Using Dynamic Features for Automatic Cervical Precancer Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Dynamic Image Dataset
2.1.1. Smartphone as an Acquisition Device
2.1.2. Labels
2.2. Classifier
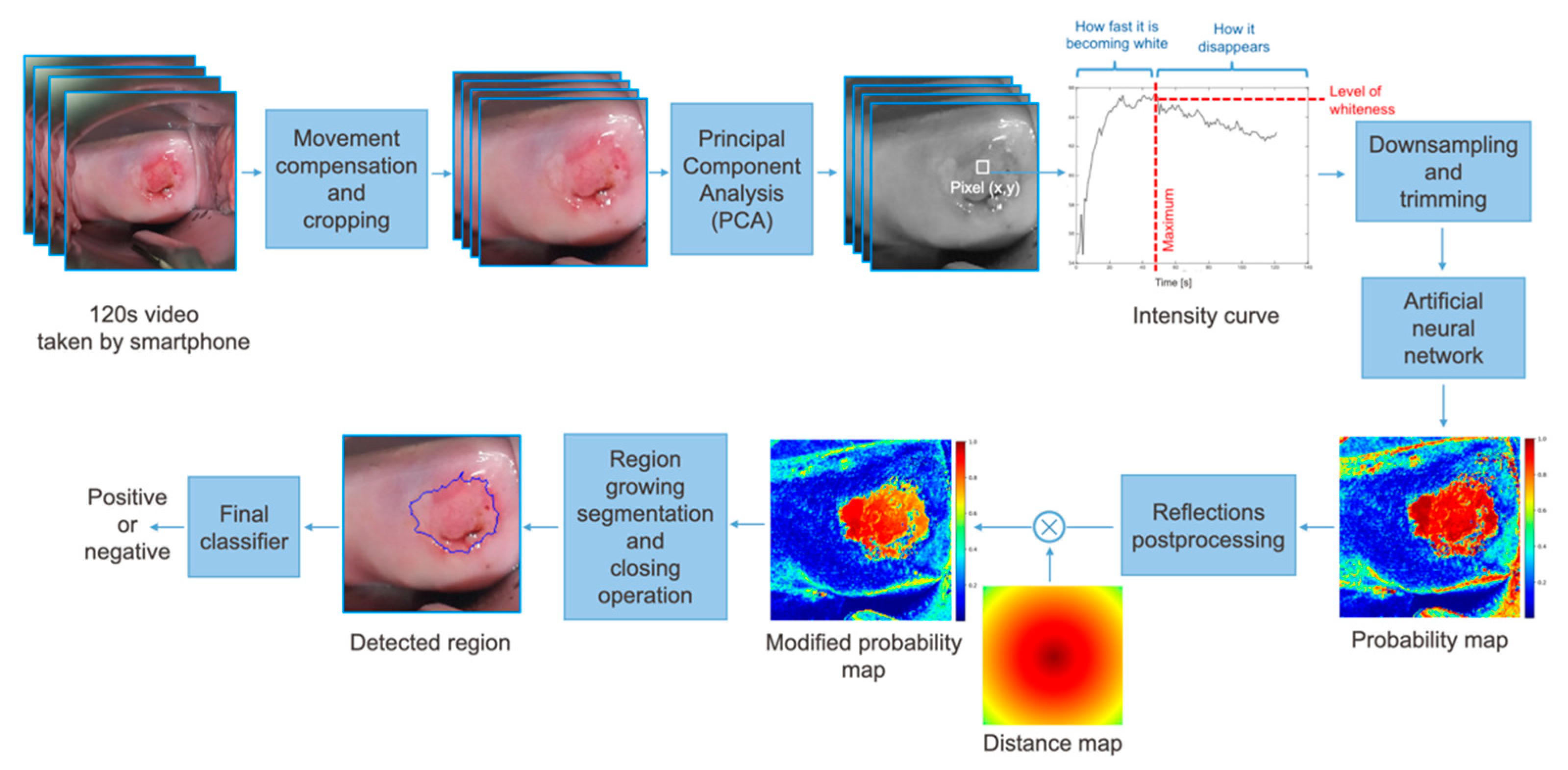
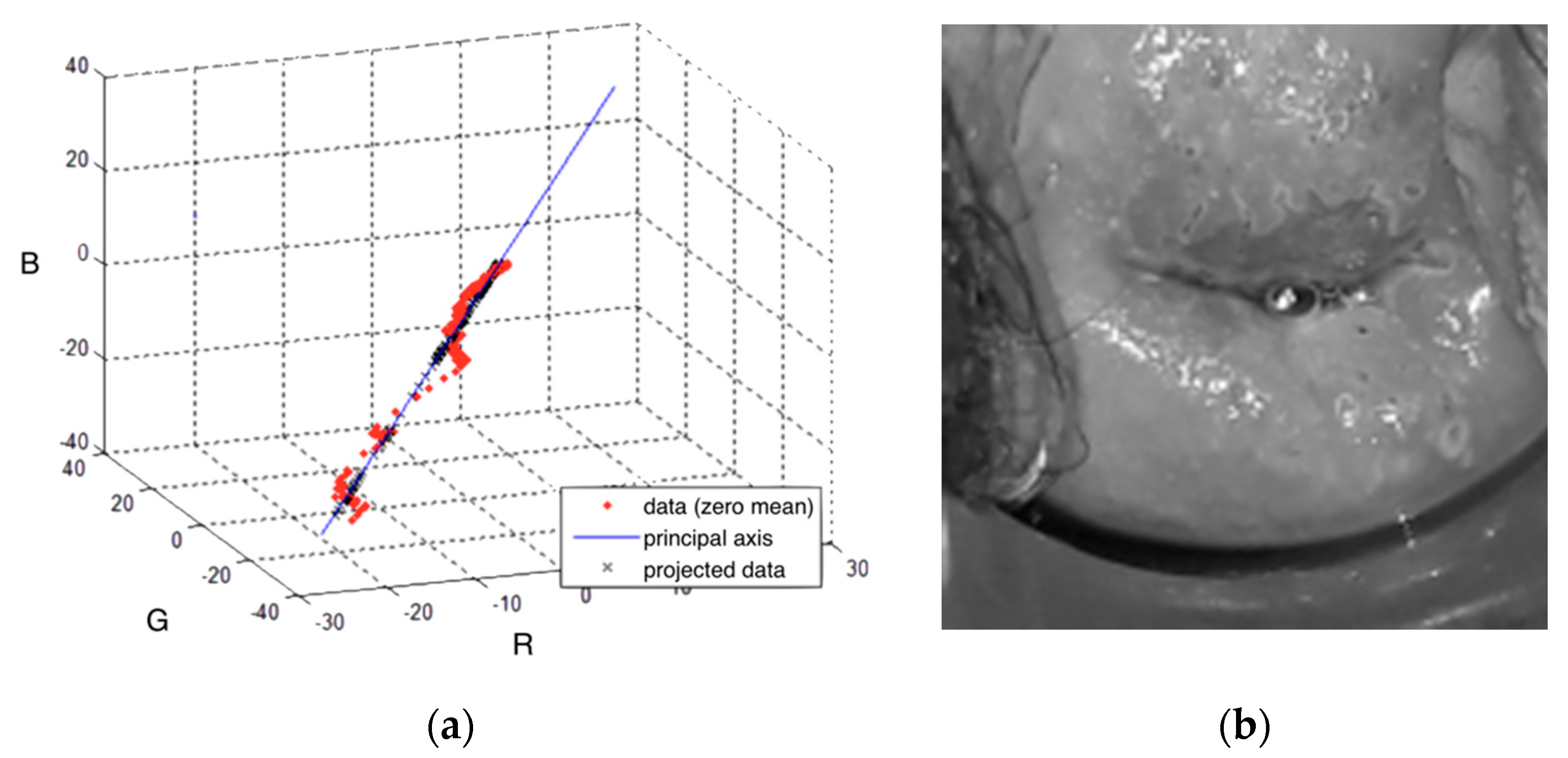
2.2.1. Preprocessing
2.2.2. Neural Network Architecture
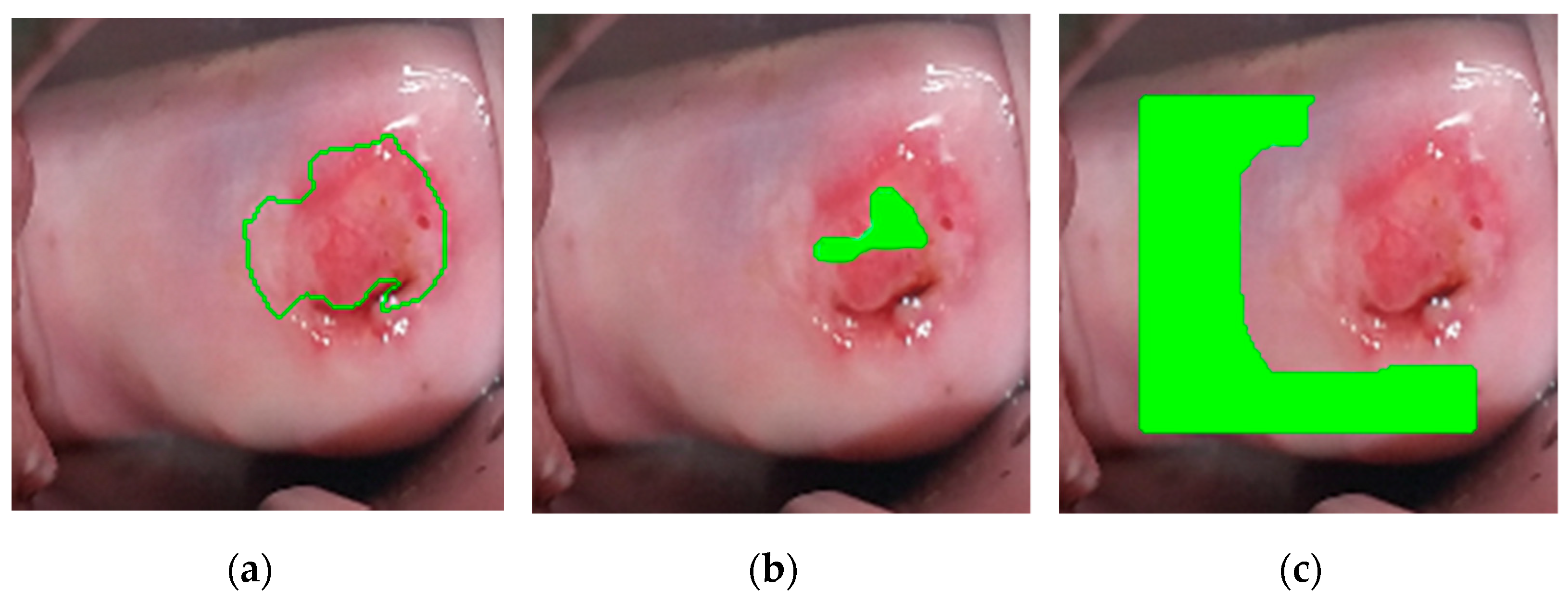
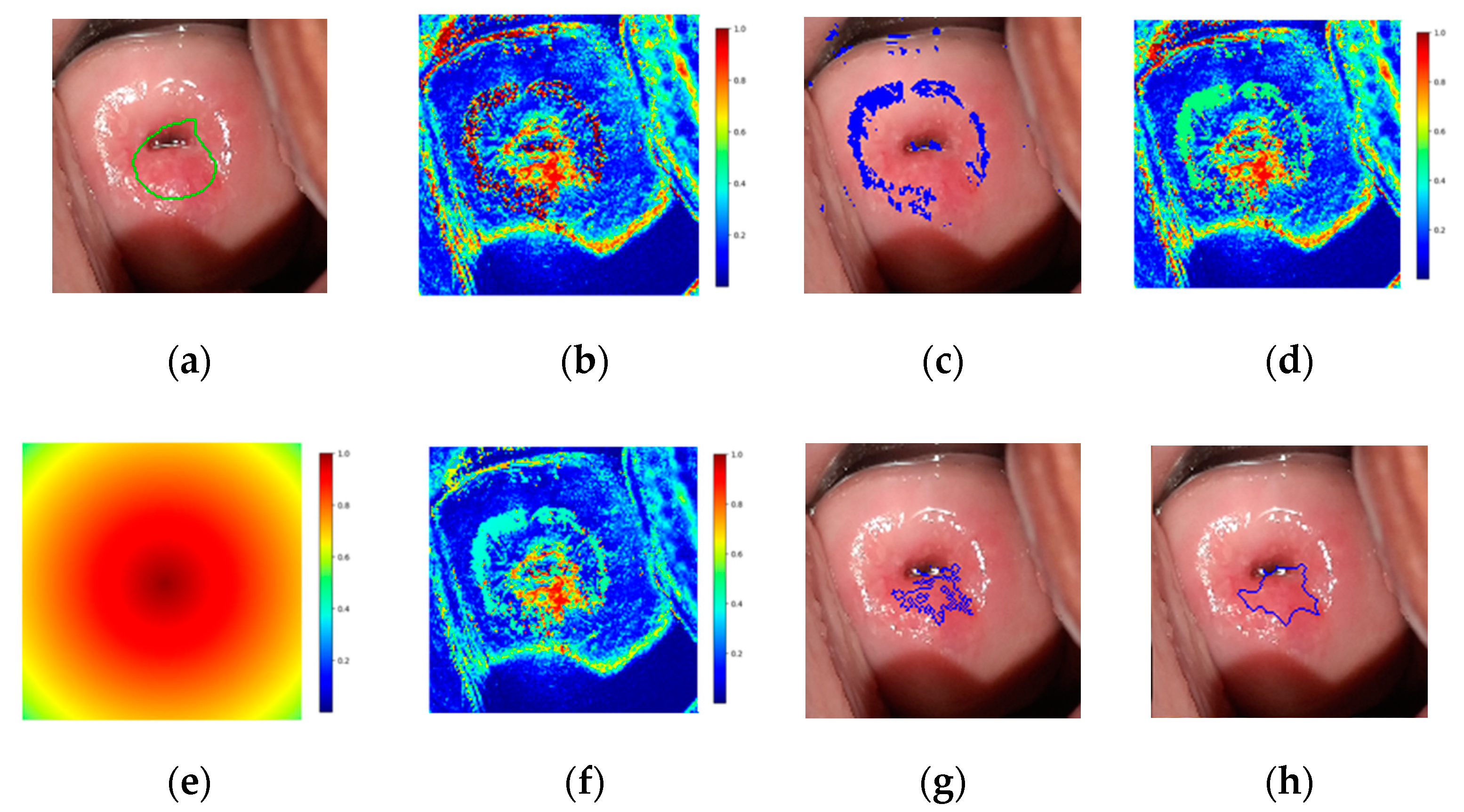
2.2.3. Postprocessing
- The first seed is randomly selected from the highest values of the modified probability map and added to the region.
- The pixel’s four adjacent neighbors are separately analyzed. Any neighbor is considered to lie on the affected region if they satisfy two criteria:
- Homogeneity criterion: the difference between the seed probability and the neighbor should be less than a fixed threshold.
- Minimum probability criterion: the probability of the neighbor should be above a threshold.
- The newly added pixels are compared to their own neighbors under the same criteria.
- Steps 2 and 3 are repeated until the criteria are not met for any neighbor of the pixels lying on the region.
- A new seed is selected such that it has the highest probability and has not been identified as part of the region before.
- The procedure is repeated until the predefined maximum number of seeds has been distributed.
