A Current and Newly Proposed Artificial Intelligence Algorithm for Reading Small Bowel Capsule Endoscopy
Abstract
:1. Introduction
2. Application of Artificial Intelligence (AI) into the Reading of Small Bowel Capsule Endoscopy (SBCE)
2.1. Automatic Detection of Small Bowel Lesions
2.2. Automatic Classification of Small Bowel Cleanliness
2.3. Automatic Compartmentalization of Small Bowel
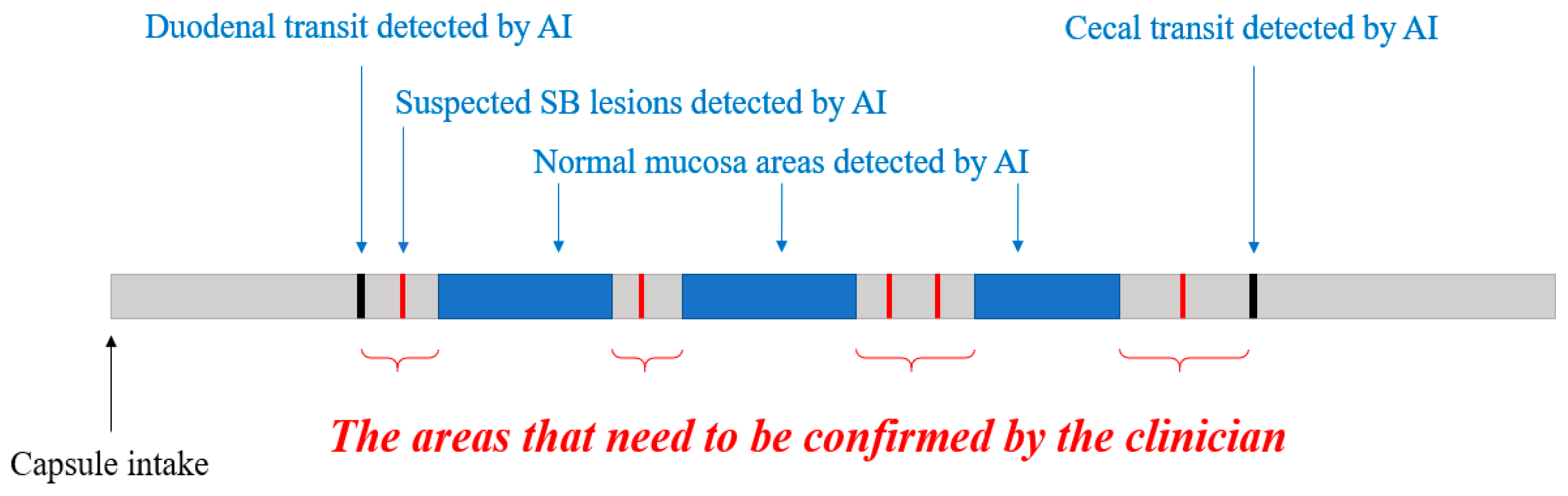
3. New Proposals for Using an AI Algorithm in Clinical Practice
3.1. Automatic Filtering of Normal Images
3.2. Automatic Reconfirmation of Small Bowel Lesion
3.3. New Consensus on the Small Bowel Lesions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iddan, G.; Meron, G.; Glukhovsky, A.; Swain, P. Wireless capsule endoscopy. Nature 2000, 405, 417. [Google Scholar] [CrossRef]
- Committee, A.T.; Wang, A.; Banerjee, S.; Barth, B.A.; Bhat, Y.M.; Chauhan, S.; Gottlieb, K.T.; Konda, V.; Maple, J.T.; Murad, F.; et al. Wireless capsule endoscopy. Gastrointest. Endosc. 2013, 78, 805–815. [Google Scholar] [CrossRef]
- Pennazio, M.; Spada, C.; Eliakim, R.; Keuchel, M.; May, A.; Mulder, C.J.; Rondonotti, E.; Adler, S.N.; Albert, J.; Baltes, P.; et al. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) Clinical Guideline. Endoscopy 2015, 47, 352–376. [Google Scholar] [CrossRef] [Green Version]
- Enns, R.A.; Hookey, L.; Armstrong, D.; Bernstein, C.N.; Heitman, S.J.; Teshima, C.; Leontiadis, G.I.; Tse, F.; Sadowski, D. Clinical Practice Guidelines for the Use of Video Capsule Endoscopy. Gastroenterology 2017, 152, 497–514. [Google Scholar] [CrossRef] [Green Version]
- Kwack, W.G.; Lim, Y.J. Current Status and Research into Overcoming Limitations of Capsule Endoscopy. Clin. Endosc. 2016, 49, 8–15. [Google Scholar] [CrossRef]
- Ou, G.; Shahidi, N.; Galorport, C.; Takach, O.; Lee, T.; Enns, R. Effect of longer battery life on small bowel capsule endoscopy. World J. Gastroenterol. 2015, 21, 2677–2682. [Google Scholar] [CrossRef]
- Tacchino, R.M. Bowel length: Measurement, predictors, and impact on bariatric and metabolic surgery. Surg. Obes. Relat. Dis. 2015, 11, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.J.; Lee, O.Y.; Jeen, Y.T.; Lim, C.Y.; Cheung, D.Y.; Cheon, J.H.; Ye, B.D.; Song, H.J.; Kim, J.S.; Do, J.H.; et al. Indications for Detection, Completion, and Retention Rates of Small Bowel Capsule Endoscopy Based on the 10-Year Data from the Korean Capsule Endoscopy Registry. Clin. Endosc. 2015, 48, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Rondonotti, E.; Spada, C.; Adler, S.; May, A.; Despott, E.J.; Koulaouzidis, A.; Panter, S.; Domagk, D.; Fernandez-Urien, I.; Rahmi, G.; et al. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) Technical Review. Endoscopy 2018, 50, 423–446. [Google Scholar] [CrossRef] [Green Version]
