Pulmonary Hypertension in Association with Lung Disease: Quantitative CT and Artificial Intelligence to the Rescue? State-of-the-Art Review
Abstract
1. Introduction
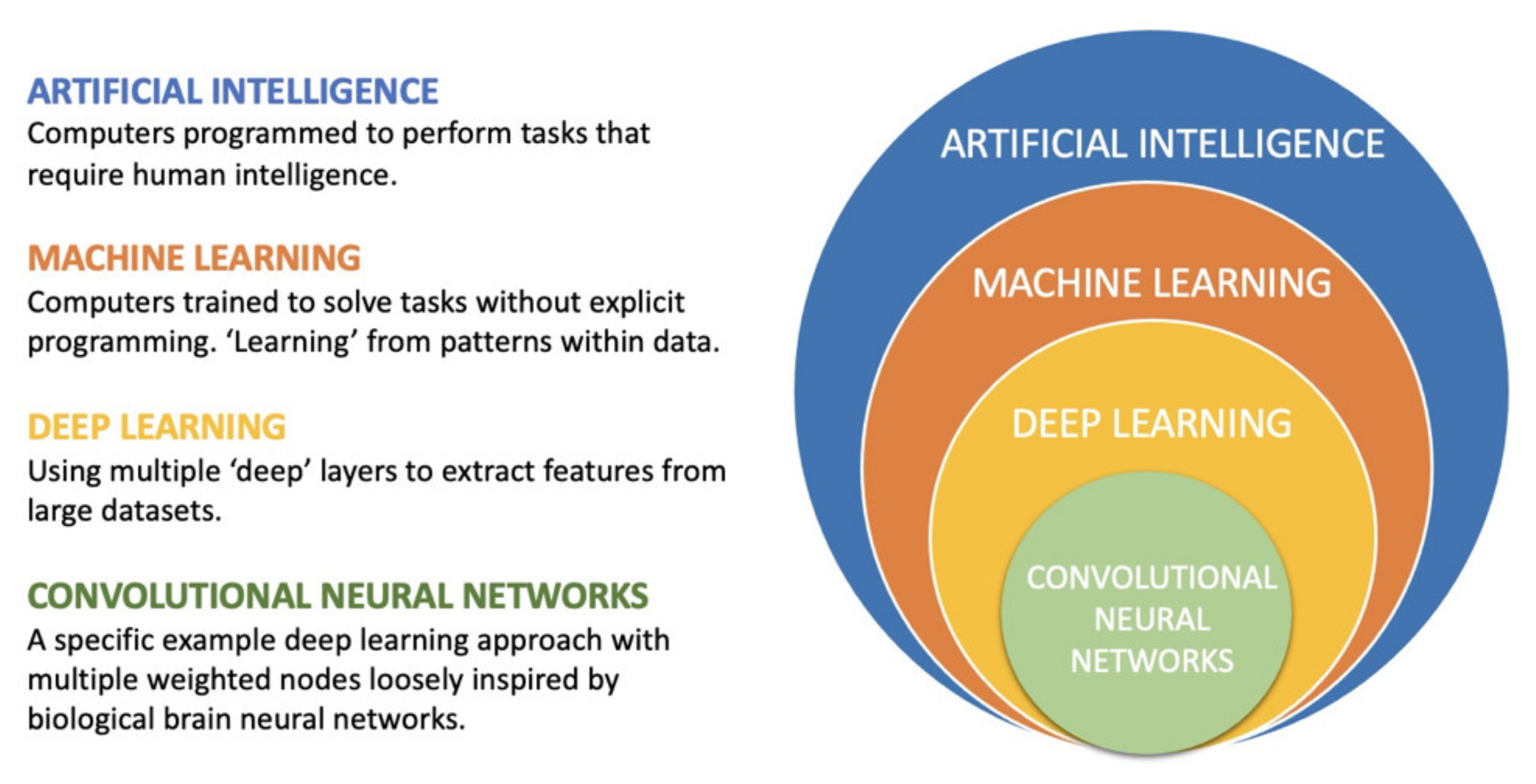
2. AI, Machine Learning and Deep Learning
2.1. Definitions
2.2. Supervised vs Unsupervised Learning
2.3. General vs. Narrow Intelligence
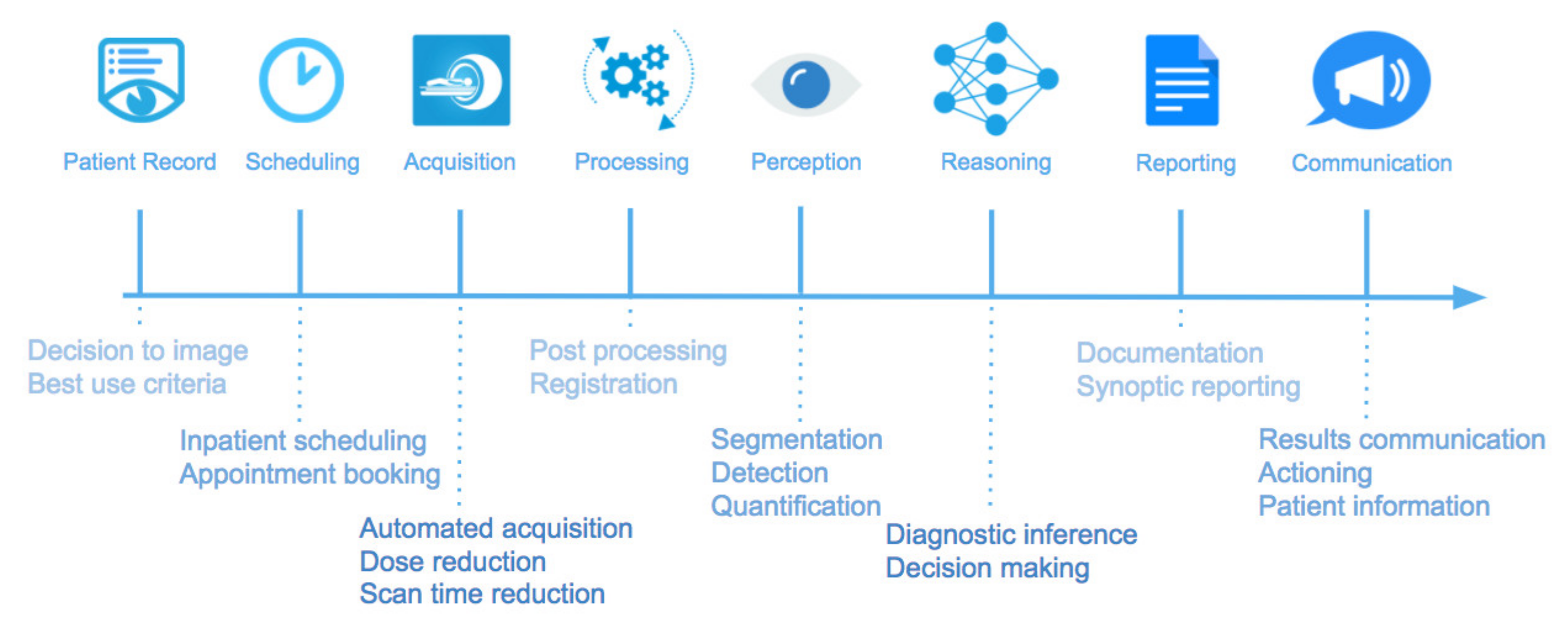
3. Machine Learning in Chest CT
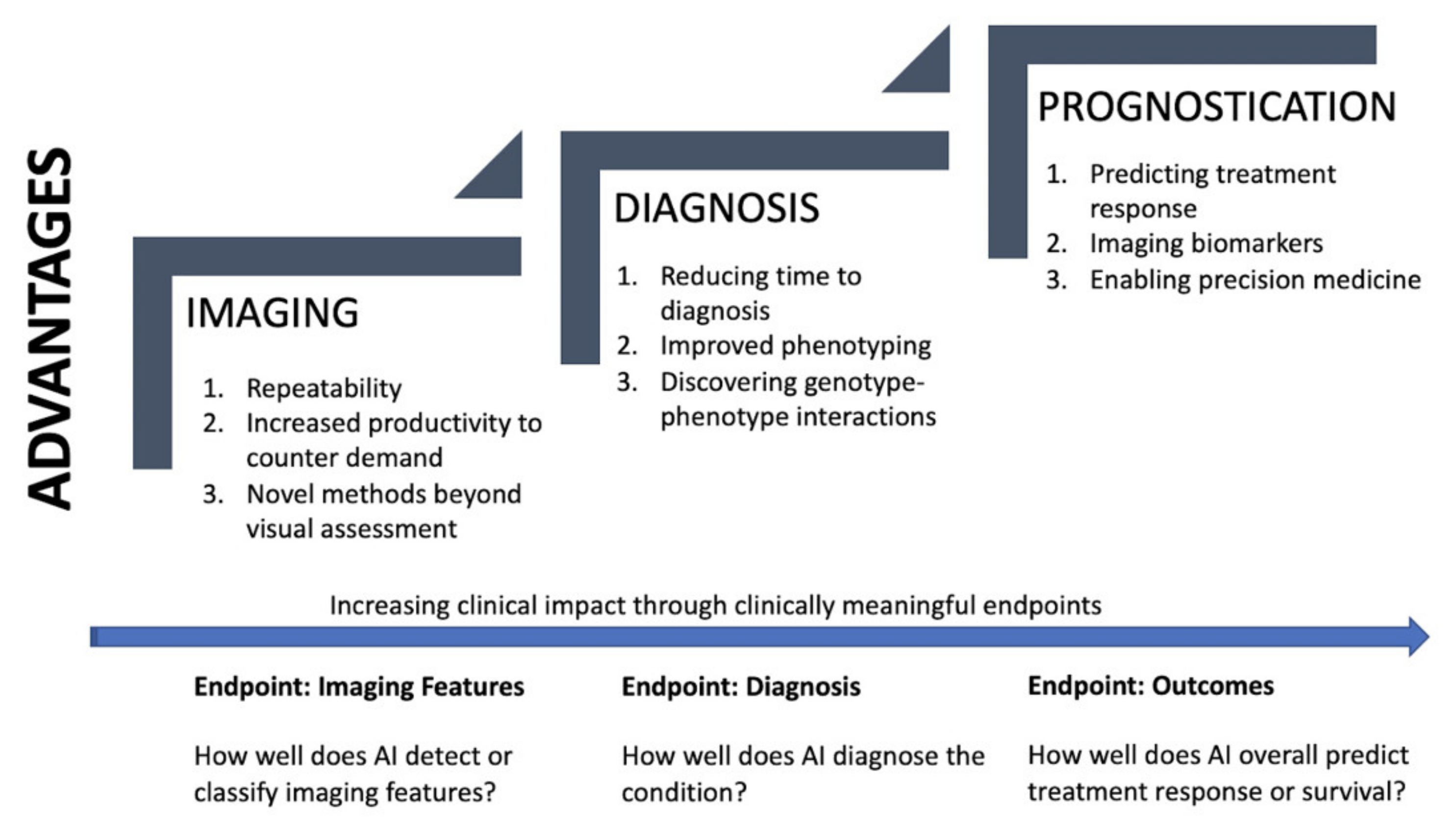
4. Promise of Quantitative Chest CT in PH
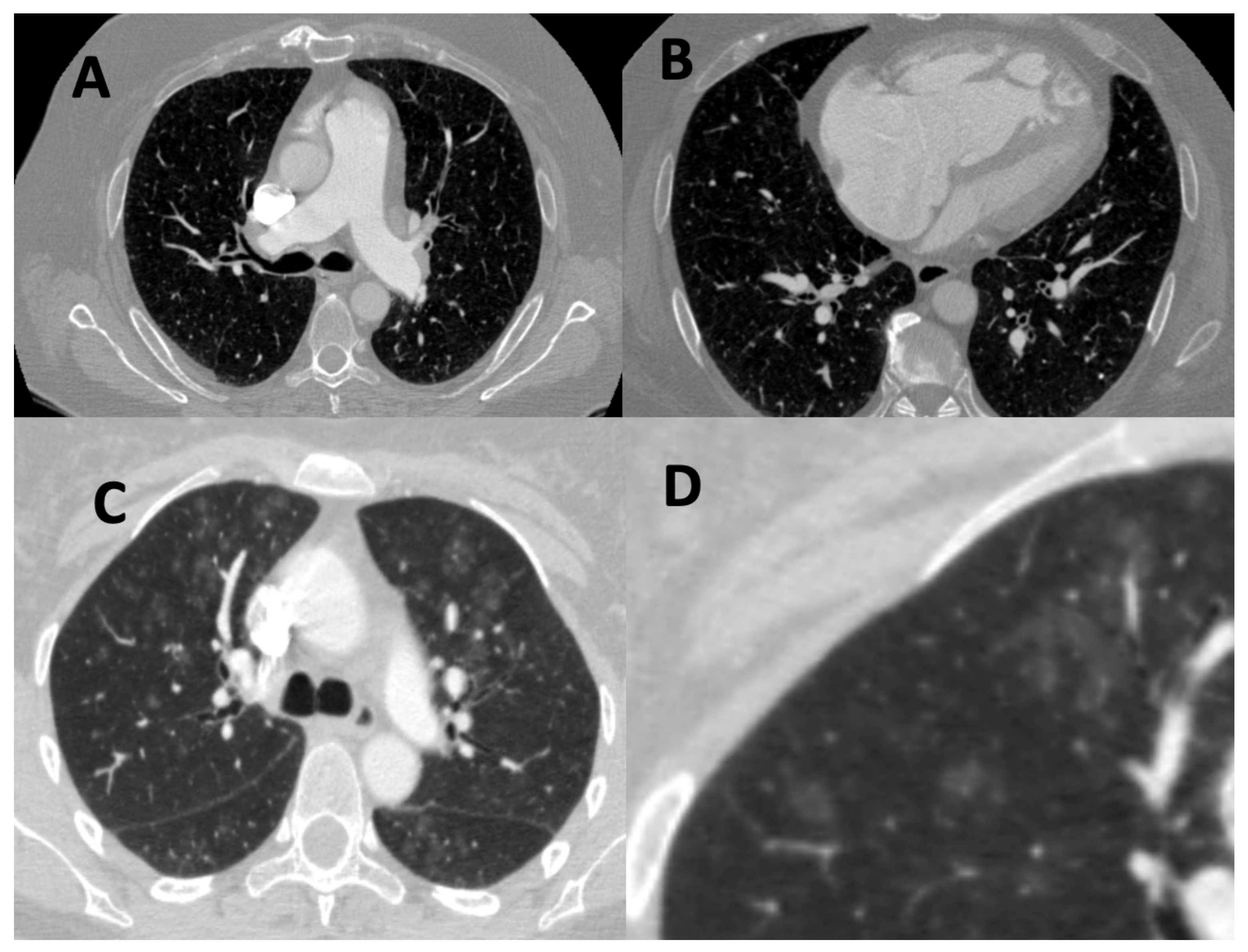
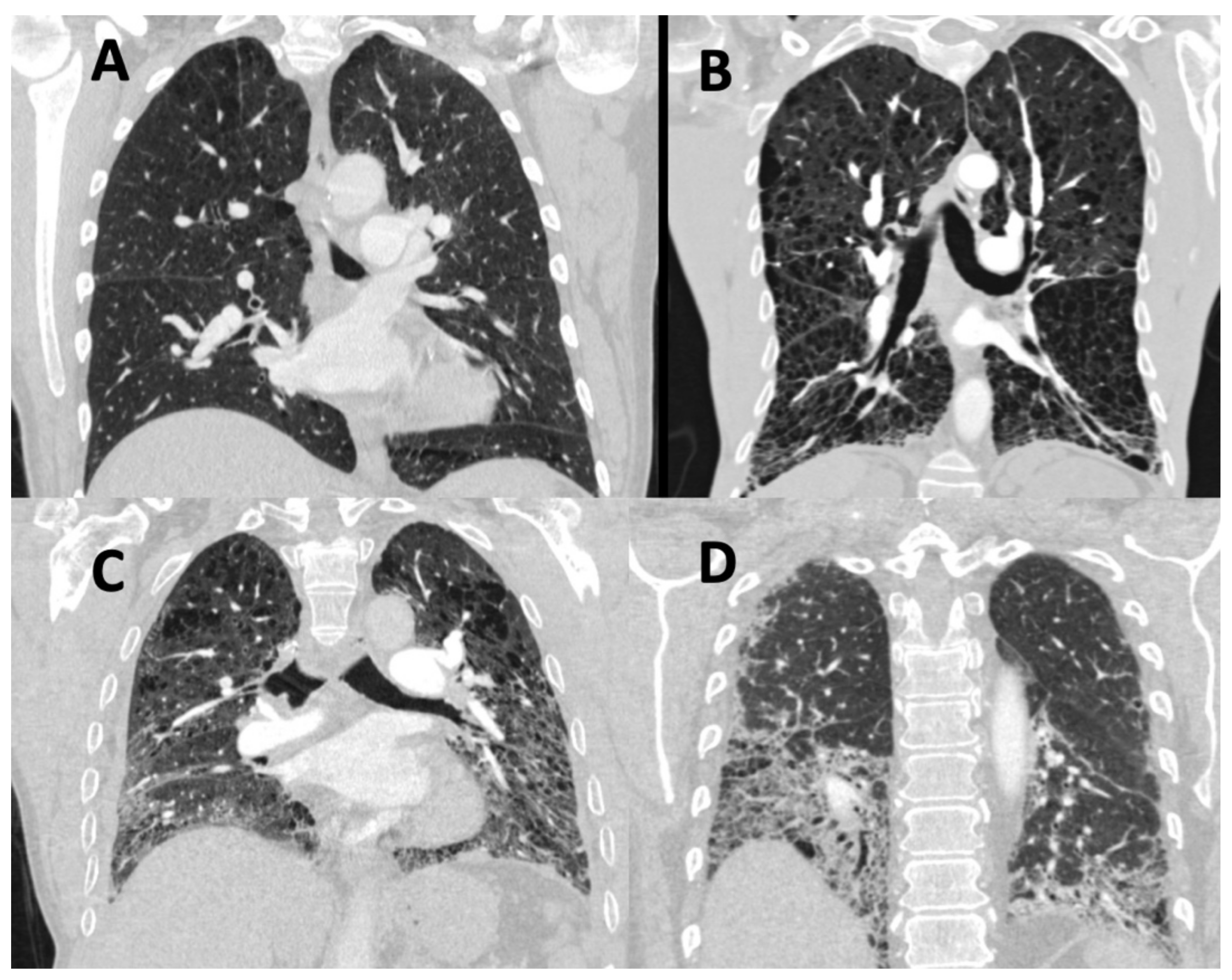
4.1. Imaging
4.1.1. Repeatability
4.1.2. Increased Efficiency to Counter Rising Demand
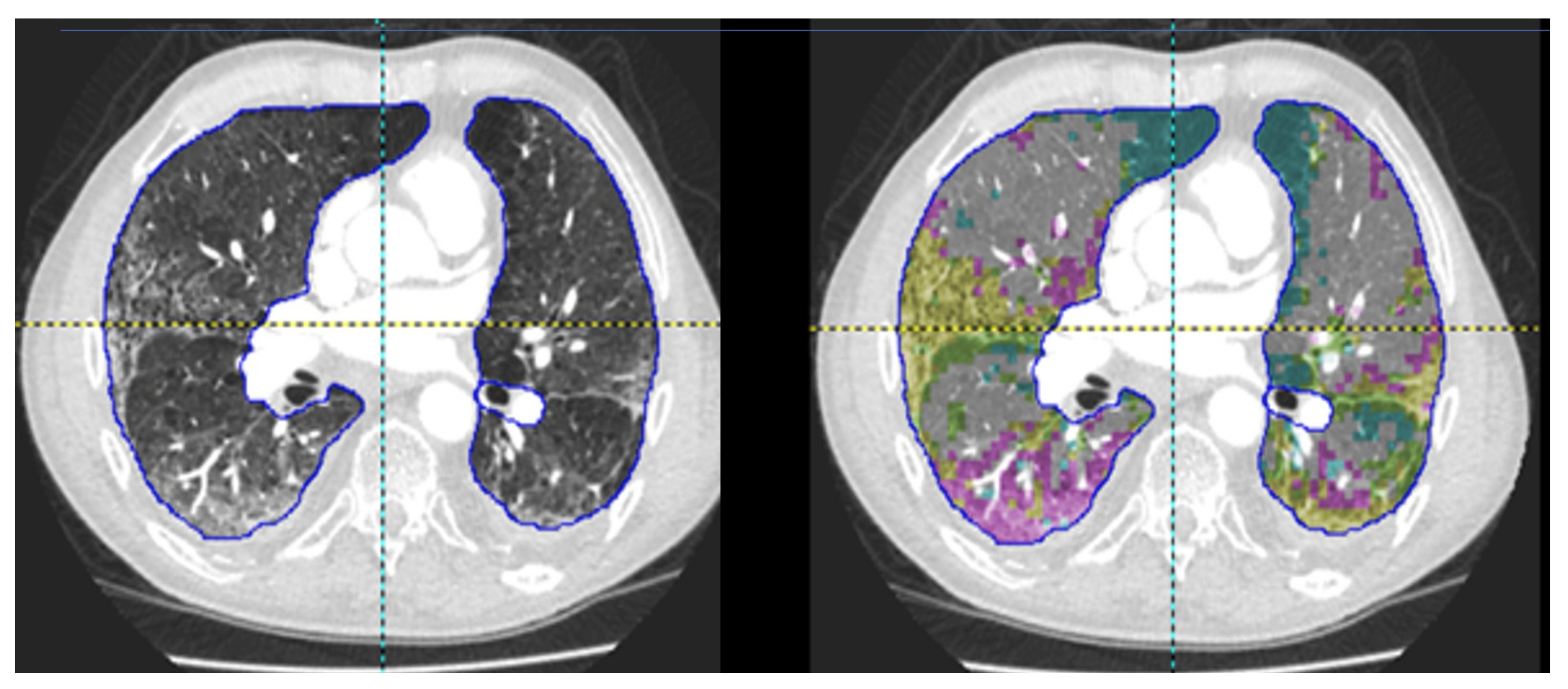
4.1.3. Novel Methodologies beyond Visual Assessment
