Usefulness of BATF3 Immunohistochemistry in Diagnosing Classical Hodgkin Lymphoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue and Cell Lines
2.2. Immunohistochemistry
2.3. Western Blotting
2.4. Statistical Analysis
3. Results
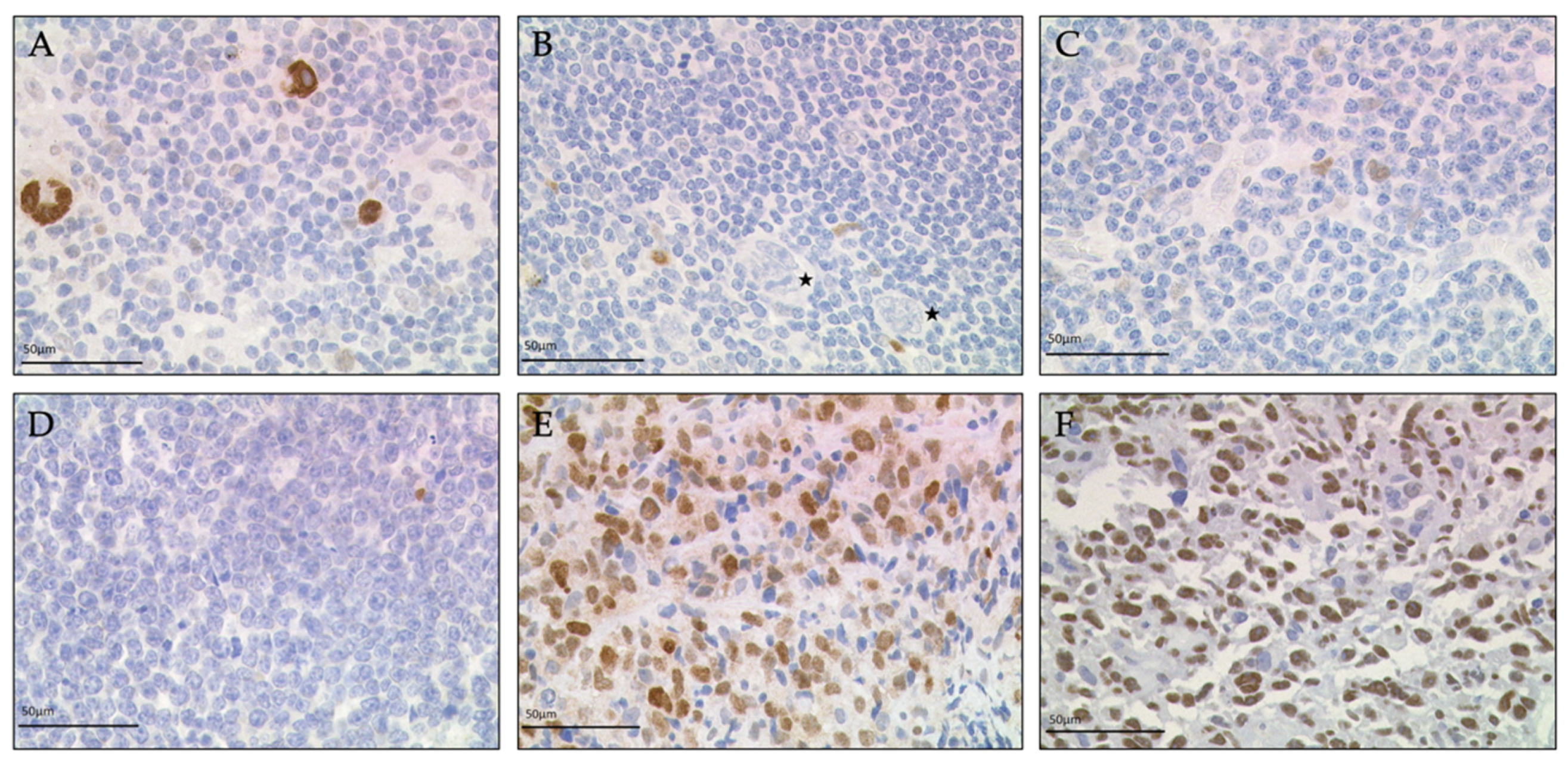
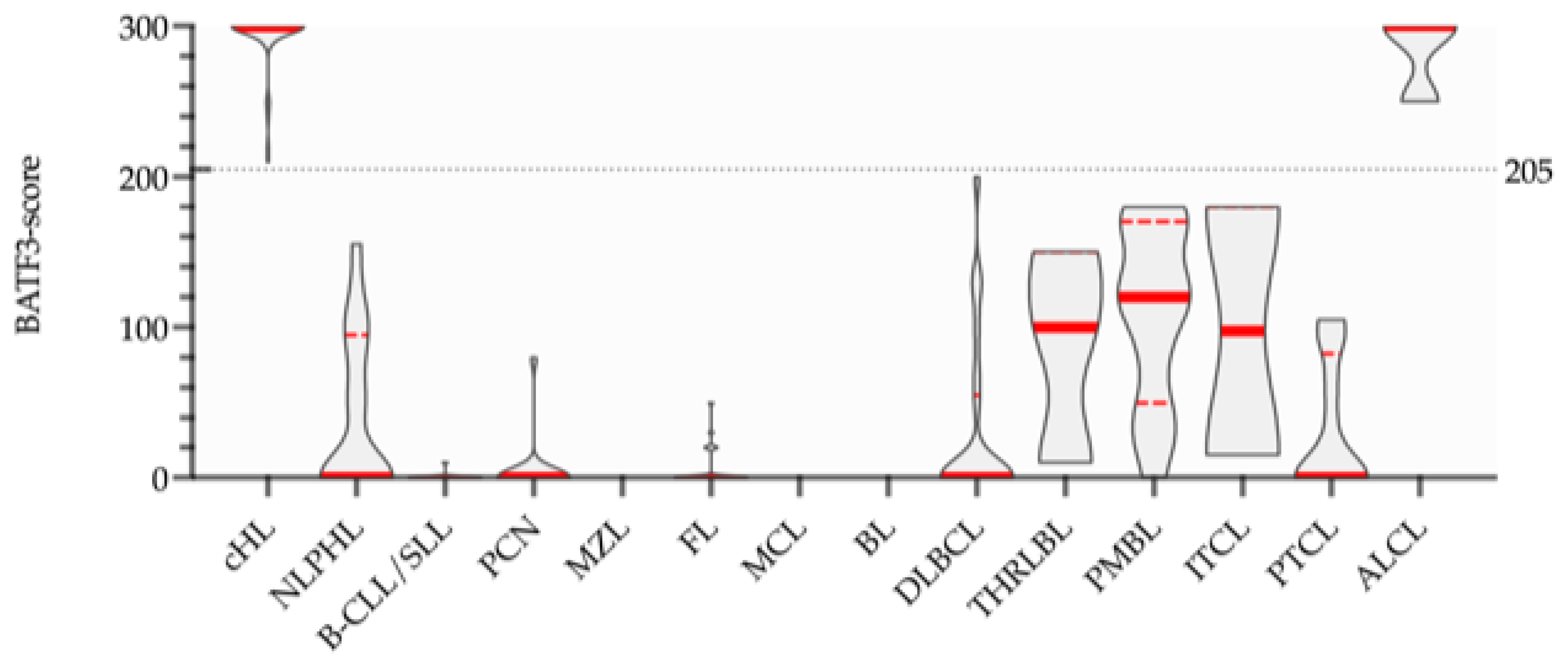
3.1. Immunohistochemistry
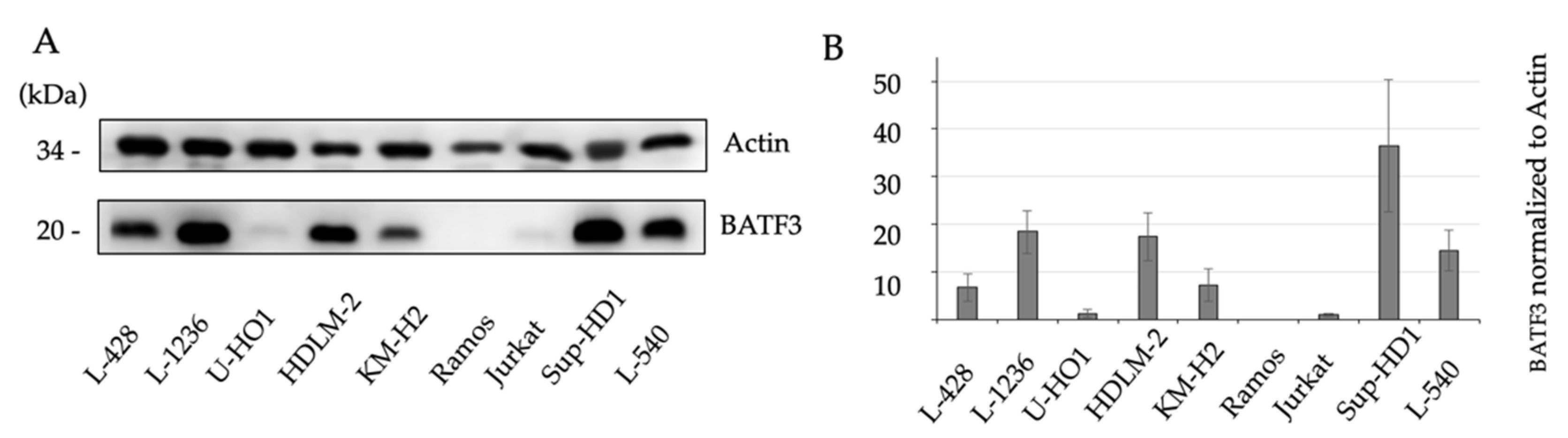
3.2. Western Blotting
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Kanzler, H.; Küppers, R.; Hansmann, M.L.; Rajewsky, K. Hodgkin and Reed-Sternberg cells in Hodgkin’s disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J. Exp. Med. 1996, 184, 1495–1505. [Google Scholar] [CrossRef]
- Küppers, R.; Rajewsky, K.; Zhao, M.; Simons, G.; Laumann, R.; Fischer, R.; Hansmann, M.L. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc. Natl. Acad. Sci. USA 1994, 91, 10962–10966. [Google Scholar] [CrossRef]
- Schwering, I.; Bräuninger, A.; Klein, U.; Jungnickel, B.; Tinguely, M.; Diehl, V.; Hansmann, M.L.; Dalla-Favera, R.; Rajewsky, K.; Küppers, R. Loss of the B-lineage-specific gene expression program in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 2003, 101, 1505–1512. [Google Scholar] [CrossRef]
- Schmid, C.; Pan, L.; Diss, T.; Isaacson, P.G. Expression of B-cell antigens by Hodgkin’s and Reed-Sternberg cells. Am. J. Pathol. 1991, 139, 701–707. [Google Scholar]
- Torlakovic, E.; Tierens, A.; Dang, H.D.; Delabie, J. The transcription factor PU.1, necessary for B-cell development is expressed in lymphocyte predominance, but not classical Hodgkin’s disease. Am. J. Pathol. 2001, 159, 1807–1814. [Google Scholar] [CrossRef]
- Re, D.; Müschen, M.; Ahmadi, T.; Wickenhauser, C.; Staratschek-Jox, A.; Holtick, U.; Diehl, V.; Wolf, J. Oct-2 and Bob-1 deficiency in Hodgkin and Reed Sternberg cells. Cancer Res. 2001, 61, 2080–2084. [Google Scholar]