- D’Halluin, P.N.; Delvaux, M.; Lapalus, M.G.; Sacher-Huvelin, S.; Ben Soussan, E.; Heyries, L.; Filoche, B.; Saurin, J.C.; Gay, G.; Heresbach, D. Does the “Suspected Blood Indicator” improve the detection of bleeding lesions by capsule endoscopy? Gastrointest. Endosc. 2005, 61, 243–249. [Google Scholar] [CrossRef]
- Yung, D.E.; Sykes, C.; Koulaouzidis, A. The validity of suspected blood indicator software in capsule endoscopy: A systematic review and meta-analysis. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Shiotani, A.; Honda, K.; Kawakami, M.; Murao, T.; Matsumoto, H.; Tarumi, K.; Kusunoki, H.; Hata, J.; Haruma, K. Evaluation of RAPID((R)) 5 Access software for examination of capsule endoscopies and reading of the capsule by an endoscopy nurse. J. Gastroenterol. 2011, 46, 138–142. [Google Scholar] [CrossRef]
- Oh, D.J.; Kim, K.S.; Lim, Y.J. A New Active Locomotion Capsule Endoscopy under Magnetic Control and Automated Reading Program. Clin. Endosc. 2020, 53, 395–401. [Google Scholar] [CrossRef]
- Phillips, F.; Beg, S. Video capsule endoscopy: Pushing the boundaries with software technology. Transl. Gastroenterol. Hepatol. 2021, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Meng, M.Q. A deep convolutional neural network for bleeding detection in Wireless Capsule Endoscopy images. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2016, 2016, 639–642. [Google Scholar] [CrossRef]
- Aoki, T.; Yamada, A.; Aoyama, K.; Saito, H.; Tsuboi, A.; Nakada, A.; Niikura, R.; Fujishiro, M.; Oka, S.; Ishihara, S.; et al. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest. Endosc. 2019, 89, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xing, Y.; Zhang, L.; Gao, H.; Zhang, H. A systematic evaluation and optimization of automatic detection of ulcers in wireless capsule endoscopy on a large dataset using deep convolutional neural networks. Phys. Med. Biol. 2019, 64, 235014. [Google Scholar] [CrossRef] [PubMed]
- Klang, E.; Barash, Y.; Margalit, R.Y.; Soffer, S.; Shimon, O.; Albshesh, A.; Ben-Horin, S.; Amitai, M.M.; Eliakim, R.; Kopylov, U. Deep learning algorithms for automated detection of Crohn’s disease ulcers by video capsule endoscopy. Gastrointest. Endosc. 2020, 91, 606–613. [Google Scholar] [CrossRef]
- Barash, Y.; Azaria, L.; Soffer, S.; Margalit Yehuda, R.; Shlomi, O.; Ben-Horin, S.; Eliakim, R.; Klang, E.; Kopylov, U. Ulcer severity grading in video capsule images of patients with Crohn’s disease: An ordinal neural network solution. Gastrointest. Endosc. 2021, 93, 187–192. [Google Scholar] [CrossRef]