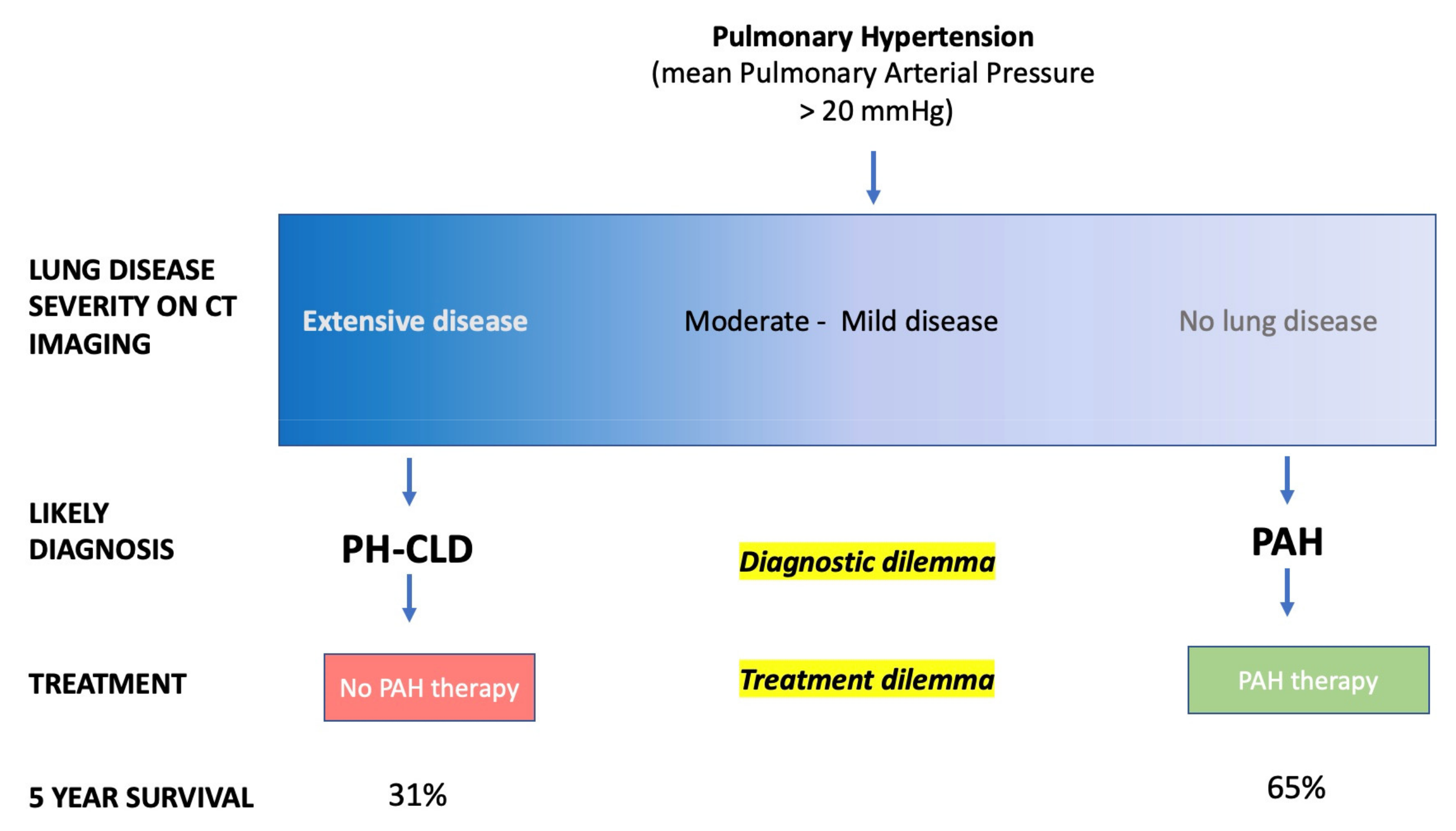
4.2. Diagnosis
4.2.1. Reducing Time to Diagnosis and Error
4.2.2. Improving Phenotyping and Classification
4.2.3. Discovering Genotype-Phenotype Associations
4.3. Prognostication
4.3.1. Predicting Treatment Response and Survival
4.3.2. Imaging Biomarkers as Clinical Endpoints
4.3.3. Enabling Precision Medicine and Big Data Analysis
5. Limitations, Challenges, and Solutions
5.1. Limitations in Quantitative CT Research
5.1.1. Variations in Data
5.1.2. Inspiration, Breathing and Motion Artefacts
5.1.3. Lack of Studies Involving Intravenous Contrast
5.2. Current Challenges Facing Machine Learning Research
5.2.1. High-Quality Training Data Is Hard to Obtain
5.2.2. Lack of External Multi Centre Validation and Prospective Studies
