2.1. Medical Data and Preprocessing
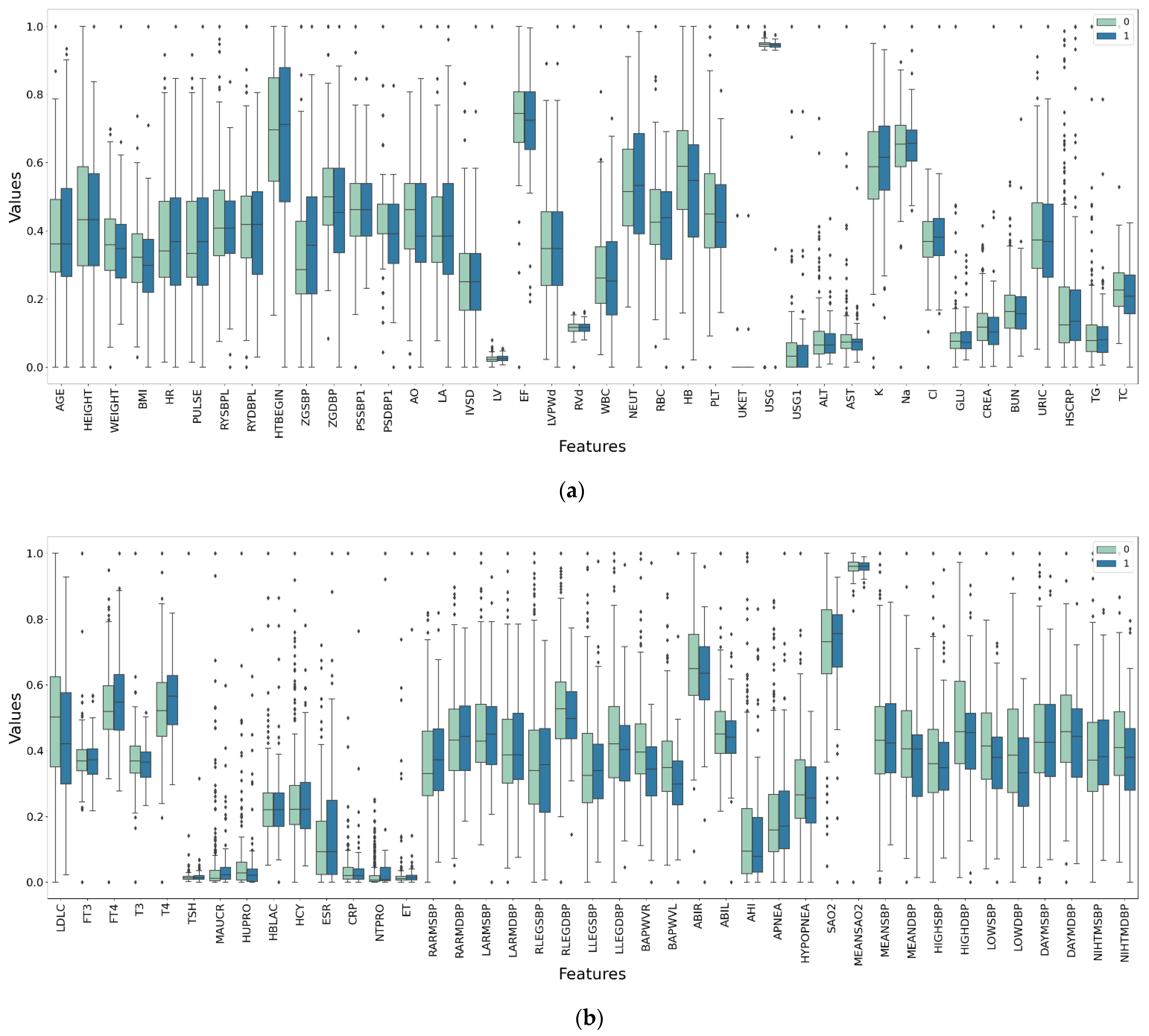
The medical data used in this study were obtained from a hypertension center of a tertiary-grade A class hospital in Beijing. The hospital collected data from 1357 patients with hypertension from September 2012 to December 2016. The patients come from various regions in China. The data set were divided into two parts. One part was the medical examination data and related survey data (i.e., characteristic data) during the patient admission. The other part involved the data on whether the outcomes occurred in the patients (i.e., the labeled data: yes/no or 1/0) marked by the hospital staff during the follow-up period after the patient was discharged. Characteristic data included baseline data, limb blood pressure, ambulatory blood pressure, echocardiography, heart failure, and other categories—a total of 132 examination indicators. The outcomes involved complications of the four target organs: heart, brain, kidney, and fundus.