- Leenhardt, R.; Vasseur, P.; Li, C.; Saurin, J.C.; Rahmi, G.; Cholet, F.; Becq, A.; Marteau, P.; Histace, A.; Dray, X.; et al. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest. Endosc. 2019, 89, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Tsuboi, A.; Oka, S.; Aoyama, K.; Saito, H.; Aoki, T.; Yamada, A.; Matsuda, T.; Fujishiro, M.; Ishihara, S.; Nakahori, M.; et al. Artificial intelligence using a convolutional neural network for automatic detection of small-bowel angioectasia in capsule endoscopy images. Dig. Endosc. 2020, 32, 382–390. [Google Scholar] [CrossRef]
- Aoki, T.; Yamada, A.; Kato, Y.; Saito, H.; Tsuboi, A.; Nakada, A.; Niikura, R.; Fujishiro, M.; Oka, S.; Ishihara, S.; et al. Automatic detection of blood content in capsule endoscopy images based on a deep convolutional neural network. J. Gastroenterol. Hepatol. 2020, 35, 1196–1200. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Aoki, T.; Aoyama, K.; Kato, Y.; Tsuboi, A.; Yamada, A.; Fujishiro, M.; Oka, S.; Ishihara, S.; Matsuda, T.; et al. Automatic detection and classification of protruding lesions in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest. Endosc. 2020, 92, 144–151. [Google Scholar] [CrossRef]
- Ding, Z.; Shi, H.; Zhang, H.; Meng, L.; Fan, M.; Han, C.; Zhang, K.; Ming, F.; Xie, X.; Liu, H.; et al. Gastroenterologist-Level Identification of Small-Bowel Diseases and Normal Variants by Capsule Endoscopy Using a Deep-Learning Model. Gastroenterology 2019, 157, 1044–1054. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Yamada, A.; Kato, Y.; Saito, H.; Tsuboi, A.; Nakada, A.; Niikura, R.; Fujishiro, M.; Oka, S.; Ishihara, S.; et al. Automatic detection of various abnormalities in capsule endoscopy videos by a deep learning-based system: A multicenter study. Gastrointest. Endosc. 2021, 93, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.; Lee, H.H.; Park, C.; Tama, B.A.; Kim, J.S.; Cheung, D.Y.; Chung, W.C.; Cho, Y.S.; Lee, K.M.; Choi, M.G.; et al. Improved classification and localization approach to small bowel capsule endoscopy using convolutional neural network. Dig. Endosc. 2021, 33, 598–607. [Google Scholar] [CrossRef]
- Park, J.; Hwang, Y.; Nam, J.H.; Oh, D.J.; Kim, K.B.; Song, H.J.; Kim, S.H.; Kang, S.H.; Jung, M.K.; Jeong Lim, Y. Artificial intelligence that determines the clinical significance of capsule endoscopy images can increase the efficiency of reading. PLoS ONE 2020, 15, e0241474. [Google Scholar] [CrossRef]
- Otani, K.; Nakada, A.; Kurose, Y.; Niikura, R.; Yamada, A.; Aoki, T.; Nakanishi, H.; Doyama, H.; Hasatani, K.; Sumiyoshi, T.; et al. Automatic detection of different types of small-bowel lesions on capsule endoscopy images using a newly developed deep convolutional neural network. Endoscopy 2020, 52, 786–791. [Google Scholar] [CrossRef]
- Aoki, T.; Yamada, A.; Aoyama, K.; Saito, H.; Fujisawa, G.; Odawara, N.; Kondo, R.; Tsuboi, A.; Ishibashi, R.; Nakada, A.; et al. Clinical usefulness of a deep learning-based system as the first screening on small-bowel capsule endoscopy reading. Dig. Endosc. 2020, 32, 585–591. [Google Scholar] [CrossRef]