5.2.3. ‘Black Box’ and Interpretability
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Galiè, N.; Humbert, M.; Vachiery, J.-L.; Gibbs, S.; Lang, I.; Torbicki, A.; Simonneau, G.; Peacock, A.; Vonk Noordegraaf, A.; Beghetti, M.; et al. 2015 ESC/ERS Guidelines for the diagnosis and treatment of pulmonary hypertension: The Joint Task Force for the Diagnosis and Treatment of Pulmonary Hypertension of the European Society of Cardiology (ESC) and the European Respiratory Society (ERS): Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC), International Society for Heart and Lung Transplantation (ISHLT). Eur. Heart J. 2016, 37, 67–119. [Google Scholar] [CrossRef] [PubMed]
- Hurdman, J.; Condliffe, R.; Elliot, C.A.; Swift, A.; Rajaram, S.; Davies, C.; Hill, C.; Hamilton, N.; Armstrong, I.J.; Billings, C.; et al. Pulmonary hypertension in COPD: Results from the ASPIRE registry. Eur. Respir. J. 2013, 41, 1292–1301. [Google Scholar] [CrossRef]
- Seeger, W.; Adir, Y.; Barberà, J.A.; Champion, H.; Coghlan, J.G.; Cottin, V.; De Marco, T.; Galiè, N.; Ghio, S.; Gibbs, S.; et al. Pulmonary hypertension in chronic lung diseases. J. Am. Coll. Cardiol. 2013, 62, D109–D116. [Google Scholar] [CrossRef]
- Elia, D.; Caminati, A.; Zompatori, M.; Cassandro, R.; Lonati, C.; Luisi, F.; Pelosi, G.; Provencher, S.; Harari, S. Pulmonary hypertension and chronic lung disease: Where are we headed? Eur. Respir. Rev. 2019, 28. [Google Scholar] [CrossRef]