- Steimle-Grauer, S.A.; Tinguely, M.; Seada, L.; Fellbaum, C.; Hansmann, M.L. Expression patterns of transcription factors in progressively transformed germinal centers and Hodgkin lymphoma. Virchows Arch. 2003, 442, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, G.; Song, J.Y.; Tzankov, A.; Dirnhofer, S.; Heinze, G.; Kohl, M.; Traverse-Glehen, A.; Eberle, F.C.; Hanson, J.C.; Raffeld, M.A.; et al. Aberrant T-cell antigen expression in classical Hodgkin lymphoma is associated with decreased event-free survival and overall survival. Blood 2013, 121, 1795–1804. [Google Scholar] [CrossRef] [PubMed]
- Sorg, U.R.; Morse, T.M.; Patton, W.N.; Hock, B.D.; Angus, H.B.; Robinson, B.A.; Colls, B.M.; Hart, D.N. Hodgkin’s cells express CD83, a dendritic cell lineage associated antigen. Pathology 1997, 29, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Fromm, J.R. Flow cytometric analysis of CD123 is useful for immunophenotyping classical Hodgkin lymphoma. Cytom. B Clin. Cytom. 2011, 80, 91–99. [Google Scholar] [CrossRef]
- Schwab, U.; Stein, H.; Gerdes, J.; Lemke, H.; Kirchner, H.; Schaadt, M.; Diehl, V. Production of a monoclonal antibody specific for Hodgkin and Sternberg-Reed cells of Hodgkin's disease and a subset of normal lymphoid cells. Nature 1982, 299, 65–67. [Google Scholar] [CrossRef]
- Stein, H.; Uchánska-Ziegler, B.; Gerdes, J.; Ziegler, A.; Wernet, P. Hodgkin and Sternberg-Reed cells contain antigens specific to late cells of granulopoiesis. Int. J. Cancer 1982, 29, 283–290. [Google Scholar] [CrossRef]
- Carbone, A.; Gloghini, A.; Aldinucci, D.; Gattei, V.; Dalla-Favera, R.; Gaidano, G. Expression pattern of MUM1/IRF4 in the spectrum of pathology of Hodgkin's disease. Br. J. Haematol. 2002, 117, 366–372. [Google Scholar] [CrossRef]