- Spada, C.; McNamara, D.; Despott, E.J.; Adler, S.; Cash, B.D.; Fernandez-Urien, I.; Ivekovic, H.; Keuchel, M.; McAlindon, M.; Saurin, J.C.; et al. Performance measures for small-bowel endoscopy: A European Society of Gastrointestinal Endoscopy (ESGE) Quality Improvement Initiative. United Eur. Gastroenterol. J. 2019, 7, 614–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Lim, Y.J.; Park, J.; Shim, K.N.; Yang, D.H.; Chun, J.; Kim, J.S.; Lee, H.S.; Chun, H.J.; Research Group for Capsule Endoscopy/Small Bowel, E. Changes in performance of small bowel capsule endoscopy based on nationwide data from a Korean Capsule Endoscopy Registry. Korean J. Intern. Med. 2020, 35, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Ponte, A.; Pinho, R.; Rodrigues, A.; Carvalho, J. Review of small-bowel cleansing scales in capsule endoscopy: A panoply of choices. World J. Gastrointest. Endosc. 2016, 8, 600–609. [Google Scholar] [CrossRef]
- Van Weyenberg, S.J.; De Leest, H.T.; Mulder, C.J. Description of a novel grading system to assess the quality of bowel preparation in video capsule endoscopy. Endoscopy 2011, 43, 406–411. [Google Scholar] [CrossRef] [Green Version]
- Dray, X.; Houist, G.; Le Mouel, J.P.; Saurin, J.C.; Vanbiervliet, G.; Leandri, C.; Rahmi, G.; Duburque, C.; Kirchgesner, J.; Leenhardt, R.; et al. Prospective evaluation of third-generation small bowel capsule endoscopy videos by independent readers demonstrates poor reproducibility of cleanliness classifications. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101612. [Google Scholar] [CrossRef] [PubMed]
- Noorda, R.; Nevarez, A.; Colomer, A.; Pons Beltran, V.; Naranjo, V. Automatic evaluation of degree of cleanliness in capsule endoscopy based on a novel CNN architecture. Sci. Rep. 2020, 10, 17706. [Google Scholar] [CrossRef]
- Leenhardt, R.; Souchaud, M.; Houist, G.; Le Mouel, J.P.; Saurin, J.C.; Cholet, F.; Rahmi, G.; Leandri, C.; Histace, A.; Dray, X. A neural network-based algorithm for assessing the cleanliness of small bowel during capsule endoscopy. Endoscopy 2020. [Google Scholar] [CrossRef]
- Nam, J.H.; Hwang, Y.; Oh, D.J.; Park, J.; Kim, K.B.; Jung, M.K.; Lim, Y.J. Development of a deep learning-based software for calculating cleansing score in small bowel capsule endoscopy. Sci. Rep. 2021, 11, 4417. [Google Scholar] [CrossRef]
- ASGE Standards of Practice Committee; Fisher, L.; Lee Krinsky, M.; Anderson, M.A.; Appalaneni, V.; Banerjee, S.; Ben-Menachem, T.; Cash, B.D.; Decker, G.A.; Fanelli, R.D.; et al. The role of endoscopy in the management of obscure GI bleeding. Gastrointest. Endosc. 2010, 72, 471–479. [Google Scholar] [CrossRef]