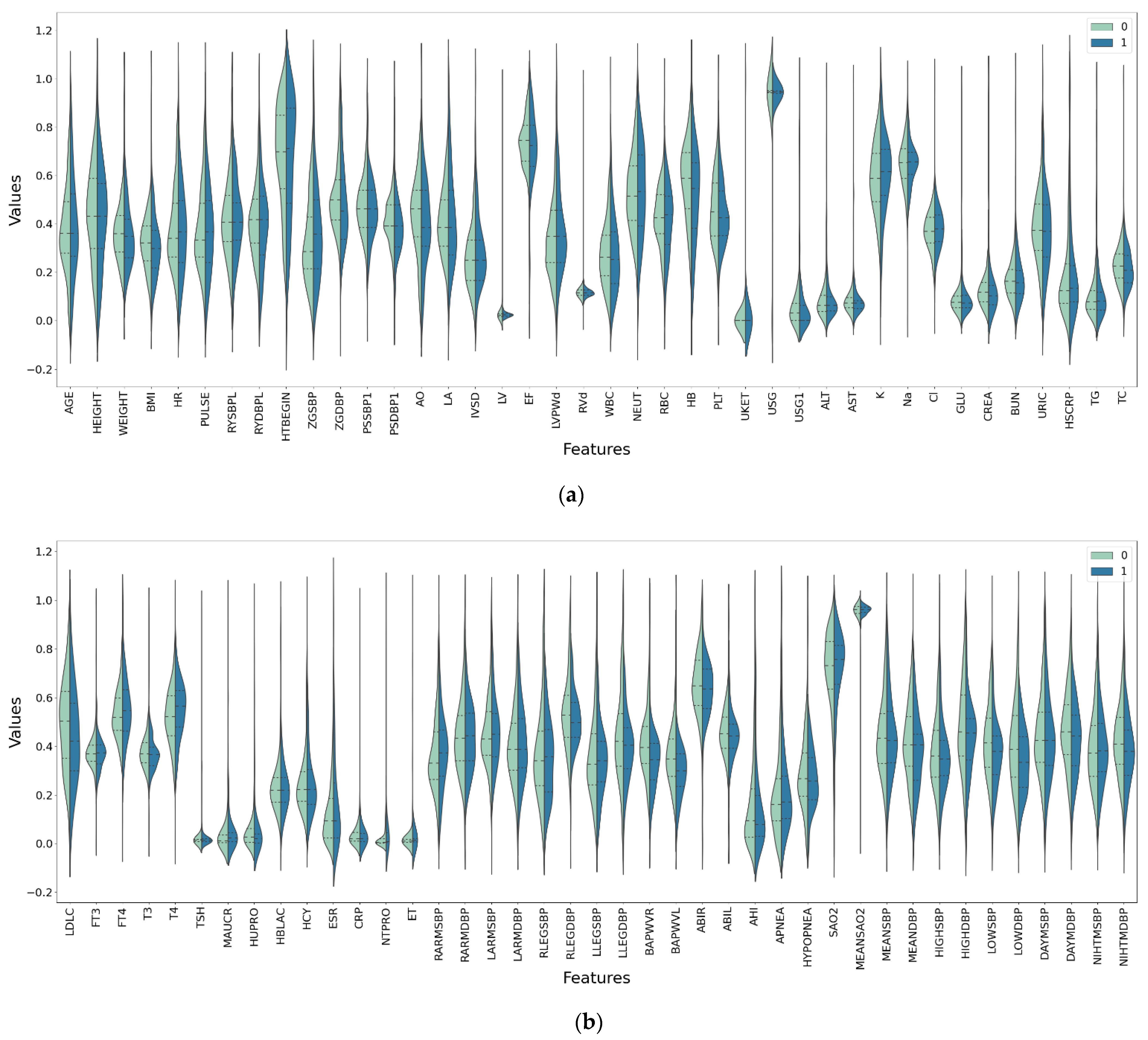
Table 1 shows the name, medical description, data type, mean value, standard deviation, and data distribution range of some medical indicators of the data set.
Table A1 in
Appendix A is a list of all the medical features that are considered in this study.
There are impurities in the original data. (1) There are a large number of missing values in the original data set, and some attributes have missing values of more than 90%. (2) There are some abnormal values, which exceed the regular distribution interval of the attribute. (3) For different physical examination indicators, the attribute dimensional units are different.