3. Results
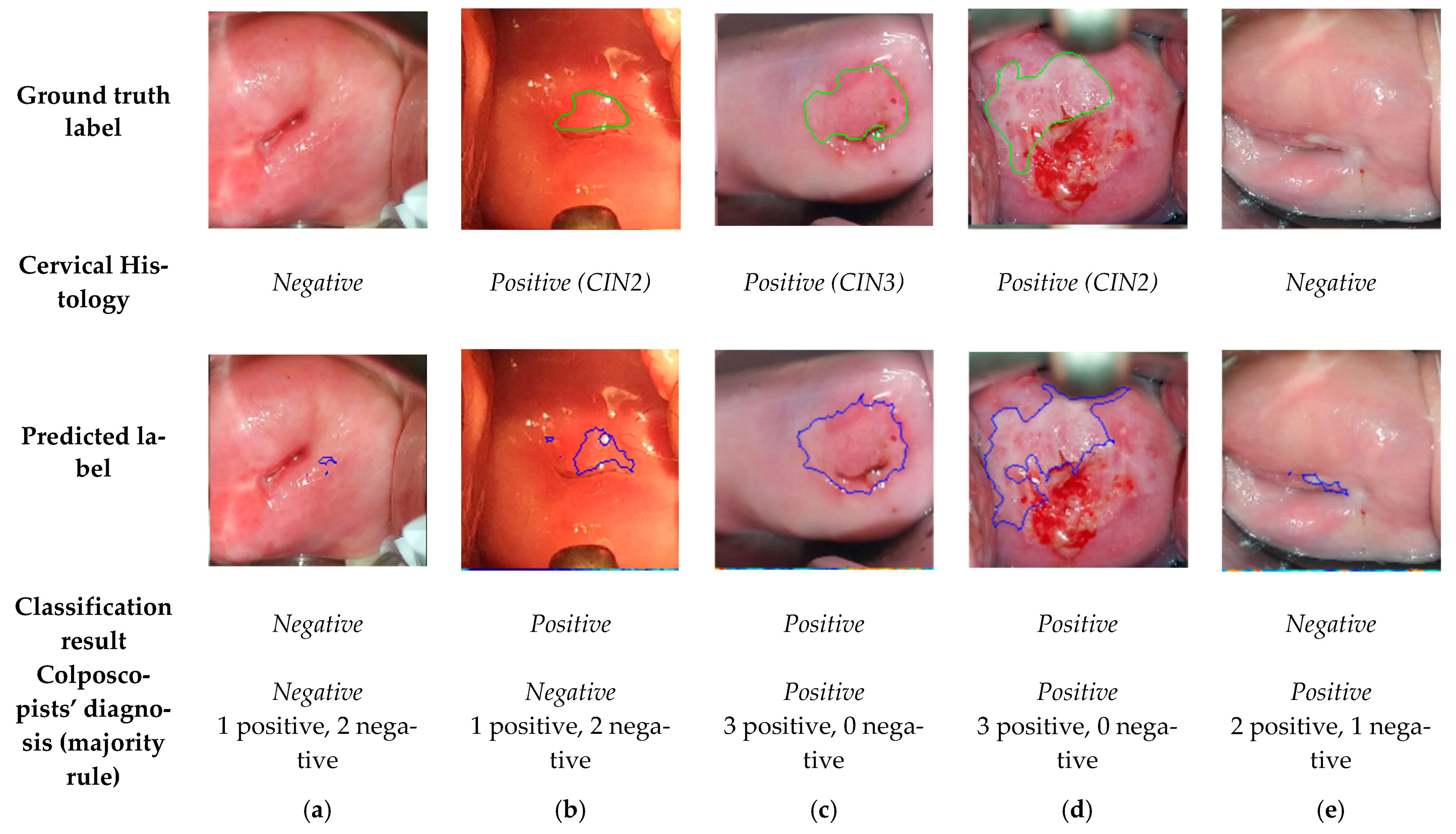
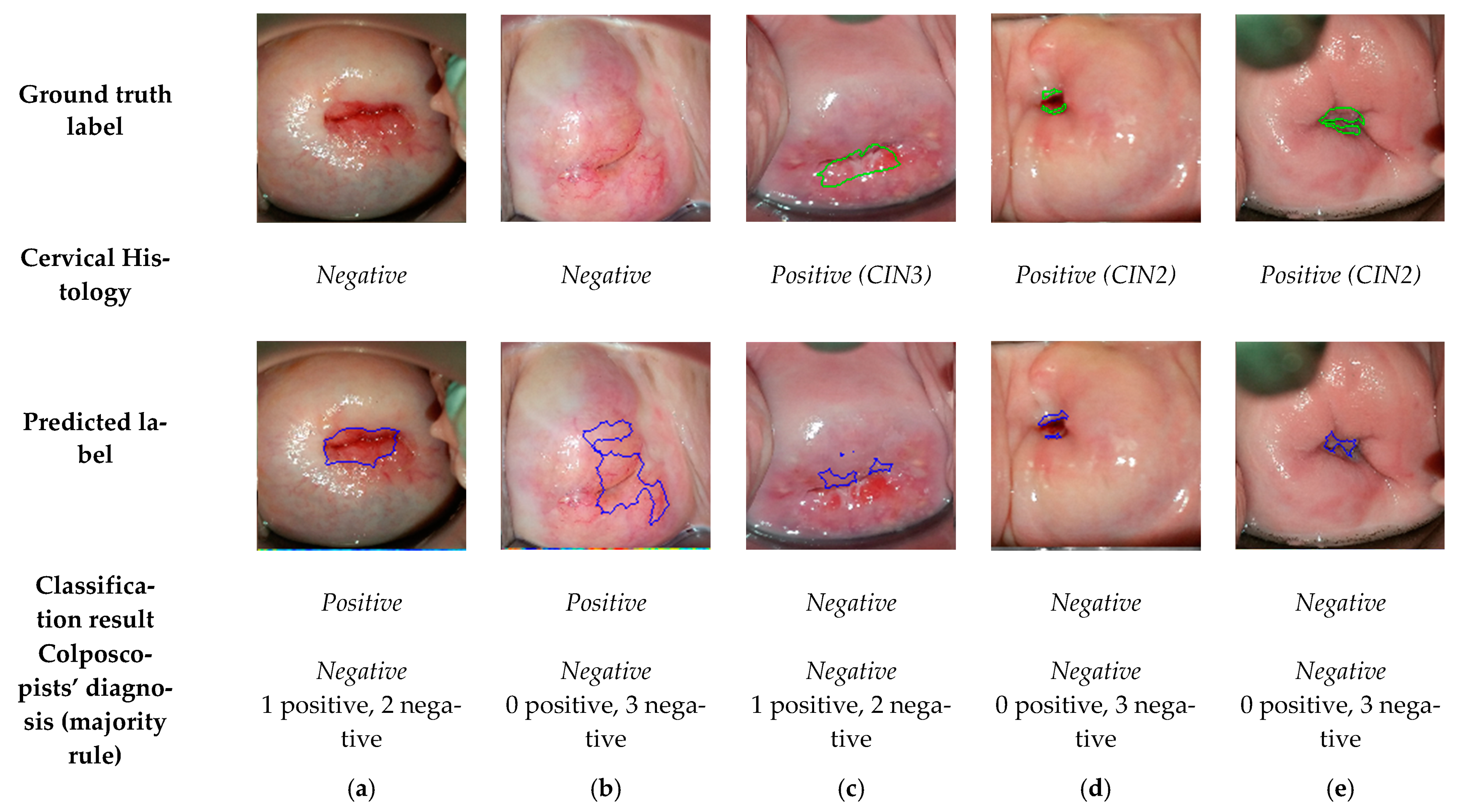
3.1. Comparision of the Final Sequence Classification, Colposcopists’ Classification, and Histology Results
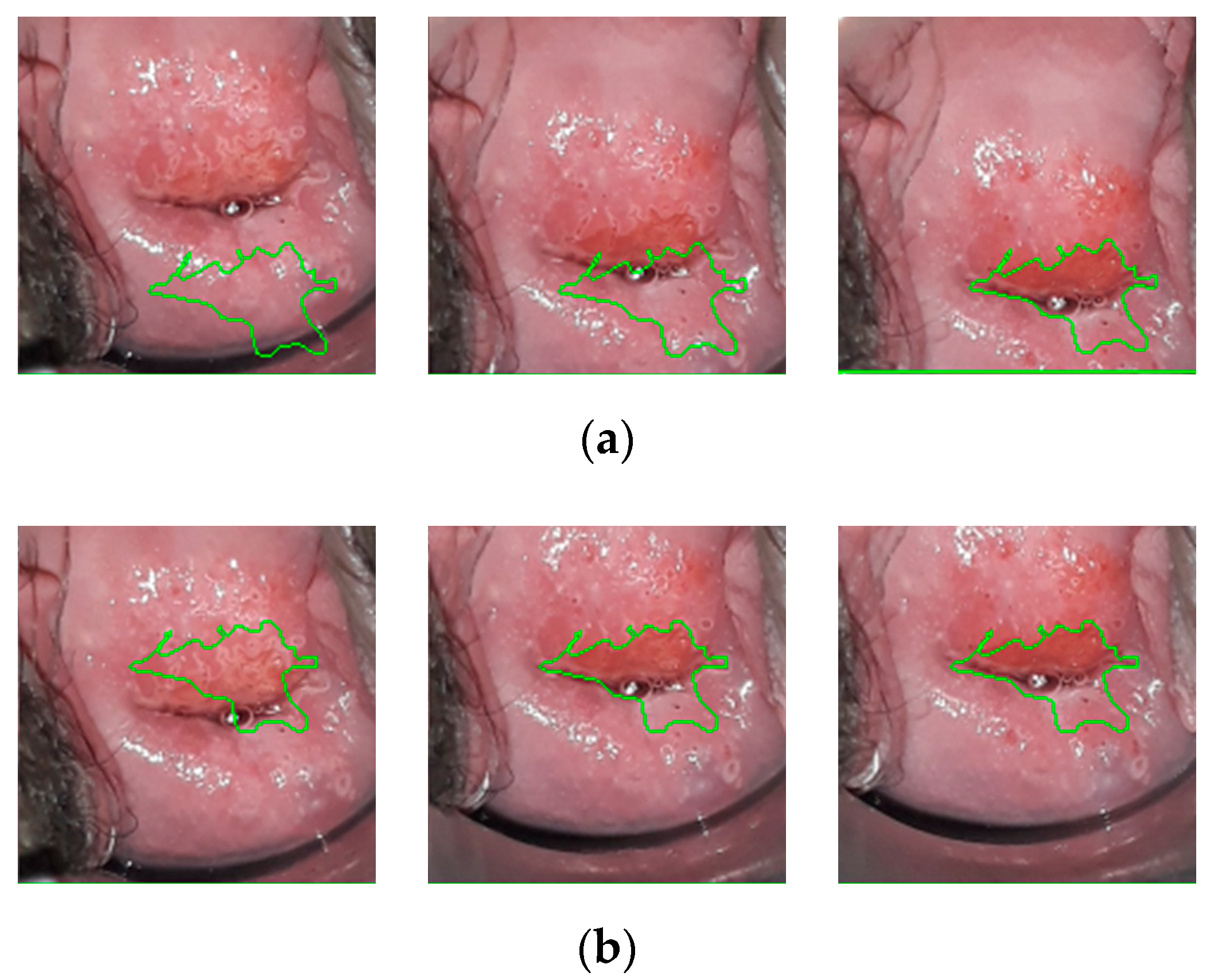
3.2. Comparision of the Predicted Lesions by the Algorithm and Annotations by Colposcopists during VIA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, caac.21660. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Guidelines for Screening and Treatment of Precancerous Lesions for Cervical Cancer Prevention; WHO: Geneva, Switzerland, 2013. [Google Scholar]
- Bigoni, J.; Gundar, M.; Tebeu, P.M.; Bongoe, A.; Schäfer, S.; Fokom-Domgue, J.; Catarino, R.; Tincho, E.F.; Bougel, S.; Vassilakos, P.; et al. Cervical cancer screening in sub-Saharan Africa: A randomized trial of VIA versus cytology for triage of HPV-positive women. Int. J. Cancer 2015, 137, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Gravitt, P.E.; Paul, P.; Katki, H.A.; Vendantham, H.; Ramakrishna, G.; Sudula, M.; Kalpana, B.; Ronnett, B.M.; Vijayaraghavan, K.; Shah, K.S. Effectiveness of VIA, pap, and HPV DNA testing in a cervical cancer screening program in a Peri-Urban community in Andhra Pradesh, India. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Ajenifuja, K.O.; Gage, J.C.; Adepiti, A.C.; Wentzensen, N.; Eklund, C.; Reilly, M.; Hutchinson, M.; Burk, R.D.; Schiffman, M. A population-based study of visual inspection with acetic acid (VIA) for cervical screening in rural nigeria. Int. J. Gynecol. Cancer 2013, 23, 507–512. [Google Scholar] [CrossRef]
- Etherington, I.J.; Luesley, D.M.; Shafi, M.I.; Dunn, J.; Hiller, L.; Jordan, J.A. Observer variability among colposcopists from the West Midlands region. BJOG Int. J. Obstet. Gynaecol. 1997, 104, 1380–1384. [Google Scholar] [CrossRef]
- Devi, M.A.; Ravi, S.; Vaishnavi, J.; Punitha, S. Classification of Cervical Cancer Using Artificial Neural Networks. Procedia Comput. Sci. 2016, 89, 465–472. [Google Scholar] [CrossRef]
- Fernandes, K.; Cardoso, J.S.; Fernandes, J. Automated Methods for the Decision Support of Cervical Cancer Screening Using Digital Colposcopies. IEEE Access 2018, 6, 33910–33927. [Google Scholar] [CrossRef]
- Jusman, Y.; Ng, S.C.; Abu Osman, N.A. Intelligent screening systems for cervical cancer. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef]
- Chitra, B.; Kumar, S.S. Recent advancement in cervical cancer diagnosis for automated screening: A detailed review. J. Ambient. Intell. Humaniz. Comput. 2021, 1, 3. [Google Scholar] [CrossRef]
- Taha, B.; Dias, J.; Werghi, N. Classification of cervical-cancer using pap-smear images: A convolutional neural network approach. In Proceedings of the Medical Image Understanding and Analysis (MIUA), Edinburgh, UK, 11–13 July 2017. [Google Scholar]
- Mariarputham, E.J.; Stephen, A. Nominated texture based cervical cancer classification. Comput. Math. Methods Med. 2015, 2015. [Google Scholar] [CrossRef]
