | |
| No | NA | 10 (40.0%) | |
| Unknown | NA | 4 (16.0%) |
| Metabolite | Mode a | m/z b | rt (min) c | Train Set d | Test Set | |||
|---|---|---|---|---|---|---|---|---|
| VIP e | p-Value | FC f | p-Value | FC f | ||||
| Histidine g | Neg | 154.061 | 0.86 | 2.06 | 7.36 × 10−5 | 0.64 | 2.27 × 10−3 | 0.46 |
| Uracil | Pos | 113.035 | 2.16 | 1.52 | 1.21 × 10−4 | 0.59 | 1.09 × 10−1 | 0.74 |
| Biliverdin | Pos | 639.407 | 11 | 1.34 | 2.23 × 10−4 | 0.36 | 1.33 × 10−1 | 0.5 |
| Pyrroline hydroxycarboxylic acid g | Pos | 130.05 | 5.16 | 2.05 | 4.42 × 10−4 | 1.77 | 1.66 × 10−2 | 3.76 |
| Bilirubin | Pos | 585.27 | 14.42 | 1.31 | 6.79 × 10−4 | 0.63 | 5.03 × 10−1 | 0.81 |
| Phenylalanine g | Pos | 166.086 | 4.57 | 2.10 | 1.31 × 10−3 | 1.73 | 2.72 × 10−2 | 1.74 |
| Uridine g | Neg | 243.063 | 2.12 | 1.06 | 2.87 × 10−3 | 0.62 | 1.27 × 10−2 | 0.48 |
| Kynurenine | Pos | 209.092 | 3.96 | 1.38 | 3.46 × 10−3 | 1.69 | 5.67 × 10−2 | 1.9 |
| Malic acid | Neg | 133.013 | 1.15 | 1.45 | 3.66 × 10−3 | 0.65 | 5.61 × 10−2 | 0.53 |
| Androsterone sulfate g | Neg | 369.176 | 6.72 | 1.27 | 5.62 × 10−3 | 0.48 | 3.66 × 10−2 | 0.34 |
| Tauroursodeoxycholic acid | Neg | 498.292 | 7.53 | 1.52 | 6.26 × 10−3 | 2.82 | 1.63 × 10−1 | 0.58 |
| 3-Guanidinopropanoate g | Neg | 130.061 | 0.97 | 1.14 | 7.81 × 10−3 | 0.58 | 2.07 × 10−2 | 0.37 |
| Taurocholic acid | Neg | 514.287 | 6.92 | 1.82 | 8.26 × 10−3 | 3.39 | 5.49 × 10−1 | 0.6 |
| 5-Hydroxyindoleacetic acid g | Pos | 192.065 | 3.96 | 1.16 | 1.69 × 10−2 | 1.51 | 4.85 × 10−2 | 1.93 |
| Oleamide g | Pos | 282.279 | 12.16 | 1.73 | 2.11 × 10−2 | 3.35 | 2.25 × 10−2 | 4.34 |
| Glycocholic acid | Neg | 464.304 | 6.71 | 1.01 | 2.16 × 10−2 | 2.07 | 8.21 × 10−1 | 1.18 |
| Dehydroepiandrosterone g | Neg | 367.16 | 7.28 | 1.12 | 2.59 × 10−2 | 0.55 | 1.88 × 10−2 | 0.33 |
| Prasterone sulfate g | Neg | 367.16 | 7.28 | 1.12 | 2.59 × 10−2 | 0.55 | 1.88 × 10−2 | 0.33 |
| Creatine | Pos | 132.077 | 0.97 | 1.32 | 2.85 × 10−2 | 0.55 | 8.72 × 10−1 | 1.07 |
| Dihydroxybenzoic acid | Neg | 153.019 | 5.08 | 1.12 | 4.96 × 10−2 | 0.63 | 3.33 × 10−1 | 0.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, N.; Yang, C.; Zhou, S.; Song, S.; Jin, Y.; Wang, D.; Liu, J.; Gao, Y.; Yang, H.; Mao, W.; et al. Combination of Plasma-Based Metabolomics and Machine Learning Algorithm Provides a Novel Diagnostic Strategy for Malignant Mesothelioma. Diagnostics 2021, 11, 1281. https://doi.org/10.3390/diagnostics11071281
Li N, Yang C, Zhou S, Song S, Jin Y, Wang D, Liu J, Gao Y, Yang H, Mao W, et al. Combination of Plasma-Based Metabolomics and Machine Learning Algorithm Provides a Novel Diagnostic Strategy for Malignant Mesothelioma. Diagnostics. 2021; 11(7):1281. https://doi.org/10.3390/diagnostics11071281
Chicago/Turabian StyleLi, Na, Chenxi Yang, Sicheng Zhou, Siyu Song, Yuyao Jin, Ding Wang, Junping Liu, Yun Gao, Haining Yang, Weimin Mao, and et al. 2021. "Combination of Plasma-Based Metabolomics and Machine Learning Algorithm Provides a Novel Diagnostic Strategy for Malignant Mesothelioma" Diagnostics 11, no. 7: 1281. https://doi.org/10.3390/diagnostics11071281
APA StyleLi, N., Yang, C., Zhou, S., Song, S., Jin, Y., Wang, D., Liu, J., Gao, Y., Yang, H., Mao, W., & Chen, Z. (2021). Combination of Plasma-Based Metabolomics and Machine Learning Algorithm Provides a Novel Diagnostic Strategy for Malignant Mesothelioma. Diagnostics, 11(7), 1281. https://doi.org/10.3390/diagnostics11071281