The deletion and mean interpolation are used to deal with missing values. Features and samples with missing values exceeding 50% are directly deleted; for deleted data sets, missing values are interpolated according to the mean value of attributes. Outliers are directly deleted. In this study, the maximum and minimum standardization methods are used to unify the dimensions.
2.2. Gain Sequence Forward Tabu Search Feature Selection (GSFTS-FS)
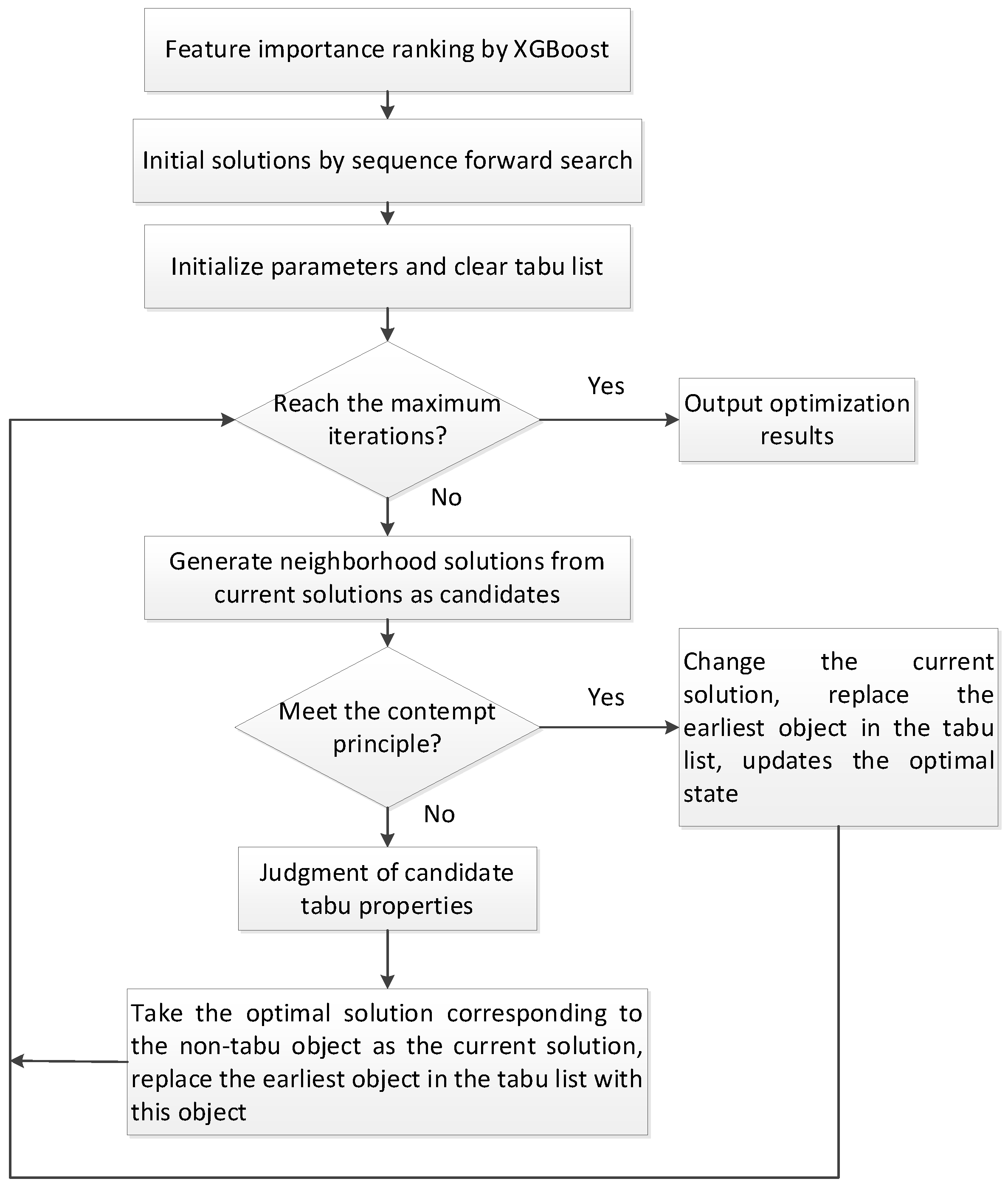
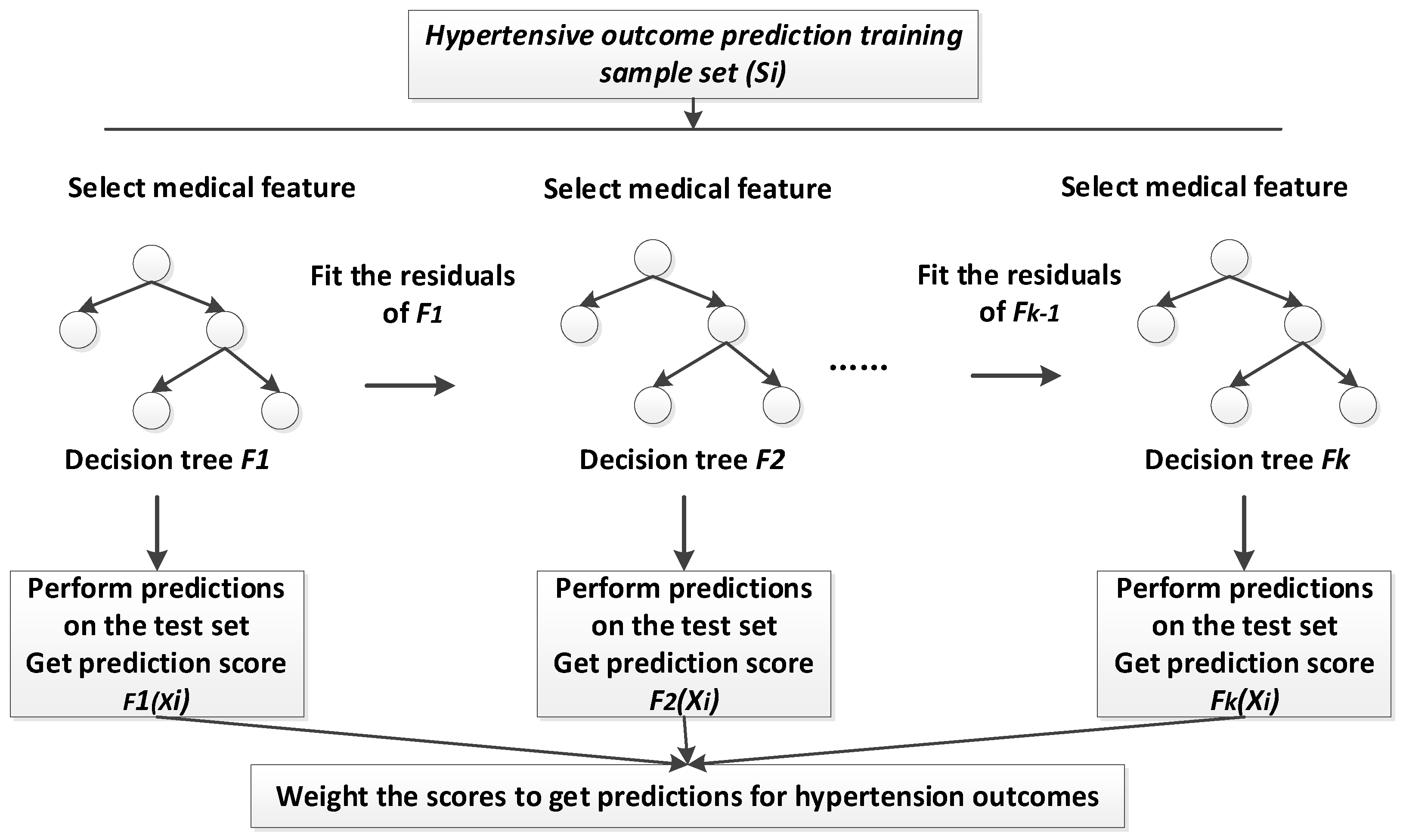
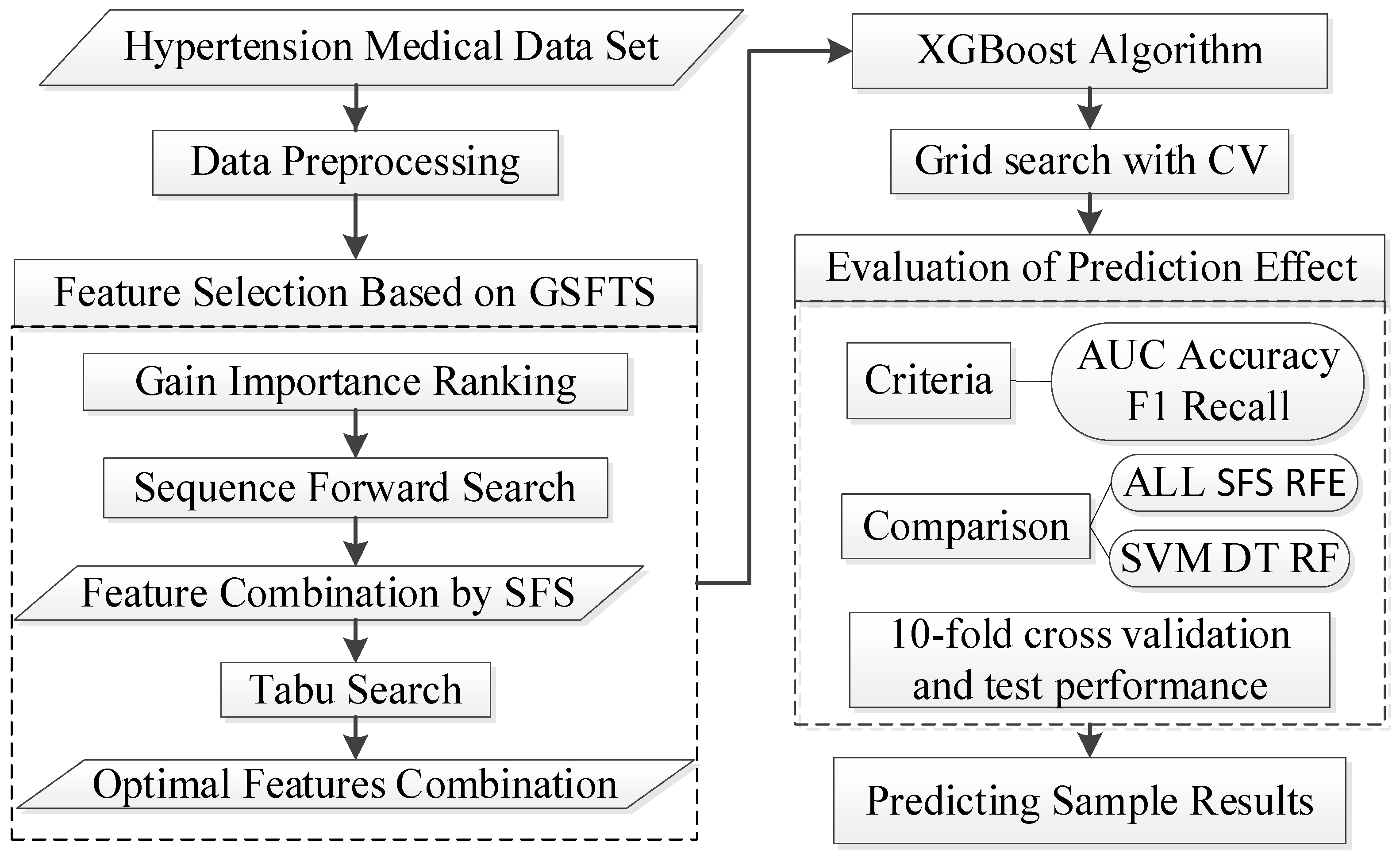
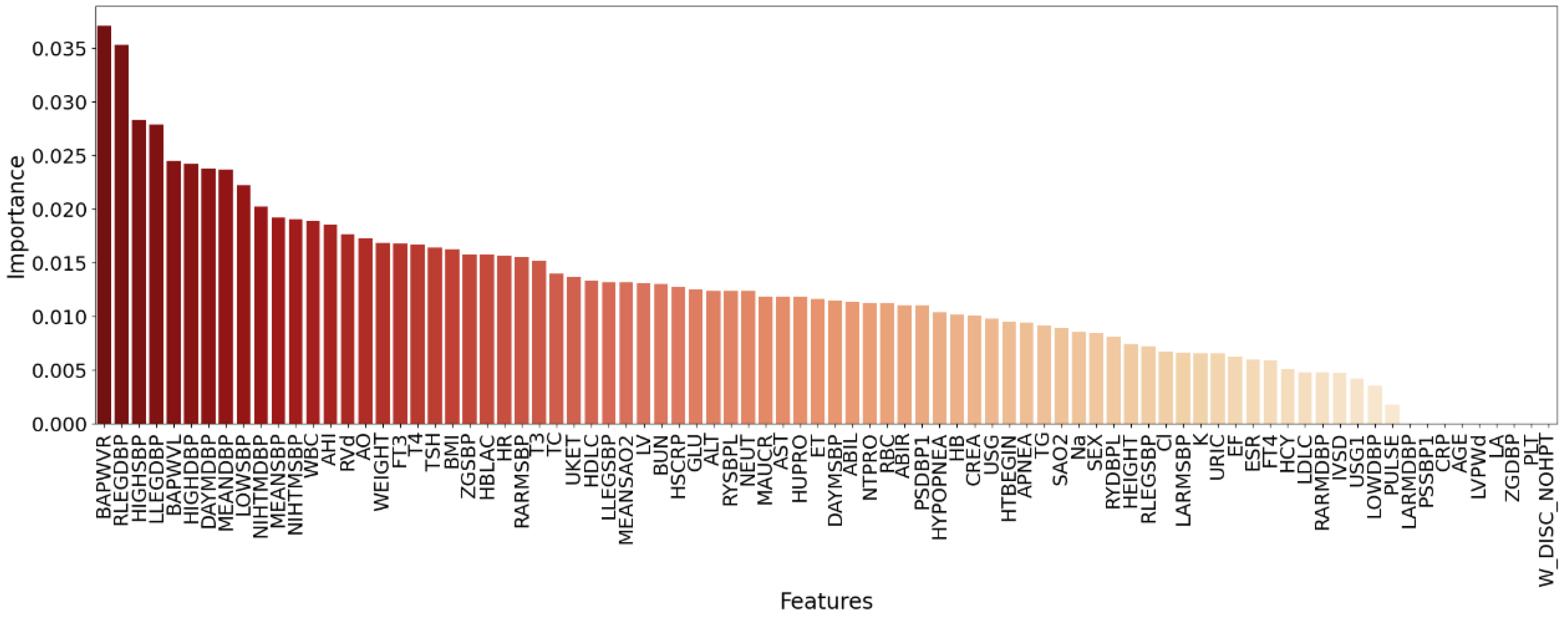
In this study, we proposed a new medical feature selection (FS) strategy called gain sequence forward tabu search (GSFTS). GSFTS-FS is a wrapper feature selection method. It takes the performance of the prediction model as a criterion and objective function to evaluate the quality of the selected feature subset. It is mainly divided into three steps. First, XGBoost rank and score feature importance based on the average gain. Second, sequence forward search based on the ranking is performed to obtain initial feature combinations. Finally, the selected feature combination is further optimized by tabu search algorithm. The basic steps of the GSFTS-FS algorithm are shown in
Figure 1.
