The Effect of Genomic DNA Contamination on the Detection of Circulating Long Non-Coding RNAs: The Paradigm of MALAT1
Abstract
:1. Introduction
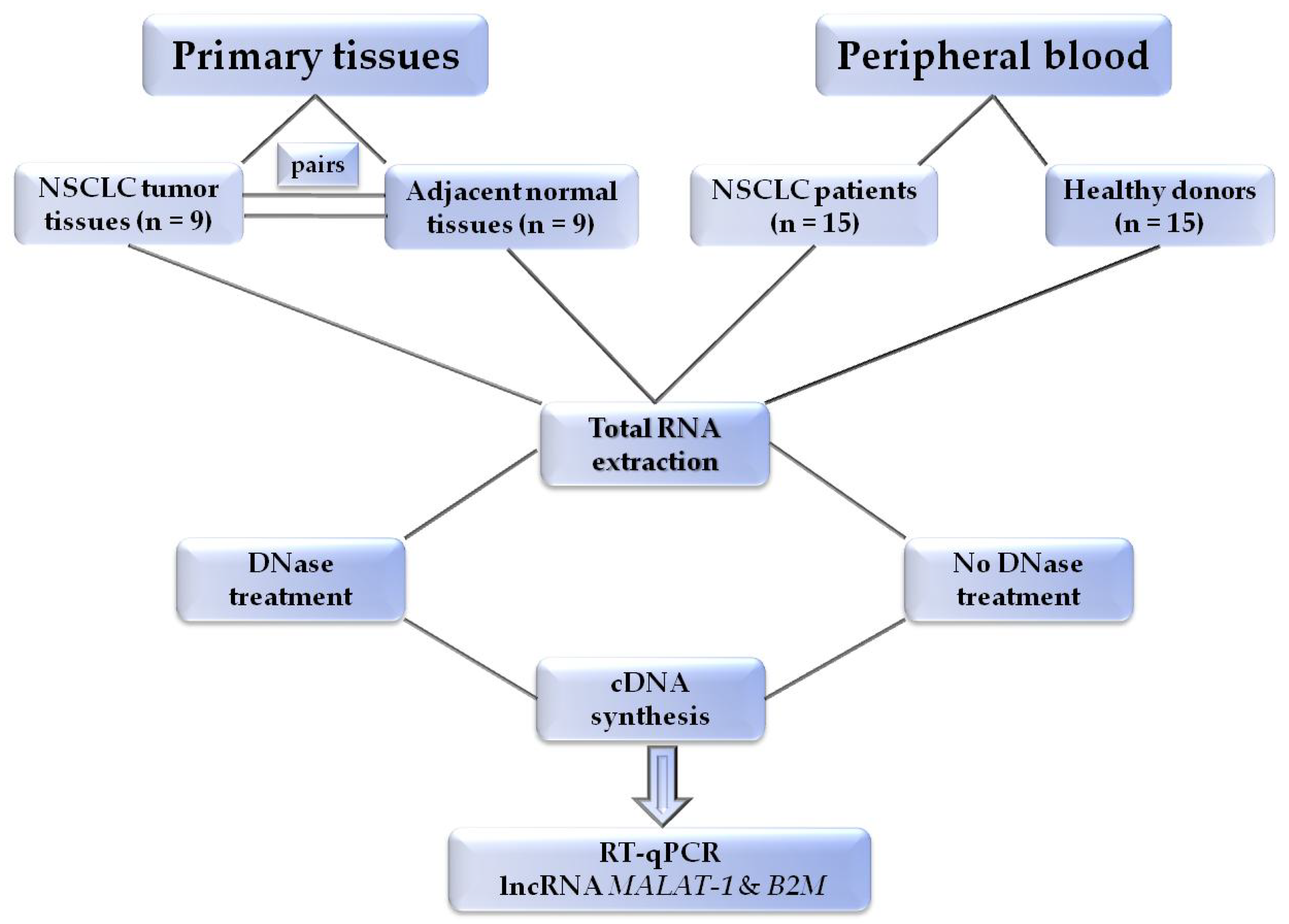
2. Materials and Methods
2.1. Patients and Samples
2.2. Plasma Preparation
2.3. RNA Extraction
2.4. DNAse Treatment
2.5. cDNA Synthesis
2.6. RT-qPCR Assay
2.7. Normalization of Data
2.8. Statistical Analysis
3. Results
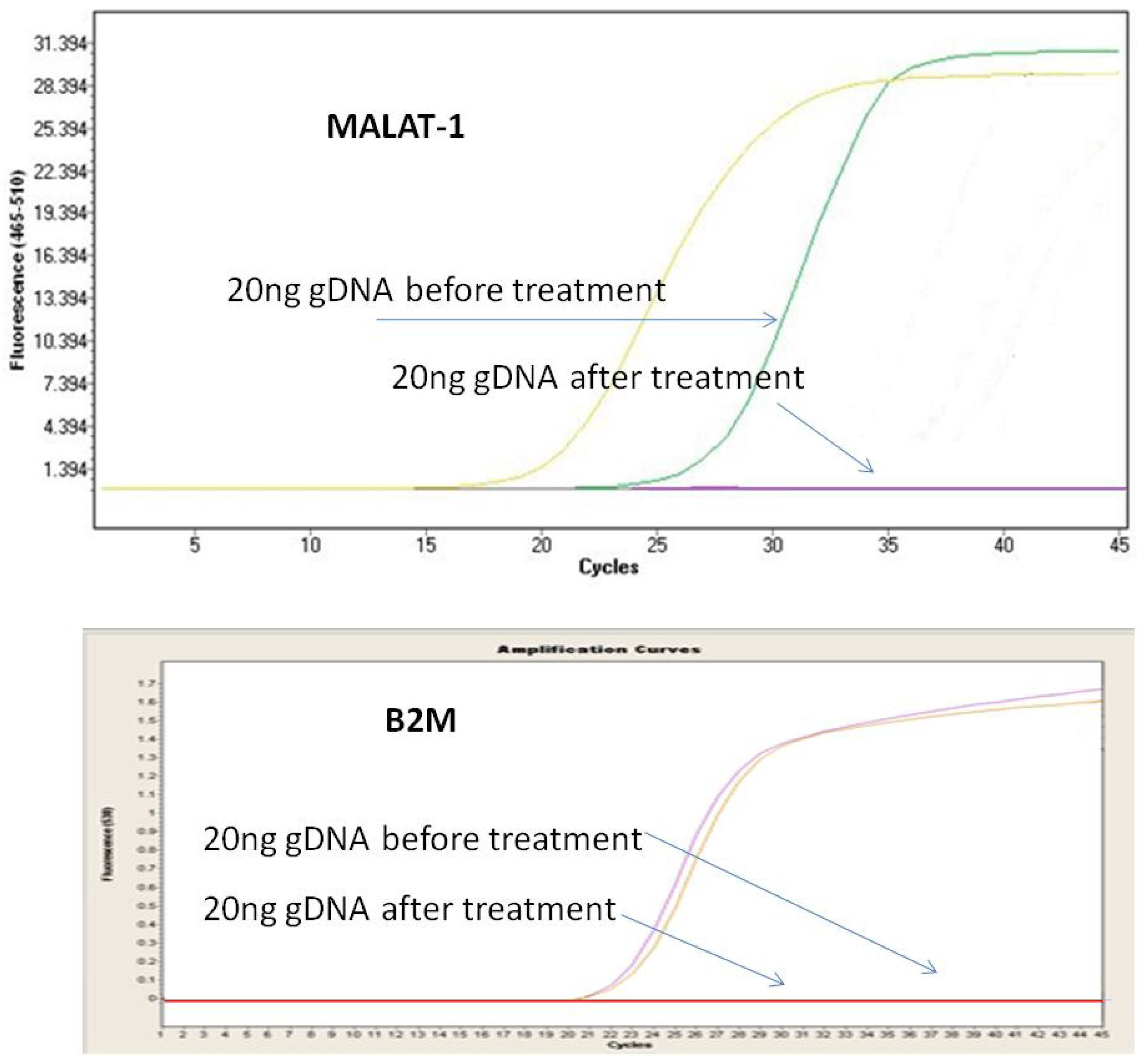
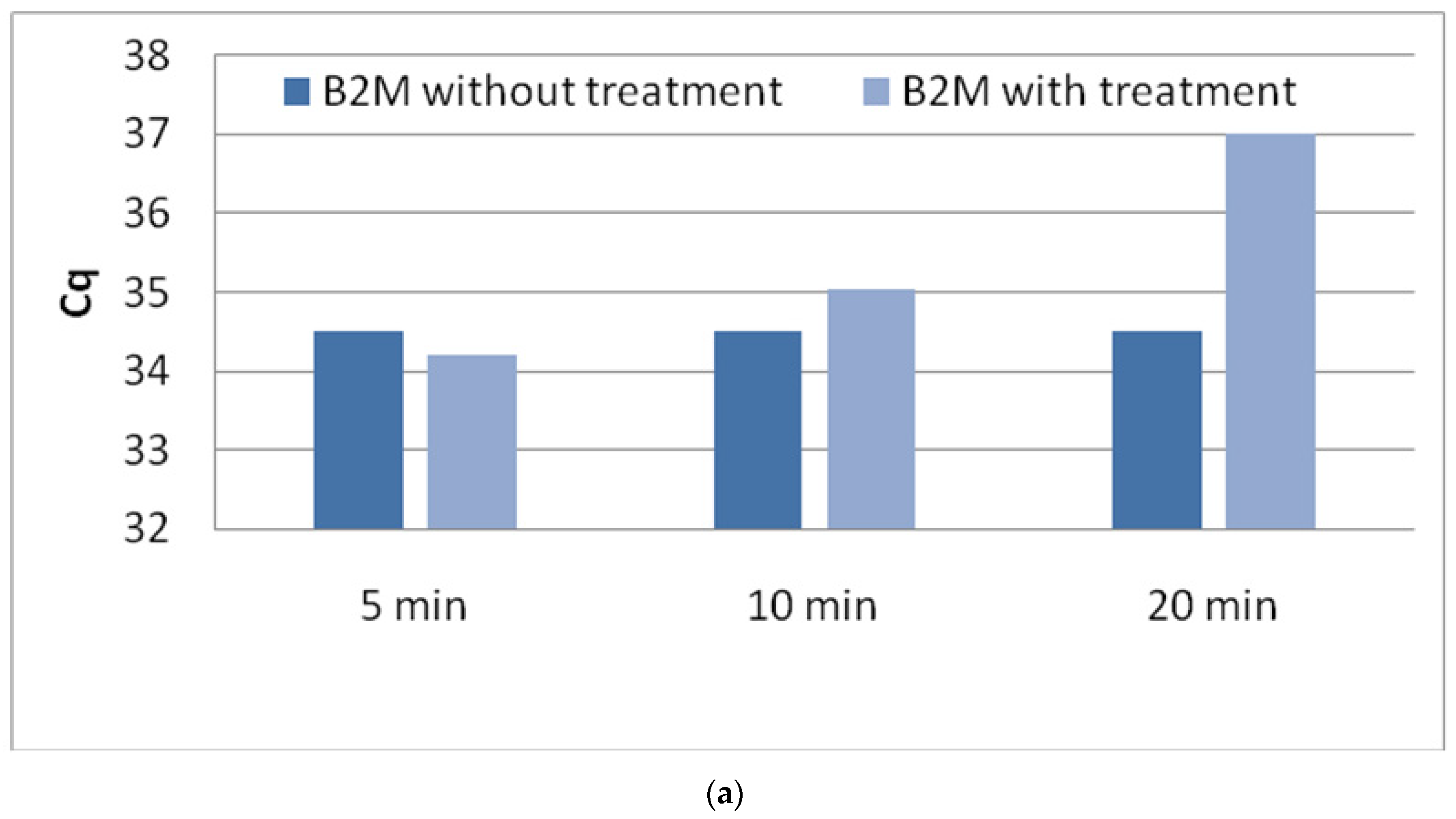
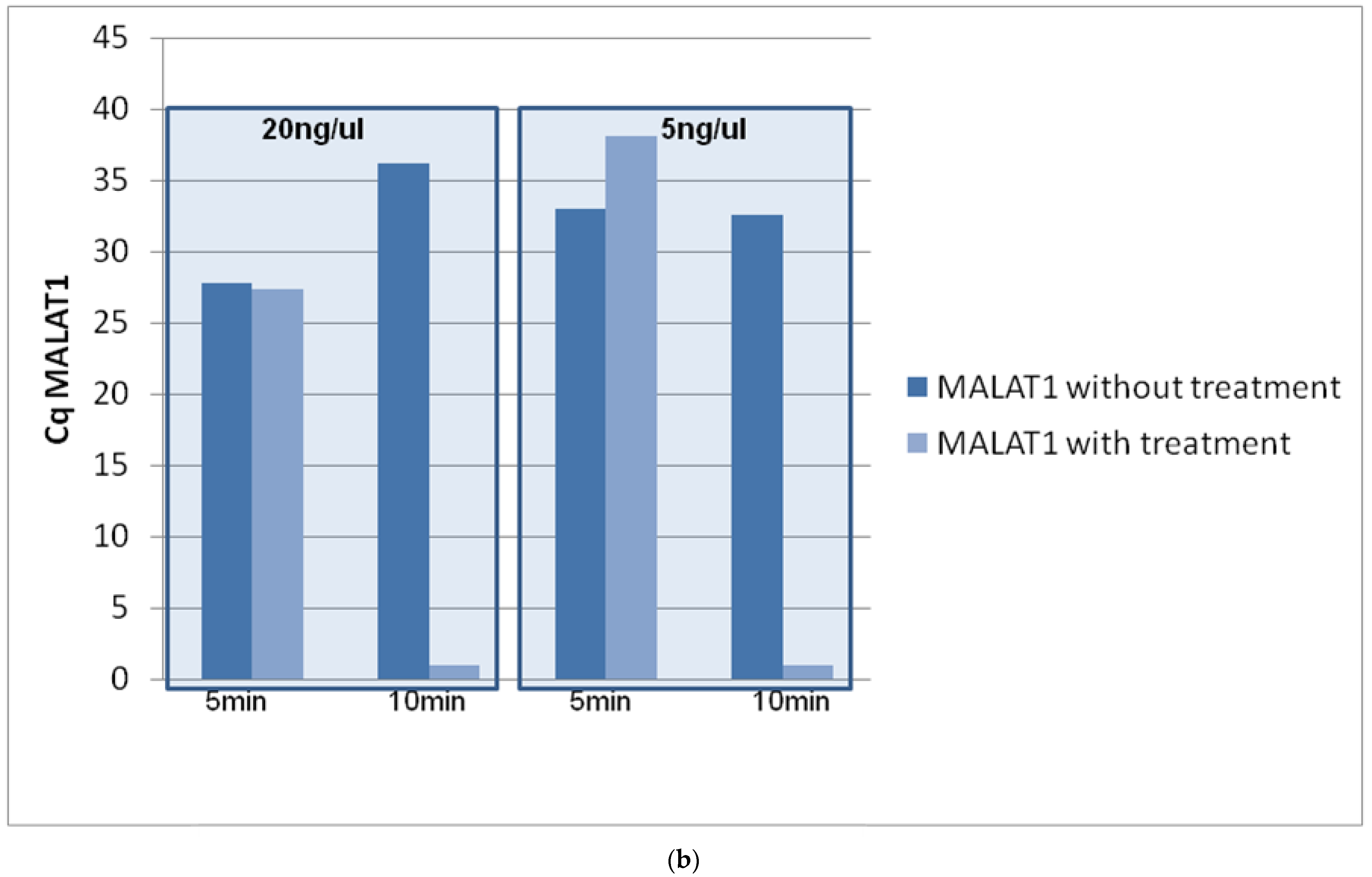
3.1. Optimization of DNase Treatment Conditions
3.1.1. Enzyme Incubation
3.1.2. Concentration of gDNA
3.1.3. Repeatability of the Procedure
3.2. False-Positive Results on MALAT1 Expression in Clinical Samples
3.2.1. NSCLC Primary Tissues
3.2.2. ccfRNA in Plasma
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [Green Version]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of longnoncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.C.; Awasthee, N.; Rai, V.; Chava, S.; Gunda, V.; Challagundla, K.B. Long non-coding RNAs and nuclear factor-κB crosstalk in cancer and other human diseases. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188316. [Google Scholar] [CrossRef] [PubMed]
- Bassett, A.R.; Akhtar, A.; Barlow, D.P.; Bird, A.P.; Brockdorff, N.; Duboule, D.; Ephrussi, A.; Ferguson-Smith, A.C.; Gingeras, T.R.; Haerty, W.; et al. Considerations when investigating lncRNA function in vivo. Elife 2014, 3, e03058. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, J.; Yin, Y.; Meng, Q.; Lyu, Y. The role of EMT-related lncRNA in the process of triple-negative breast cancer metastasis. Biosci. Rep. 2021, 41. [Google Scholar] [CrossRef]
- Acha-Sagredo, A.; Uko, B.; Pantazi, P.; Bediaga, N.G.; Moschandrea, C.; Rainbow, L.; Marcus, M.W.; Davies, M.P.A.; Field, J.K.; Liloglou, T. Long non-coding RNA dysregulation is a frequent event in non-small cell lung carcinoma pathogenesis. Br. J. Cancer 2020, 122, 1050–1058. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Yu, Y.; Huang, H.; Xu, A.; Xu, H.; Zhou, Y. Expression Level and Clinical Significance of NKILA in Human Cancers: A Systematic Review and Meta-Analysis. BioMed. Res. Int. 2020, 2020, 1–9. [Google Scholar] [CrossRef]