- Tominaga, K.; Sato, H.; Yokomichi, H.; Tsuchiya, A.; Yoshida, T.; Kawata, Y.; Mizusawa, T.; Yokoyama, J.; Terai, S. Variation in small bowel transit time on capsule endoscopy. Ann. Transl. Med. 2020, 8, 348. [Google Scholar] [CrossRef] [PubMed]
- Rondonotti, E.; Herrerias, J.M.; Pennazio, M.; Caunedo, A.; Mascarenhas-Saraiva, M.; de Franchis, R. Complications, limitations, and failures of capsule endoscopy: A review of 733 cases. Gastrointest. Endosc. 2005, 62, 712–716. [Google Scholar] [CrossRef]
- Gan, T.; Liu, S.; Yang, J.; Zeng, B.; Yang, L. A pilot trial of Convolution Neural Network for automatic retention-monitoring of capsule endoscopes in the stomach and duodenal bulb. Sci. Rep. 2020, 10, 4103. [Google Scholar] [CrossRef] [Green Version]
- Soffer, S.; Klang, E.; Shimon, O.; Nachmias, N.; Eliakim, R.; Ben-Horin, S.; Kopylov, U.; Barash, Y. Deep learning for wireless capsule endoscopy: A systematic review and meta-analysis. Gastrointest. Endosc. 2020, 92, 831–839.e8. [Google Scholar] [CrossRef]
- Dray, X.; Iakovidis, D.; Houdeville, C.; Jover, R.; Diamantis, D.; Histace, A.; Koulaouzidis, A. Artificial intelligence in small bowel capsule endoscopy—Current status, challenges and future promise. J. Gastroenterol. Hepatol. 2021, 36, 12–19. [Google Scholar] [CrossRef]
- Kyriakos, N.; Karagiannis, S.; Galanis, P.; Liatsos, C.; Zouboulis-Vafiadis, I.; Georgiou, E.; Mavrogiannis, C. Evaluation of four time-saving methods of reading capsule endoscopy videos. Eur. J. Gastroenterol. Hepatol. 2012, 24, 1276–1280. [Google Scholar] [CrossRef]
- Hosoe, N.; Watanabe, K.; Miyazaki, T.; Shimatani, M.; Wakamatsu, T.; Okazaki, K.; Esaki, M.; Matsumoto, T.; Abe, T.; Kanai, T.; et al. Evaluation of performance of the Omni mode for detecting video capsule endoscopy images: A multicenter randomized controlled trial. Endosc. Int. Open 2016, 4, 878–882. [Google Scholar] [CrossRef]
- Beg, S.; Wronska, E.; Araujo, I.; Gonzalez Suarez, B.; Ivanova, E.; Fedorov, E.; Aabakken, L.; Seitz, U.; Rey, J.F.; Saurin, J.C.; et al. Use of rapid reading software to reduce capsule endoscopy reading times while maintaining accuracy. Gastrointest. Endosc. 2020, 91, 1322–1327. [Google Scholar] [CrossRef]
- Saurin, J.C.; Delvaux, M.; Gaudin, J.L.; Fassler, I.; Villarejo, J.; Vahedi, K.; Bitoun, A.; Canard, J.M.; Souquet, J.C.; Ponchon, T.; et al. Diagnostic value of endoscopic capsule in patients with obscure digestive bleeding: Blinded comparison with video push-enteroscopy. Endoscopy 2003, 35, 576–584. [Google Scholar] [CrossRef]
- Korman, L.Y.; Delvaux, M.; Gay, G.; Hagenmuller, F.; Keuchel, M.; Friedman, S.; Weinstein, M.; Shetzline, M.; Cave, D.; de Franchis, R. Capsule endoscopy structured terminology (CEST): Proposal of a standardized and structured terminology for reporting capsule endoscopy procedures. Endoscopy 2005, 37, 951–959. [Google Scholar] [CrossRef]