Based on the concept of the GSFTS algorithm, the specific process is as follows:
1. Feature importance ranking.
We first build an initial classifier (XGBoost) and fit the data. We calculate the average information gain across all split points in XGBoost of each feature to rank all feature importance. The higher the gain, the greater the feature contribution and the higher the importance.
2. Initial Solution by gain-based sequence forward search.
The traditional tabu search algorithm has two shortcomings: (1) Strong dependence on the initial solution. A good initial solution helps the search to reach the optimal solution quickly, while a bad initial solution often makes the search difficult or impossible to reach the optimal solution. (2) The running time of the algorithm is greatly affected by the initial solution. A better initial solution can push the search move closer to the optimal solution with fewer iterations, thereby reducing the search time. The search with poor initial solution needs many iterations to get close to the optimal solution, which prolongs the search time.
Aiming to provide a better initial optimal solution for tabu search, we proposed a new Sequence Forward Search based on feature importance ranking by information Gain (gain-based SFS). Suppose the feature importance is ranked as (Fa, Fb, Fc…), specific steps are as follows:
(1) Add the feature Fa, which ranks first in importance, to the feature subset S. The current subset is S′ = {Fa}, the dimension of the subset is i = 1, and the classification accuracy on the training set is selected as the evaluation function f.
(2) Calculate the evaluation function score under the current feature subset f(S′).
(3) According to the order of feature importance, the feature with ranking i + 1 is added to feature subset S′.
(4) Calculate the evaluation function score f(S′) under the current feature subset, if the score drops, stop searching; if the score rises, repeat step (3).
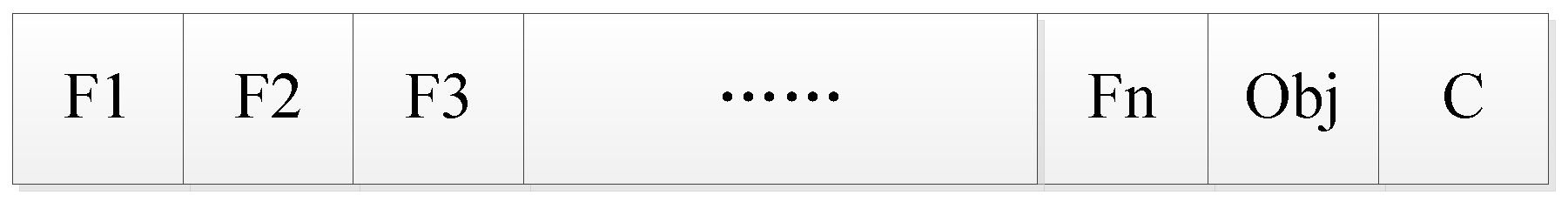
3. Encoding.
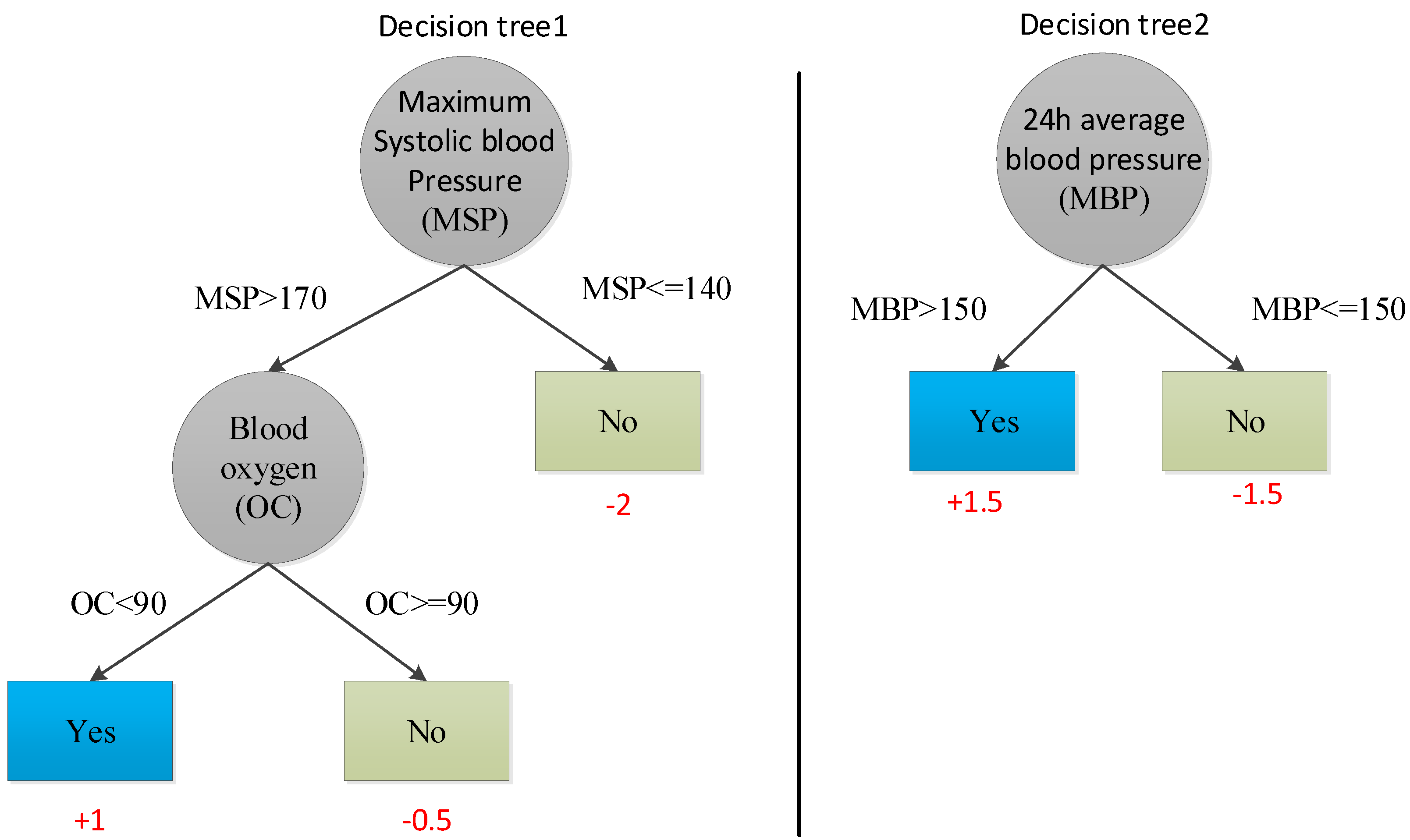
We propose a coding structure as shown in
Figure 2 before tabu search. It consists of three parts. The first part F1, F2, F3… Fn represents each feature in an n-dimensional medical feature set by a 0/1-bit string. If the feature is in the feature subset, then Fi (i ∈ [1, n]) is 1, or else it is 0. The second part is the objective function (accuracy, precision, recall, F1, and AUC), and the third part is the selected classification algorithm.
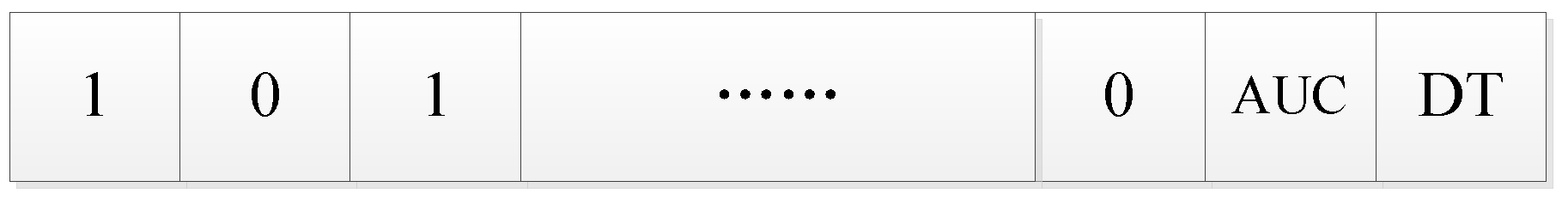
Figure 3 is an example of an initial solution after encoding.
4. Neighborhood feasible solution.
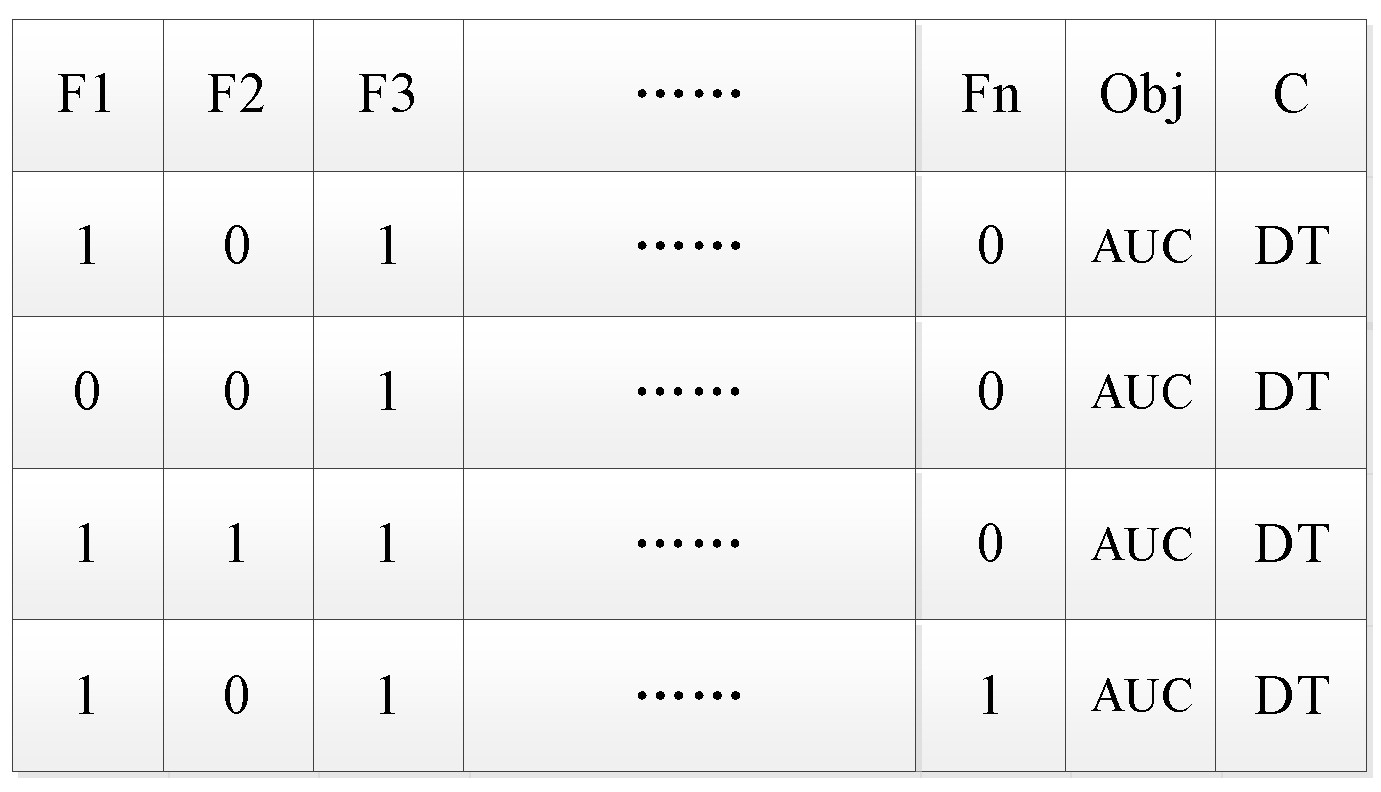
It is important to generate a neighborhood feasible solution based on the current solution. The specific method is to randomly select the feature code in the initial solution. If the feature number is 0, add the feature (the code is changed to 1); if the feature number is 1, then delete the feature (the code is changed to 0). Each neighborhood feasible solution differs from the initial solution by only one feature code. Then a specified number of neighborhood feasible solutions are generated. The number of feasible solutions in the neighborhood is the candidate set length.