- Guo, L.; Tang, Y.; Wang, Y.; Xu, H. Prognostic Value of lncRNA NEAT1 as a New Biomarker in Digestive System Tumors: A Systematic Study and Meta-analysis. Expert Rev. Mol. Diagn. 2021, 21, 91–99. [Google Scholar] [CrossRef]
- Soghli, N.; Yousefi, T.; Abolghasemi, M.; Qujeq, D. NORAD, a critical long non-coding RNA in human cancers. Life Sci. 2021, 264, 118665. [Google Scholar] [CrossRef] [PubMed]
- Amirinejad, R.; Rezaei, M.; Shirvani-Farsani, Z. An update on long intergenic noncoding RNA p21: A regulatory molecule with various significant functions in cancer. Cell Biosci. 2020, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SRsplicing factor phosphorylation. Mol. Cell. 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Sun, H.; Wang, Y.; Yang, X.; Meng, Q.; Yang, H.; Zhu, H.; Tang, W.; Li, X.; Aschner, M.; et al. MALAT1 rs664589 Polymorphism Inhibits Binding to miR-194-5p, Contributing to Colorectal Cancer Risk, Growth, and Metastasis. Cancer Res. 2019, 79, 5432–5441. [Google Scholar] [CrossRef] [Green Version]
- Pei, C.; Gong, X.; Zhang, Y. LncRNA MALAT-1 promotes growth and metastasis of epithelial ovarian cancer via sponging microrna-22. Am. J. Transl. Res. 2020, 12, 6977–6987. [Google Scholar]
- Wang, K.; Zhao, Y.; Wang, Y.-M. LncRNA MALAT1 Promotes Survival of Epithelial Ovarian Cancer Cells by Downregulating miR-145-5p. Cancer Manag. Res. 2020, 12, 11359–11369. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, M.; Zhang, Z. Lncrna malat1 modulates oxaliplatin resistance of gastric cancer via sponging mir-22-3p. Onco Targets Ther. 2020, 13, 1343–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amodio, N.; Stamato, M.A.; Juli, G.; Morelli, E.; Fulciniti, M.; Manzoni, M.; Taiana, E.; Agnelli, L.; Cantafio, M.E.G.; Romeo, E.; et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia 2018, 32, 1948–1957. [Google Scholar] [CrossRef] [Green Version]