- Sokouti, B.; Haghipour, S.; Tabrizi, A.D. A framework for diagnosing cervical cancer disease based on feedforward MLP neural network and ThinPrep histopathological cell image features. Neural Comput. Appl. 2014, 24, 221–232. [Google Scholar] [CrossRef]
- Wu, M.; Yan, C.; Liu, H.; Liu, Q.; Yin, Y. Automatic classification of cervical cancer from cytological images by using convolutional neural network. Biosci. Rep. 2018, 38, 20181769. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, J.; Choi, H.J.; Lee, B.D.; Lee, K.N. Diagnosing cervical cell images using pre-trained convolutional neural network as feature extractor. In Proceedings of the 2017 IEEE International Conference on Big Data and Smart Computing (BigComp), Jeju, Korea, 13–16 February 2017. [Google Scholar]
- Almubarak, H.A.; Stanley, R.J.; Long, R.; Antani, S.; Thoma, G.; Zuna, R.; Frazier, S.R. Convolutional Neural Network Based Localized Classification of Uterine Cervical Cancer Digital Histology Images. Procedia Comput. Sci. 2017, 114, 281–287. [Google Scholar] [CrossRef]
- Xiang, Y.; Sun, W.; Pan, C.; Yan, M.; Yin, Z.; Liang, Y. A novel automation-assisted cervical cancer reading method based on convolutional neural network. Biocybern. Biomed. Eng. 2020, 40, 611–623. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, Q.; Zhou, H.; Xia, C.; Wang, J. Automated Prediction of Cervical Precancer Based on Deep Learning. In Proceedings of the Chinese Intelligent Systems Conference 2020, Shenzhen, China, 24–25 October 2020. [Google Scholar]
- Miyagi, Y.; Takehara, K.; Miyake, T. Application of deep learning to the classification of uterine cervical squamous epithelial lesion from colposcopy images. Mol. Clin. Oncol. 2019, 11, 583–589. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization; Buiu, C.; Dănăilă, V.R.; Răduţă, C.N.; Guo, P.; Singh, S.; Xue, Z.; Long, R.; Antani, S.; Das, A. World Health Organization; Buiu, C.; Dănăilă, V.R.; Răduţă, C.N.; Guo, P.; Singh, S.; Xue, Z.; Long, R.; Antani, S.; Das, A.; et al. Preprocessing for automating early detection of cervical cancer. In Proceedings of the 2019 IEEE EMBS International Conference on Biomedical and Health Informatics, Chicago, IL, USA, 19–22 May 2019; Volume 8, pp. 597–600. [Google Scholar]
- Buiu, C.; Dănăilă, V.R.; Răduţă, C.N. MobileNetV2 ensemble for cervical precancerous lesions classification. Processes 2020, 8, 595. [Google Scholar] [CrossRef]
- Asiedu, M.N.; Simhal, A.; Chaudhary, U.; Mueller, J.L.; Lam, C.T.; Schmitt, J.W.; Venegas, G.; Sapiro, G.; Ramanujam, N. Development of Algorithms for Automated Detection of Cervical Pre-Cancers with a Low-Cost, Point-of-Care, Pocket Colposcope. IEEE Trans. Biomed. Eng. 2019, 66, 2306–2318. [Google Scholar] [CrossRef]
- Asiedu, M.N.; Skerrett, E.; Sapiro, G.; Ramanujam, N. Combining multiple contrasts for improving machine learning-based classification of cervical cancers with a low-cost point-of-care Pocket colposcope. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society 2020, Montreal, QC, Canada, 20–24 July 2020; pp. 1148–1151. [Google Scholar]