- Lai, L.H.; Wong, G.L.; Chow, D.K.; Lau, J.Y.; Sung, J.J.; Leung, W.K. Inter-observer variations on interpretation of capsule endoscopies. Eur. J. Gastroenterol. Hepatol. 2006, 18, 283–286. [Google Scholar] [CrossRef]

| Author (CNN System) | Lesion Categories (Trained Images) Validation and/or Test (Images) | Results |
|---|---|---|
| Ding et al. [24] (ResNet) | 2 normal variants lymphangiectasia, lymphatic follicular hyperplasia 8 abnormal lesions inflammation, ulcer, bleeding, polyp vascular disease, protruding lesion, diverticulum, parasite (Total 158,235 trained images) | 1. Overall sensitivity 99%. 2. Overall specificity 100%. 3. Shorter reading times than conventional reading. (p < 0.001) |
| 5000 cases (113,268,334 images) | ||
| Aoki et al. [25] (SSD + ResNET50) | Mucosal breaks (5360 images) Angioectasia (2237 images) Protruding lesions (30,584 images) Blood content (6503 images) | 1. Detection rate was 100%, 97%, 99% and 100% for each lesion. |
| 379 cases (5,050,226 images) | ||
| Otani et al. [28] (RetinaNet) | Erosions and ulcers (398 images) Vascular lesions (538 images) Tumors (4590 images) | 1. AUC 0.996 at inflamed 2. AUC 0.950 at vascular 3. AUC 0.950 at tumors |
| 29 cases (14,867 images) in external validation | ||
| Park et al. [27] (Inception-Resnet-V2) | Inflamed mucosa Atypical vascularity, or bleeding (Total 60,000 images) | 1. Overall AUC 0.998. 2. Shorter the reading time for trainees (p = 0.029) |
| 20 cases (210,100 images) in external validation | ||
| Hwang et al. [26] (VGGNet and Grad-CAM) | Hemorrhagic lesions Ulcerative lesions (Total 3778 images 1) | 1. Overall AUC 0.9957 2. Sensitivity 96.95% 3. Specificity 97.13% |
| 162 cases (5760 images) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, D.J.; Hwang, Y.; Lim, Y.J. A Current and Newly Proposed Artificial Intelligence Algorithm for Reading Small Bowel Capsule Endoscopy. Diagnostics 2021, 11, 1183. https://doi.org/10.3390/diagnostics11071183
Oh DJ, Hwang Y, Lim YJ. A Current and Newly Proposed Artificial Intelligence Algorithm for Reading Small Bowel Capsule Endoscopy. Diagnostics. 2021; 11(7):1183. https://doi.org/10.3390/diagnostics11071183
Chicago/Turabian StyleOh, Dong Jun, Youngbae Hwang, and Yun Jeong Lim. 2021. "A Current and Newly Proposed Artificial Intelligence Algorithm for Reading Small Bowel Capsule Endoscopy" Diagnostics 11, no. 7: 1183. https://doi.org/10.3390/diagnostics11071183
APA StyleOh, D. J., Hwang, Y., & Lim, Y. J. (2021). A Current and Newly Proposed Artificial Intelligence Algorithm for Reading Small Bowel Capsule Endoscopy. Diagnostics, 11(7), 1183. https://doi.org/10.3390/diagnostics11071183