- Gong, N.; Teng, X.; Li, J.; Liang, X.-J. Antisense Oligonucleotide-Conjugated Nanostructure-Targeting lncRNA MALAT1 Inhibits Cancer Metastasis. ACS Appl. Mater. Interfaces 2018, 11, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Tiansheng, G.; Junming, H.; Xiaoyun, W.; Peixi, C.; Shaoshan, D.; Qianping, C. lncRNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 Promotes Proliferation and Invasion of Non-Small Cell Lung Cancer Cells via Down-Regulating miR-202 Expression. Cell J. 2019, 22, 375–385. [Google Scholar]
- Guo, F.; Yu, F.; Wang, J.; Li, Y.; Li, Y.; Li, Z.; Zhou, Q. Expression of MALAT1 in the peripheral whole blood of patients with lung cancer. Biomed. Rep. 2015, 3, 309–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Weng, Y.; He, F.; Liang, D.; Cai, L. LncRNA MALAT-1 competitively regulates miR-124 to promote EMT and development of non-small-cell lung cancer. Anti-Cancer Drugs 2018, 29, 628–636. [Google Scholar] [CrossRef]
- Zhang, R.; Xia, Y.; Wang, Z.; Zheng, J.; Chen, Y.; Li, X.; Wang, Y.; Ming, H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2017, 490, 406–414. [Google Scholar] [CrossRef]
- Lv, P.; Yang, S.; Liu, W.; Qin, H.; Tang, X.; Wu, F.; Liu, Z.; Gao, H.; Liu, X. Circulating plasma lncRNAs as novel markers of EGFR mutation status and monitors of epidermal growth factor receptor-tyrosine kinase inhibitor therapy. Thorac. Cancer 2020, 11, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhao, W.; Zhang, L.; Wang, L.; Wang, J.; Wan, Z.; Hong, Y.; Yu, L. MALAT1-miR-101-SOX9 feedback loop modulates the chemo-resistance of lung cancer cell to DDP via Wnt signaling pathway. Oncotarget 2017, 8, 94317–94329. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Su, Z.-Z.; Shen, Q.-M. Long non-coding RNA MALAT1 regulates proliferation, apoptosis, migration and invasion via miR-374b-5p/SRSF7 axis in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1853–1862. [Google Scholar]