- Matti, R.; Gupta, V.; Sa, D.; Sebag, C.; Peterson, C.W.; Levitz, D. Introduction of mobile colposcopy as a primary screening tool for different socioeconomic populations in urban India. Pan Asian J. Obs. Gyn. 2019, 2, 4–11. [Google Scholar]
- Peterson, C.; Rose, D.; Mink, J.; Levitz, D. Real-Time Monitoring and Evaluation of a Visual-Based Cervical Cancer Screening Program Using a Decision Support Job Aid. Diagnostics 2016, 6, 20. [Google Scholar] [CrossRef]
- Xue, Z.; Novetsky, A.P.; Einstein, M.H.; Marcus, J.Z.; Befano, B.; Guo, P.; Demarco, M.; Wentzensen, N.; Long, L.R.; Schiffman, M.; et al. A demonstration of automated visual evaluation of cervical images taken with a smartphone camera. Int. J. Cancer 2020, 147, 2416–2423. [Google Scholar] [CrossRef]
- Hu, L.; Bell, D.; Antani, S.; Xue, Z.; Yu, K.; Horning, M.P.; Gachuhi, N.; Wilson, B.; Jaiswal, M.S.; Befano, B.; et al. An Observational Study of Deep Learning and Automated Evaluation of Cervical Images for Cancer Screening. J. Natl. Cancer Inst. 2019, 111, 923–932. [Google Scholar] [CrossRef]
- Soutter, W.P.; Diakomanolis, E.; Lyons, D.; Ghaem-Maghami, S.; Ajala, T.; Haidopoulos, D.; Doumplis, D.; Kalpaktsoglou, C.; Sakellaropoulos, G.; Soliman, S.; et al. Dynamic spectral imaging: Improving colposcopy. Clin. Cancer Res. 2009, 15, 1814–1820. [Google Scholar] [CrossRef][Green Version]
- Harris, K.E.; Lavin, P.T.; Akin, M.D.; Papagiannakis, E.; Denardis, S. Rate of detecting CIN3+ among patients with ASC-US using digital colposcopy and dynamic spectral imaging. Oncol. Lett. 2020, 20, 17. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, A.; Founta, C.; Papagiannakis, E.; Naik, R.; Fisher, A. Standardized Digital Colposcopy with Dynamic Spectral Imaging for Conservative Patient Management. Case Rep. Obstet. Gynecol. 2017, 2017, 1–5. [Google Scholar] [CrossRef]
- Budithi, S.; Peevor, R.; Pugh, D.; Papagiannakis, E.; Durman, A.; Banu, N.; Alalade, A.; Leeson, S. Evaluating Colposcopy with Dynamic Spectral Imaging During Routine Practice at Five Colposcopy Clinics in Wales: Clinical Performance. Gynecol. Obstet. Investig. 2018, 83, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Cholkeri-Singh, A.; Lavin, P.T.; Olson, C.G.; Papagiannakis, E.; Weinberg, L. Digital Colposcopy with Dynamic Spectral Imaging for Detection of Cervical Intraepithelial Neoplasia 2+ in Low-Grade Referrals: The IMPROVE-COLPO Study. J. Low. Genit. Tract Dis. 2018, 22, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, J.; Xue, P.; Tang, C.; Chang, J.; Chu, C.; Ma, K.; Li, Q.; Zheng, Y.; Qiao, Y. Computer-Aided Cervical Cancer Diagnosis Using Time-Lapsed Colposcopic Images. IEEE Trans. Med. Imaging 2020, 39, 3403–3415. [Google Scholar] [CrossRef]
- Yue, Z.; Ding, S.; Zhao, W.; Wang, H.; Ma, J.; Zhang, Y.; Zhang, Y. Automatic CIN Grades Prediction of Sequential Cervigram Image Using LSTM with Multistate CNN Features. IEEE J. Biomed. Health Inform. 2020, 24, 844–854. [Google Scholar] [CrossRef]