- Krenacs, L.; Himmelmann, A.W.; Quintanilla-Martinez, L.; Fest, T.; Riva, A.; Wellmann, A.; Bagdi, E.; Kehrl, J.H.; Jaffe, E.S.; Raffeld, M. Transcription factor B-cell-specific activator protein (BSAP) is differentially expressed in B cells and in subsets of B-cell lymphomas. Blood 1998, 92, 1308–1316. [Google Scholar] [CrossRef]
- Weiss, L.M. Atypical Phenotypes in Classical Hodgkin Lymphoma. Surg. Pathol. Clin. 2013, 6, 729–742. [Google Scholar] [CrossRef] [PubMed]
- Weiser, C.; Petkova, M.V.; Rengstl, B.; Döring, C.; von Laer, D.; Hartmann, S.; Küppers, R.; Hansmann, M.L.; Newrzela, S. Ectopic expression of transcription factor BATF3 induces B-cell lymphomas in a murine B-cell transplantation model. Oncotarget 2018, 9, 15942–15951. [Google Scholar] [CrossRef] [PubMed]
- Schwering, I.; Bräuninger, A.; Distler, V.; Jesdinsky, J.; Diehl, V.; Hansmann, M.L.; Rajewsky, K.; Küppers, R. Profiling of Hodgkin's lymphoma cell line L1236 and germinal center B cells: Identification of Hodgkin's lymphoma-specific genes. Mol. Med. 2003, 9, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Brune, V.; Tiacci, E.; Pfeil, I.; Doring, C.; Eckerle, S.; van Noesel, C.J.; Klapper, W.; Falini, B.; von Heydebreck, A.; Metzler, D.; et al. Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J. Exp. Med. 2008, 205, 2251–2268. [Google Scholar] [CrossRef]
- Lollies, A.; Hartmann, S.; Schneider, M.; Bracht, T.; Weiss, A.L.; Arnolds, J.; Klein-Hitpass, L.; Sitek, B.; Hansmann, M.L.; Kuppers, R.; et al. An oncogenic axis of STAT-mediated BATF3 upregulation causing MYC activity in classical Hodgkin lymphoma and anaplastic large cell lymphoma. Leukemia 2018, 32, 92–101. [Google Scholar] [CrossRef]