- Liu, J.; Tian, W.; Zhang, W.; Jia, Y.; Yang, X.; Wang, Y.; Zhang, J. MicroRNA-142-3p/MALAT1 inhibits lung cancer progression through repressing β-catenin expression. Biomed. Pharmacother. 2019, 114, 108847. [Google Scholar] [CrossRef]
- Ren, S.; Wang, F.; Shen, J.; Sun, Y.; Xu, W.; Lu, J.; Wei, M.; Xu, C.; Wu, C.; Zhang, Z.; et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur. J. Cancer 2013, 49, 2949–2959. [Google Scholar] [CrossRef]
- Hao, T.; Wang, Z.; Yang, J.; Zhang, Y.; Shang, Y.; Sun, J. MALAT1 knockdown inhibits prostate cancer progression by regulating miR-140/BIRC6 axis. Biomed. Pharmacother. 2020, 123, 109666. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Piao, H.-L.; Kim, B.-J.; Yao, F.; Han, Z.; Wang, Y.; Xiao, Z.; Siverly, A.N.; Lawhon, S.E.; Ton, B.N.; et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 2018, 50, 1705–1715. [Google Scholar] [CrossRef]
- Stone, J.K.; Kim, J.-H.; Vukadin, L.; Richard, A.; Giannini, H.K.; Lim, S.-T.S.; Tan, M.; Ahn, E.-Y.E. Hypoxia induces cancer cell-specific chromatin interactions and increases MALAT1 expression in breast cancer cells. J. Biol. Chem. 2019, 294, 11213–11224. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Zhang, Y.; Fu, Y.; Gong, H.; Guo, J.; Wu, K.; Jia, Q.; Ding, X. Long non-coding RNA MALAT1 regulates BLCAP mRNA expression through binding to miR-339-5p and promotes poor prognosis in breast cancer. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [Green Version]
- Zhu, K.; Ren, Q.; Zhao, Y. lncRNA MALAT1 overexpression promotes proliferation, migration and invasion of gastric cancer by activating the PI3K/AKT pathway. Oncol. Lett. 2019, 17, 5335–5342. [Google Scholar] [CrossRef] [Green Version]
- Shao, G.; Zhao, Z.; Zhao, W.; Hu, G.; Zhang, L.; Li, W.; Xing, C.; Zhang, X. Long non-coding RNA MALAT1 activates autophagy and promotes cell proliferation by downregulating microRNA-204 expression in gastric cancer. Oncol. Lett. 2019, 19, 805–812. [Google Scholar] [CrossRef]