- Nathan, S.D.; Barbera, J.A.; Gaine, S.P.; Harari, S.; Martinez, F.J.; Olschewski, H.; Olsson, K.M.; Peacock, A.J.; Pepke-Zaba, J.; Provencher, S.; et al. Pulmonary hypertension in chronic lung disease and hypoxia. Eur. Respir. J. 2019, 53. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.; Agusti, A.; Barberà, J.A.; Celli, B.; Criner, G.; Humbert, M.; Sin, D.D.; Voelkel, N.; Olschewski, H. Pulmonary Vascular Involvement in Chronic Obstructive Pulmonary Disease. Is There a Pulmonary Vascular Phenotype? Am. J. Respir. Crit. Care Med. 2018, 198, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Chaouat, A.; Bugnet, A.-S.; Kadaoui, N.; Schott, R.; Enache, I.; Ducoloné, A.; Ehrhart, M.; Kessler, R.; Weitzenblum, E. Severe pulmonary hypertension and chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2005, 172, 189–194. [Google Scholar] [CrossRef]
- Kiely, D.G.; Elliot, C.A.; Sabroe, I.; Condliffe, R. Pulmonary hypertension: Diagnosis and management. BMJ 2013, 346, f2028. [Google Scholar] [CrossRef] [PubMed]
- Swietlik, E.M.; Gräf, S.; Morrell, N.W. The role of genomics and genetics in pulmonary arterial hypertension. Glob. Cardiol. Sci. Pract. 2020, 2020, e202013. [Google Scholar] [CrossRef] [PubMed]
- Morrell, N.W.; Aldred, M.A.; Chung, W.K.; Elliott, C.G.; Nichols, W.C.; Soubrier, F.; Trembath, R.C.; Loyd, J.E. Genetics and genomics of pulmonary arterial hypertension. Eur. Respir. J. 2019, 53. [Google Scholar] [CrossRef]
- Lewis, R.A.; Thompson, A.A.R.; Billings, C.G.; Charalampopoulos, A.; Elliot, C.A.; Hamilton, N.; Hill, C.; Hurdman, J.; Rajaram, S.; Sabroe, I.; et al. Mild parenchymal lung disease and/or low diffusion capacity impacts survival and treatment response in patients diagnosed with idiopathic pulmonary arterial hypertension. Eur. Respir. J. 2020, 55. [Google Scholar] [CrossRef]
- Sommer, N.; Ghofrani, H.A.; Pak, O.; Bonnet, S.; Provencher, S.; Sitbon, O.; Rosenkranz, S.; Hoeper, M.M.; Kiely, D.G. Current and future treatments of pulmonary arterial hypertension. Br. J. Pharm. 2021, 178, 6–30. [Google Scholar] [CrossRef]
- NHS Digital. National Pulmonary Hypertension Audit 11th Annual Report. Available online: https://digital.nhs.uk/data-and-information/publications/statistical/national-pulmonary-hypertension-audit/11th-annual-report (accessed on 22 February 2021).