- Schleussner, N.; Merkel, O.; Costanza, M.; Liang, H.C.; Hummel, F.; Romagnani, C.; Durek, P.; Anagnostopoulos, I.; Hummel, M.; Johrens, K.; et al. The AP-1-BATF and -BATF3 module is essential for growth, survival and TH17/ILC3 skewing of anaplastic large cell lymphoma. Leukemia 2018, 32, 1994–2007. [Google Scholar] [CrossRef]
- Vrzalikova, K.; Ibrahim, M.; Vockerodt, M.; Perry, T.; Margielewska, S.; Lupino, L.; Nagy, E.; Soilleux, E.; Liebelt, D.; Hollows, R.; et al. S1PR1 drives a feedforward signalling loop to regulate BATF3 and the transcriptional programme of Hodgkin lymphoma cells. Leukemia 2018, 32, 214–223. [Google Scholar] [CrossRef]
- Tiacci, E.; Doring, C.; Brune, V.; van Noesel, C.J.; Klapper, W.; Mechtersheimer, G.; Falini, B.; Kuppers, R.; Hansmann, M.L. Analyzing primary Hodgkin and Reed-Sternberg cells to capture the molecular and cellular pathogenesis of classical Hodgkin lymphoma. Blood 2012, 120, 4609–4620. [Google Scholar] [CrossRef]
- Eckerle, S.; Brune, V.; Döring, C.; Tiacci, E.; Bohle, V.; Sundström, C.; Kodet, R.; Paulli, M.; Falini, B.; Klapper, W.; et al. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: Insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia 2009, 23, 2129–2138. [Google Scholar] [CrossRef]
- Murphy, T.L.; Tussiwand, R.; Murphy, K.M. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 2013, 13, 499–509. [Google Scholar] [CrossRef]
- Hildner, K.; Edelson, B.T.; Purtha, W.E.; Diamond, M.; Matsushita, H.; Kohyama, M.; Calderon, B.; Schraml, B.U.; Unanue, E.R.; Diamond, M.S.; et al. Batf3 deficiency reveals a critical role for CD8 alpha + dendritic cells in cytotoxic T cell immunity. Science 2008, 322, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Bundesärztekammer ZEKO. Die (Weiter-)Verwendung von Menschlichen Körpermaterialien von Verstorbenen für Zwecke Medizinischer Forschung; Bundesärztekammer: Berlin, Germany, 2003; p. 2251. [Google Scholar]