- Yiren, H.; Yingcong, Y.; Sunwu, Y.; Keqin, L.; XiaoChun, T.; Senrui, C.; Ende, C.; Xizhou, L.; Yanfan, C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol. Cancer 2017, 16, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zygalaki, E.; Tsaroucha, E.G.; Kaklamanis, L.; Lianidou, E.S. Quantitative Real-Time Reverse Transcription–PCR Study of the Expression of Vascular Endothelial Growth Factor (VEGF) Splice Variants and VEGF Receptors (VEGFR-1 and VEGFR-2) in Non–Small Cell Lung Cancer. Clin. Chem. 2007, 53, 1433–1439. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zavridou, M.; Mastoraki, S.; Strati, A.; Tzanikou, E.; Chimonidou, M.; Lianidou, E. Evaluation of Preanalytical Conditions and Implementation of Quality Control Steps for Reliable Gene Expression and DNA Methylation Analyses in Liquid Biopsies. Clin. Chem. 2018, 64, 1522–1533. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-Delta Delta C(T) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Nandwani, A.; Rathore, S.; Datta, M. LncRNAs in cancer: Regulatory and therapeutic implications. Cancer Lett. 2021, 501, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Taniue, K.; Akimitsu, N. The Functions and Unique Features of LncRNAs in Cancer Development and Tumorigenesis. Int. J. Mol. Sci. 2021, 22, 632. [Google Scholar] [CrossRef] [PubMed]
- Laurell, H.; Iacovoni, J.S.; Abot, A.; Svec, D.; Maoret, J.J.; Arnal, J.F.; Kubista, M. Correction of RT-qPCR data for genomic DNA-derived signals with ValidPrime. Nucleic Acids Res. 2012, 40, e51. [Google Scholar] [CrossRef]
- Wang, M.; Chen, X.; Yu, F.; Ding, H.; Zhang, Y.; Wang, K. Extrachromosomal Circular DNAs: Origin, formation and emerging function in Cancer. Int. J. Biol. Sci. 2021, 17, 1010–1025. [Google Scholar] [CrossRef]