- Gallay, C.; Girardet, A.; Viviano, M.; Catarino, R.; Benski, A.C.; Tran, P.L.; Ecabert, C.; Thiran, J.P.; Vassilakos, P.; Petignat, P. Cervical cancer screening in low-resource settings: A smartphone image application as an alternative to colposcopy. Int. J. Womens. Health 2017, 9, 455–461. [Google Scholar] [CrossRef]
- Levy, J.; de Preux, M.; Kenfack, B.; Sormani, J.; Catarino, R.; Tincho, E.F.; Frund, C.; Fouogue, J.T.; Vassilakos, P.; Petignat, P. Implementing the 3T-approach for cervical cancer screening in Cameroon: Preliminary results on program performance. Cancer Med. 2020, 9, 7293–7300. [Google Scholar] [CrossRef]
- Hilal, Z.; Tempfer, C.B.; Burgard, L.; Rehman, S.; Rezniczek, G.A. How long is too long? Application of acetic acid during colposcopy: A prospective study. Am. J. Obstet. Gynecol. 2020, 223, 101.e1–101.e8. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, L.M.; Tahir, N.M.; Samad, M. Video stabilization based on point feature matching technique. In Proceedings of the 2012 IEEE Control and System Graduate Research Colloquium (ICSGRC), Shah Alam, Malaysia, 16–17 July 2012; pp. 303–307. [Google Scholar]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Tebeu, P.M.; Fokom-Domgue, J.; Crofts, V.; Flahaut, E.; Catarino, R.; Untiet, S.; Vassilakos, P.; Petignat, P. Effectiveness of a two-stage strategy with HPV testing followed by visual inspection with acetic acid for cervical cancer screening in a low-income setting. Int. J. Cancer 2015, 136, E743–E750. [Google Scholar] [CrossRef] [PubMed]
- Kunckler, M.; Schumacher, F.; Kenfack, B.; Catarino, R.; Viviano, M.; Tincho, E.; Tebeu, P.M.; Temogne, L.; Vassilakos, P.; Petignat, P. Cervical cancer screening in a low-resource setting: A pilot study on an HPV-based screen-and-treat approach. Cancer Med. 2017, 6, 1752–1761. [Google Scholar] [CrossRef]
- Kudva, V.; Prasad, K.; Guruvare, S. Hybrid Transfer Learning for Classification of Uterine Cervix Images for Cervical Cancer Screening. J. Digit. Imaging 2020, 33, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Luo, Y.M.; Li, P.; Liu, P.Z.; Du, Y.Z.; Sun, P.; Dong, B.H.; Xue, H. Cervical precancerous lesions classification using pre-trained densely connected convolutional networks with colposcopy images. Biomed. Signal Process. Control 2020, 55, 101566. [Google Scholar] [CrossRef]

| Dataset Specifications | |
|---|---|
| Number of sequences available | 68 |
| Total number of discarded sequences | 24 |
| Missing histology results and VIA annotations | 2 |
| Severe movement | 16 |
| Blurriness | 2 |
| Blood flow that prevents the visualization of the cervix tissue | 3 |
| Excess of mucus that prevents the visualization of the cervix tissue | 1 |
| Total number of sequences used | 44 |
| Number of positive sequences | 29 |
| CIN3 | 18 |
| CIN2 | 11 |
| Negative sequences | 15 |
| CIN1 | 3 |
| Negative | 12 |
| Video length (seconds) | 120 |
| Number of frames | 120 |
| Number of selected positive pixels | 21,851 |
| Number of selected negative pixels | 93,725 |
| Preprocessing Parameters | |
|---|---|
| Mean principal axis | [0.3609, 0.5941, 0.7074] |
| Discarded number of frames | First 10 |