- Mader, A.; Bruderlein, S.; Wegener, S.; Melzner, I.; Popov, S.; Muller-Hermelink, H.K.; Barth, T.F.; Viardot, A.; Moller, P. U-HO1, a new cell line derived from a primary refractory classical Hodgkin lymphoma. Cytogenet. Genome Res. 2007, 119, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Mazières, J.; Brugger, W.; Cappuzzo, F.; Middel, P.; Frosch, A.; Bara, I.; Klingelschmitt, G.; Klughammer, B. Evaluation of EGFR protein expression by immunohistochemistry using H-score and the magnification rule: Re-analysis of the SATURN study. Lung Cancer 2013, 82, 231–237. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benckendorff, J.; Kuchar, J.; Leithäuser, F.; Zahn, M.; Möller, P. Usefulness of BATF3 Immunohistochemistry in Diagnosing Classical Hodgkin Lymphoma. Diagnostics 2021, 11, 1123. https://doi.org/10.3390/diagnostics11061123
Benckendorff J, Kuchar J, Leithäuser F, Zahn M, Möller P. Usefulness of BATF3 Immunohistochemistry in Diagnosing Classical Hodgkin Lymphoma. Diagnostics. 2021; 11(6):1123. https://doi.org/10.3390/diagnostics11061123
Chicago/Turabian StyleBenckendorff, Julian, Johanna Kuchar, Frank Leithäuser, Malena Zahn, and Peter Möller. 2021. "Usefulness of BATF3 Immunohistochemistry in Diagnosing Classical Hodgkin Lymphoma" Diagnostics 11, no. 6: 1123. https://doi.org/10.3390/diagnostics11061123
APA StyleBenckendorff, J., Kuchar, J., Leithäuser, F., Zahn, M., & Möller, P. (2021). Usefulness of BATF3 Immunohistochemistry in Diagnosing Classical Hodgkin Lymphoma. Diagnostics, 11(6), 1123. https://doi.org/10.3390/diagnostics11061123