- Swellam, M.; EL Magdoub, H.M.; Shawki, M.A.; Adel, M.; Hefny, M.M.; El-Shazly, S.S. Clinical impact of LncRNA XIST and LncRNA NEAT1 for diagnosis of high-risk group breast cancer patients. Curr. Probl. Cancer 2021, 2021, 100709. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Xiang, Y.; Guo, X.; Zhang, Y.; Li, C.; Xie, W.; Wu, N.; Wu, L.; Cai, T.; Ma, X.; et al. Circulating Long Noncoding RNAs Act as Diagnostic Biomarkers in Non-Small Cell Lung Cancer. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Han, X.; Zheng, Y.; Sui, J.; Bi, W. Long non-coding RNA NKILA serves as a biomarker in the early diagnosis and prognosis of patients with colorectal cancer. Oncol. Lett. 2019, 18, 2109–2117. [Google Scholar] [CrossRef] [Green Version]
- Gutschner, T.; Hämmerle, M.; Diederichs, S. MALAT1—A paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013, 91, 791–801. [Google Scholar] [CrossRef]
- Goyal, B.; Yadav, S.R.M.; Awasthee, N.; Gupta, S.; Kunnumakkara, A.B.; Gupta, S.C. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim. Biophys. ActaBioenerg. 2021, 1875, 188502. [Google Scholar] [CrossRef]
- Huang, Z.; Qin, Q.; Xia, L.; Lian, B.; Tan, Q.; Yu, Y.; Mo, Q. Significance of Oncotype DX 21-Gene Test and Expression of Long Non-Coding RNA MALAT1 in Early and Estrogen Receptor-Positive Breast Cancer Patients. Cancer Manag. Res. 2021, 13, 587–593. [Google Scholar] [CrossRef]
- Liu, X.; Huang, G.; Zhang, J.; Zhang, L.; Liang, Z. Prognostic and clinicopathological significance of long noncoding RNA MALAT-1 expression in patients with non-small cell lung cancer: A meta-analysis. PLoS ONE 2020, 15, e0240321. [Google Scholar] [CrossRef]
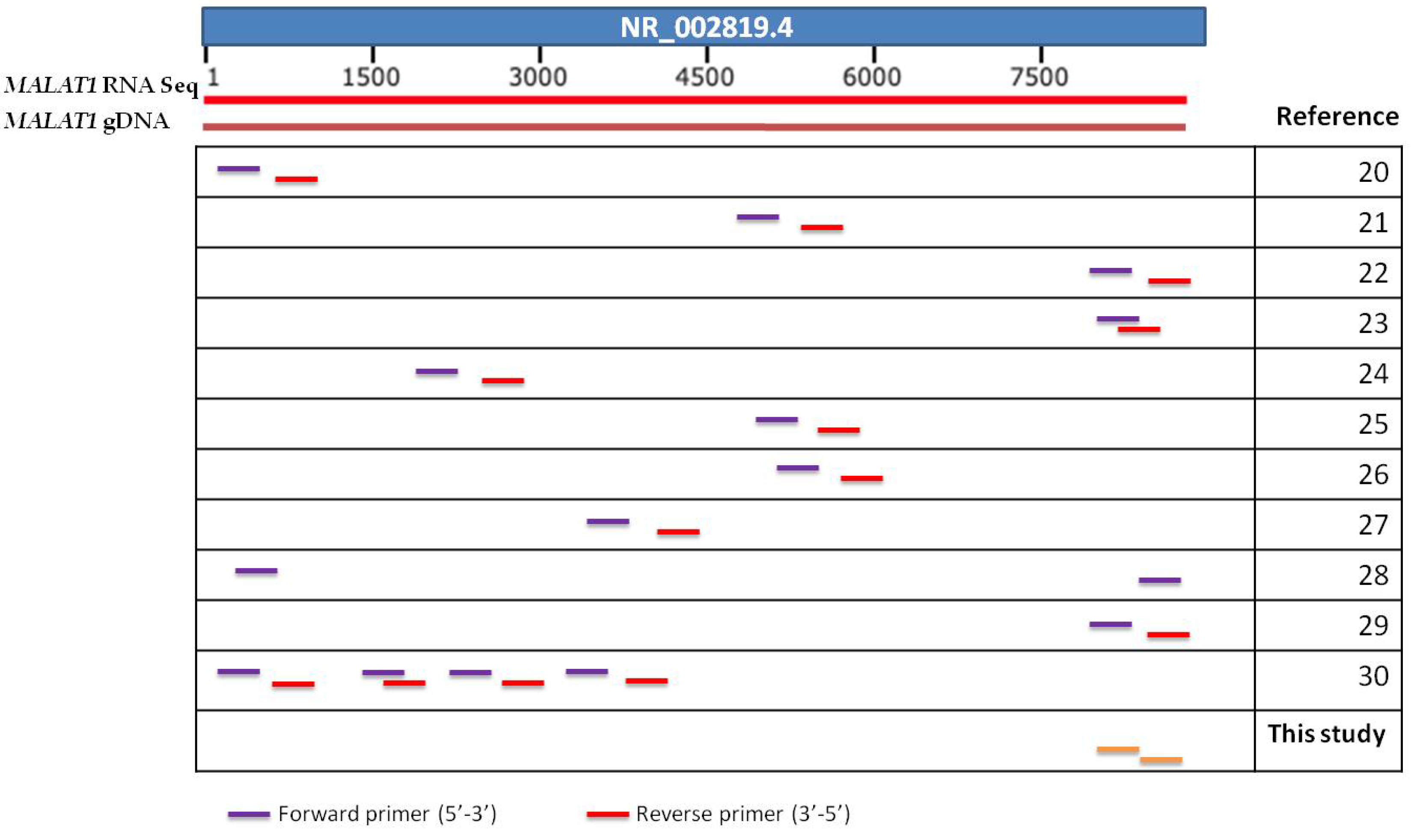
| Long Non-Coding RNA | Cancer Types | Ref. Sequence |
|---|---|---|
| MALAT1 | Lung cancer, Esophageal carcinoma, Acute myeloid leukemia, Ovarian cancer, thyroid, nerve—tibial, skin, uterus, prostate | NR_002819.4 |
| NEAT1 | Breast, lung, Prostate cancer, Head and neck squamous cell carcinoma, colon cancer, thyroid cancer | NR_028272.1 |
| DLEU | Acute myeloid leukemia, spleen, lung cancer, esophageal carcinoma, pancreatic, laryngeal, renal, cervical cancer | NR_002612.1 |
| ANRASSF1 | Breast, prostate, astric cancer | NR_109831.1 |