| Downsampling factor | 0.1 |
| Number of input features ANN | 11 |
| Range scaling coefficients used for data augmentation | [0.9, 1.15] |
| Reflections’ threshold | 0.25 |
| ANN Parameters | |
| Number of nodes | 15 |
| Number of hidden layers | 1 |
| Postprocessing Parameters | |
| Homogeneity criteria probability threshold | 0.27 |
| Probability threshold | 0.50 |
| Reflections’ threshold | 0.25 |
| Number of seeds | 5 |
| Closing cluster kernel | [7, 7] |
| Final size threshold | 450 |
| Algorithm | Colposcopist 1 | Colposcopist 2 | Colposcopist 3 | |
|---|---|---|---|---|
| True Positives | 26 | 23 | 14 | 22 |
| True Negatives | 13 | 12 | 10 | 13 |
| False Positives | 2 | 3 | 5 | 2 |
| False Negatives | 3 | 6 | 15 | 7 |
| Accuracy | 0.89 | 0.80 | 0.55 | 0.80 |
| Precision | 0.93 | 0.88 | 0.74 | 0.92 |
| Sensitivity | 0.90 | 0.79 | 0.48 | 0.76 |
| Specificity | 0.87 | 0.80 | 0.67 | 0.87 |
| Cohen’s kappa | - | 0.62 | 0.25 | 0.53 |
| Colposcopist 1 | Colposcopist 2 | Colposcopist 3 | |
|---|---|---|---|
| Accuracy | 0.92 ± 0.10 | 0.92 ± 0.08 | 0.91 ± 0.15 |
| Precision | 0.57 ± 0.28 | 0.49 ± 0.29 | 0.55 ± 0.3 |
| Sensitivity | 0.46 ± 0.25 | 0.49 ± 0.29 | 0.5 ± 0.3 |
| Specificity | 0.97 ± 0.04 | 0.96 ± 0.05 | 0.96 ± 0.04 |
| Intersection over Union (IoU) | 0.32 ± 0.19 | 0.32 ± 0.20 | 0.33 ± 0.19 |
| Accuracy | Sensitivity | Specificity | Acquisition Device | Number of Images Captured during VIA | Additional Images Used | Number of Patients | |
|---|---|---|---|---|---|---|---|
| Asiedu et al. [22] | 0.8 | 0.81 | 0.786 | Portable colposcope | 1 | VILI | 134 |
| Miyagi et al. [19] | 0.82 | 0.80 | 0.88 | Colposcope | 1 | None | 330 |
| Yue et al. [34] | 0.96 | 0.95 | 0.98 | Colposcope | 5 | VILI and green lens | 679 |
| Li et al. [33] | 0.78 | 0.78 | Not specified | Colposcope | 4 | Pre-acetic acid | 7668 |
| Kudva et al. [42] | 0.94 | 0.91 | 0.89 | Colposcope | 1 | None | 2198 |
| Zhang et al. [43] | 0.76 | 0.44 | 0.88 | Colposcope | 1 | None | 229 |
| Proposed algorithm | 0.89 | 0.9 | 0.87 | Smartphone | 120 | None | 44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viñals, R.; Vassilakos, P.; Rad, M.S.; Undurraga, M.; Petignat, P.; Thiran, J.-P. Using Dynamic Features for Automatic Cervical Precancer Detection. Diagnostics 2021, 11, 716. https://doi.org/10.3390/diagnostics11040716
Viñals R, Vassilakos P, Rad MS, Undurraga M, Petignat P, Thiran J-P. Using Dynamic Features for Automatic Cervical Precancer Detection. Diagnostics. 2021; 11(4):716. https://doi.org/10.3390/diagnostics11040716
Chicago/Turabian StyleViñals, Roser, Pierre Vassilakos, Mohammad Saeed Rad, Manuela Undurraga, Patrick Petignat, and Jean-Philippe Thiran. 2021. "Using Dynamic Features for Automatic Cervical Precancer Detection" Diagnostics 11, no. 4: 716. https://doi.org/10.3390/diagnostics11040716
APA StyleViñals, R., Vassilakos, P., Rad, M. S., Undurraga, M., Petignat, P., & Thiran, J.-P. (2021). Using Dynamic Features for Automatic Cervical Precancer Detection. Diagnostics, 11(4), 716. https://doi.org/10.3390/diagnostics11040716