Figure 4 shows the four neighborhood feasible solutions generated from the initial solution.
According to selected classifier DT and evaluation function AUC, the optimal solution of the four feasible solutions in the neighborhood is selected and regarded as the current optimal solution in the next iteration.
5. Tabu movement.
If the feature
(added or deleted) makes the neighborhood feasible solution the current optimal solution, then the feature
cannot be selected (added or deleted) in the next several rounds of T (tabu list length) iterations. For example, the third feasible solution in
Figure 4 becomes the current optimal solution because the feature F2 is added to the initial solution. Then feature F2 is added to the tabu list.
Table 2 is a tabu list with tabu length TL = 3. In the next 3 iterations, F2 cannot be added or deleted. The tabu list guarantees that the algorithm prevents searching for solutions that have been accessed, and helps to jump out of local optimal solutions.
6. Contempt principle.
Due to the existence of the tabu list, generally tabu feature will not participate in the next several rounds of search. However, when the participation of the tabu feature can make the evaluation function reach the historical optimal, the tabu feature will be amnesty, which is conducive to finding the global optimal solution. Specifically, if moving (adding/deleting) feature
can make the feasible solution better than any solution of the previous iteration, then it is allowed to add/delete the feature, even if feature
is in the tabu list. For example, if moving feature F2 in
Table 2 in the next three rounds of iteration can make the feasible solution the historically optimal solution, then remove F2 from the tabu list. The contempt principle is a method of covering tabu movement, which can avoid missing a good solution.
7. Stop rule.
We set the stop rule to a fixed number of iterations.
In summary, GSFTS-FS has the following advantages:
In gain-based-SFS, the order of adding features is arranged in order of feature importance. The more important features are prioritized and added to the feature combination until the classification algorithm reaches a certain local optimal solution. Therefore, medical features that have a significant impact on the hypertension outcomes are given priority to provide a good initial solution for the subsequent tabu search.
The tabu search optimizes the gain-based-SFS solution, which push the algorithm jump out of the local optimal solution and continue the search. The tabu search uses a tabu list to record the local optimal points that have been reached. In the next search, the information in the tabu list is used to no longer or selectively search for these points, so as to avoids converging into a local optimum.
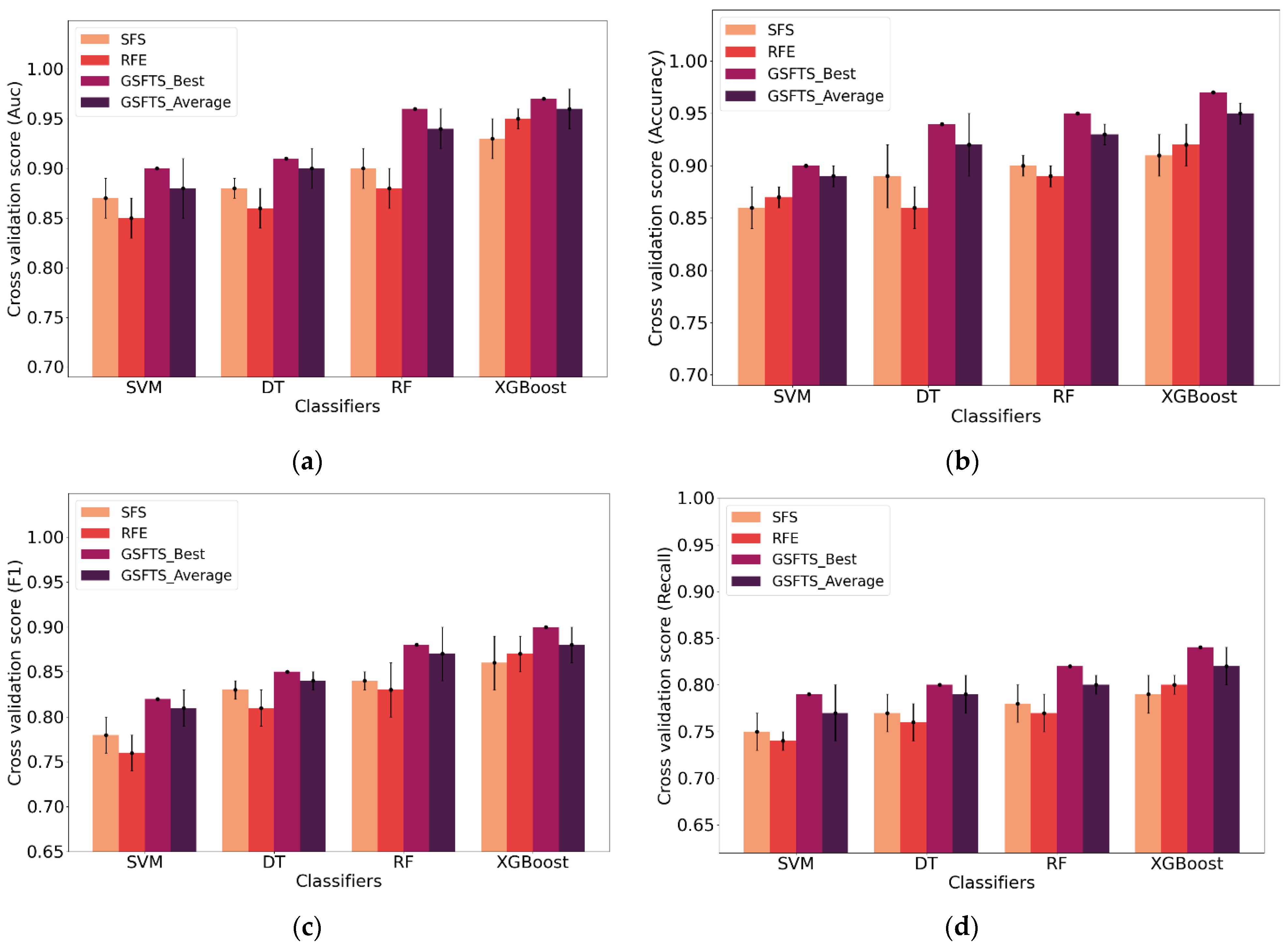
2.4. Analysis and Optimization of GSFTS-FS and XGBoost Parameters
The number of parameters to be determined in this paper is small and the value range is relatively easy to determine. Therefore, the grid search with cross validation is selected as the parameter optimization method with F1 as the evaluation index. The following parameters need to be adjusted and optimized.
(1) Candidate Set Length (CSL): the larger the length of the candidate set, the more feasible solutions can be selected in the neighborhood, and the easier it is to find the global optimal solution. However, if the length is too long, the amount of calculation will be large, and if the length is too short, it will easily fall into the local optimal solution.
(2) Tabu list length (TLL): the smaller the TLL, the larger the search range, but it is easy to repeat the search. If the TLL is too long, the calculation time will become longer.
(3) Number of iterations: the more iterations, the easier it is to find a better solution. When it reaches a certain number (saturation point), the effect will not fluctuate greatly.
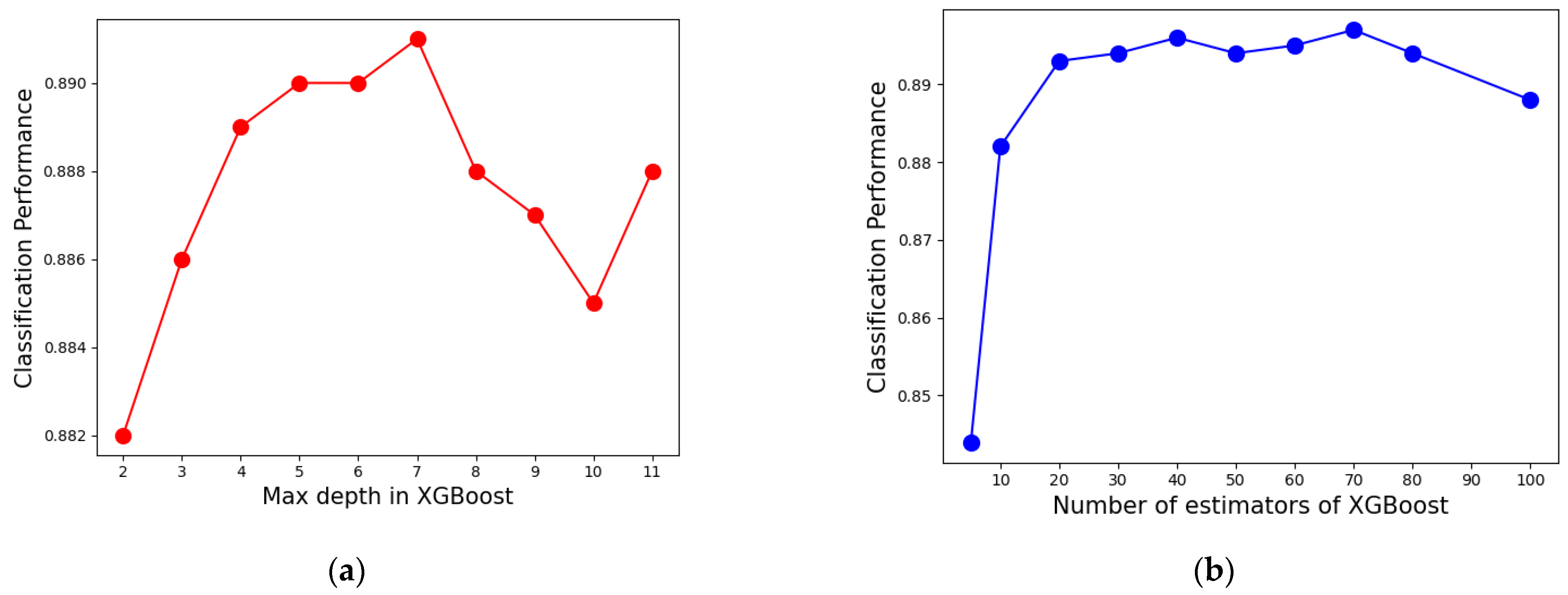
(4) Max depth of the tree in XGBoost: it is used to avoid overfitting. The larger the value, the more specific samples the model will learn.
(5) Number of estimators in XGBoost (NE): the more classifiers, the better the performance of the ensemble learning model. However, too many base classifiers will not only make it more computationally expensive and slower, but also cause overfitting.