| NKILA | Pancreatic adenocarcinoma, prostate, breast cancer, uterine carcinosarcoma, lung cancer | NR_131157.1 |
| NORAD | Lymph node metastasis, pancreatic, bladder, gastric cancer; esophageal squamous cell carcinoma, epatocellularcarcinoma, colorectal cancer | NR_027451.1 |
| KCNQ1OT1 | Esophageal carcinoma, Acute myeloid leukemia, Ovarian, Stomach adenocarcinoma | NR_002728.3 |
| CCAT2 | Colon cancer, breast cancer, hepatocellular carcinoma | NR_109834.1 |
| LincRNA-p21 | Prostate, gastric, colorectal cancer, head and neck squamous cell carcinoma, lung cancer | CD515754.1 |
| Cancer Type | Sample Origin | Number of Samples | Reference Gene | DNase Treatment | Expression of MALAT-1 | Significance | Reference |
|---|---|---|---|---|---|---|---|
| NSCLC | Tissues (tumor and adjacent) Cell Lines | 40 | GAPDH | Yes | Over-expressed | Therapeutic target | [20] |
| Plasma Tissues | 105 65 | GAPDH | No | Under-expressed | Diagnostic | [21] | |
| Tissues (tumor and adjacent) | 86 | GAPDH | No | Over-expressed | Tumor progression and development | [22] | |
| Serum (exosomes) | 77 | GAPDH | No | Over-expressed | Diagnostic, prognostic, therapeutic target | [23] | |
| Plasma | 142 | 18S rRNA | No | Over-expressed | Diagnosis of EGFR-mutant patients | [24] | |
| Cell Lines Tissues (tumor and adjacent) | 42 | RNU6B | No | Over-expressed | Therapeutic target | [25] | |
| Cell Lines Tissues (tumor and adjacent) | 30 | GAPDH or U6 | No | Over-expressed | Diagnostic | [26] | |
| Cell Lines Tissues (tumor and adjacent) | 36 | GAPDH | No | Over-expressed | Therapeutic target | [27] | |
| Prostate cancer | Plasma Tissues (tumor and adjacent) | 169 14 | β-actin | Yes | Over-expressed | Diagnostic | [28] |
| Cell Lines Tissues (tumor and adjacent) Mice | 52 | β-actin | No | Over-expressed | Therapeutic target | [29] | |
| Breast cancer | Cell Lines Mice | - | GAPDH | Yes | Under-expressed | Prognostic, Therapeutic target | [30] |
| Cell Lines Mice | - | YWHAZ | No | Under-expressed | Therapeutic target | [31] | |
| Cell Lines Clinical samples | - | GAPDH | No | Over-expressed | Prognostic | [32] | |
| Gastric cancer | Tissues Plasma | 64 | β-actin | No | Over-expressed | Prognostic, diagnostic | [33] |
| Cell Lines Tissues (tumor and adjacent) | 57 | GAPDH | No | Over-expressed | Therapeutic target | [34] | |
| Cell Lines Tissues Mice | 153 | GAPDH | No | Over-expressed | Therapeutic | [35] |
| Plasma | |||
| After DNAse treatment | Before DNAse treatment | ||
| Overexpression | Underexpression | ||
| Overexpression | 0 | 0 | |
| Underexpression | 3 (False positive) | 12 | |
| Paired t-test: 0.082 | |||
| False positives: 3/15 (20%) | |||
| Primary Tissues | |||
| After DNAse treatment | Before DNAse treatment | ||
| Overexpression | Underexpression | ||
| Overexpression | 3 | 2 | |
| Underexpression | 3 (False positive) | 1 | |
| Paired t-test: 0.681 | |||
| False positives: 3/9 (30%) | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Markou, A.N.; Smilkou, S.; Tsaroucha, E.; Lianidou, E. The Effect of Genomic DNA Contamination on the Detection of Circulating Long Non-Coding RNAs: The Paradigm of MALAT1. Diagnostics 2021, 11, 1160. https://doi.org/10.3390/diagnostics11071160
Markou AN, Smilkou S, Tsaroucha E, Lianidou E. The Effect of Genomic DNA Contamination on the Detection of Circulating Long Non-Coding RNAs: The Paradigm of MALAT1. Diagnostics. 2021; 11(7):1160. https://doi.org/10.3390/diagnostics11071160
Chicago/Turabian StyleMarkou, Athina N., Stavroula Smilkou, Emilia Tsaroucha, and Evi Lianidou. 2021. "The Effect of Genomic DNA Contamination on the Detection of Circulating Long Non-Coding RNAs: The Paradigm of MALAT1" Diagnostics 11, no. 7: 1160. https://doi.org/10.3390/diagnostics11071160
APA StyleMarkou, A. N., Smilkou, S., Tsaroucha, E., & Lianidou, E. (2021). The Effect of Genomic DNA Contamination on the Detection of Circulating Long Non-Coding RNAs: The Paradigm of MALAT1. Diagnostics, 11(7), 1160. https://doi.org/10.3390/diagnostics11071160