- Hurdman, J.; Condliffe, R.; Elliot, C.A.; Davies, C.; Hill, C.; Wild, J.M.; Capener, D.; Sephton, P.; Hamilton, N.; Armstrong, I.J.; et al. ASPIRE registry: Assessing the Spectrum of Pulmonary hypertension Identified at a REferral centre. Eur. Respir. J. 2012, 39, 945–955. [Google Scholar] [CrossRef]
- Olschewski, H.; Simonneau, G.; Galiè, N.; Higenbottam, T.; Naeije, R.; Rubin, L.J.; Nikkho, S.; Speich, R.; Hoeper, M.M.; Behr, J.; et al. Inhaled iloprost for severe pulmonary hypertension. N. Engl. J. Med. 2002, 347, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Galié, N.; Badesch, D.; Oudiz, R.; Simonneau, G.; McGoon, M.D.; Keogh, A.M.; Frost, A.E.; Zwicke, D.; Naeije, R.; Shapiro, S.; et al. Ambrisentan therapy for pulmonary arterial hypertension. J. Am. Coll. Cardiol. 2005, 46, 529–535. [Google Scholar] [CrossRef]
- Olschewski, H. The Challenge to Decide between Pulmonary Hypertension Due to Chronic Lung Disease and PAH with Chronic Lung Disease. Diagnostics 2021, 11, 311. [Google Scholar] [CrossRef] [PubMed]
- Farber, H.W.; Miller, D.P.; Poms, A.D.; Badesch, D.B.; Frost, A.E.; Muros-Le Rouzic, E.; Romero, A.J.; Benton, W.W.; Elliott, C.G.; McGoon, M.D.; et al. Five-Year outcomes of patients enrolled in the REVEAL Registry. Chest 2015, 148, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Rose, L.; Prins, K.W.; Archer, S.L.; Pritzker, M.; Weir, E.K.; Misialek, J.R.; Thenappan, T. Survival in pulmonary hypertension due to chronic lung disease: Influence of low diffusion capacity of the lungs for carbon monoxide. J. Heart Lung Transplant. 2019, 38, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Hoeper, M.M.; Meyer, K.; Rademacher, J.; Fuge, J.; Welte, T.; Olsson, K.M. Diffusion Capacity and Mortality in Patients with Pulmonary Hypertension Due to Heart Failure with Preserved Ejection Fraction. JACC Heart Fail. 2016, 4, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Sivova, N.; Launay, D.; Wémeau-Stervinou, L.; De Groote, P.; Remy-Jardin, M.; Denis, G.; Lambert, M.; Lamblin, N.; Morell-Dubois, S.; Fertin, M.; et al. Relevance of partitioning DLCO to detect pulmonary hypertension in systemic sclerosis. PLoS ONE 2013, 8, e78001. [Google Scholar] [CrossRef]
- Chandra, S.; Shah, S.J.; Thenappan, T.; Archer, S.L.; Rich, S.; Gomberg-Maitland, M. Carbon monoxide diffusing capacity and mortality in pulmonary arterial hypertension. J. Heart Lung Transplant. 2010, 29, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Benza, R.L.; Gomberg-Maitland, M.; Elliott, C.G.; Farber, H.W.; Foreman, A.J.; Frost, A.E.; McGoon, M.D.; Pasta, D.J.; Selej, M.; Burger, C.D.; et al. Predicting Survival in Patients with Pulmonary Arterial Hypertension: The REVEAL Risk Score Calculator 2.0 and Comparison with ESC/ERS-Based Risk Assessment Strategies. Chest 2019, 156, 323–337. [Google Scholar] [CrossRef]
- Hruby, J.; Butler, J. Variability of routine pulmonary function tests. Thorax 1975, 30, 548–553. [Google Scholar] [CrossRef]
- Magnussen, H.; Vaz Fragoso, C.A.; Miller, M.R.; Brusasco, V. Spirometry Variability Must Be Critically Interpreted before Negating a Clinical Diagnosis of Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2018, 197, 835–836. [Google Scholar] [CrossRef] [PubMed]
- Khalil, N.; Manganas, H.; Ryerson, C.J.; Shapera, S.; Cantin, A.M.; Hernandez, P.; Turcotte, E.E.; Parker, J.M.; Moran, J.E.; Albert, G.R.; et al. Phase 2 clinical trial of PBI-4050 in patients with idiopathic pulmonary fibrosis. Eur. Respir. J. 2019, 53. [Google Scholar] [CrossRef]
- Rajaram, S.; Swift, A.J.; Condliffe, R.; Johns, C.; Elliot, C.A.; Hill, C.; Davies, C.; Hurdman, J.; Sabroe, I.; Wild, J.M.; et al. CT features of pulmonary arterial hypertension and its major subtypes: A systematic CT evaluation of 292 patients from the ASPIRE Registry. Thorax 2015, 70, 382–387. [Google Scholar] [CrossRef]
- Swift, A.J.; Dwivedi, K.; Johns, C.; Garg, P.; Chin, M.; Currie, B.J.; Rothman, A.M.; Capener, D.; Shahin, Y.; Elliot, C.A.; et al. Diagnostic accuracy of CT pulmonary angiography in suspected pulmonary hypertension. Eur. Radiol. 2020, 30, 4918–4929. [Google Scholar] [CrossRef]
- Kiely, D.G.; Levin, D.; Hassoun, P.; Ivy, D.D.; Jone, P.-N.; Bwika, J.; Kawut, S.M.; Lordan, J.; Lungu, A.; Mazurek, J.; et al. EXPRESS: Statement on imaging and pulmonary hypertension from the Pulmonary Vascular Research Institute (PVRI). Pulm. Circ. 2019, 2045894019841990. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- RCR Position Statement on Artificial Intelligence. Available online: https://www.rcr.ac.uk/posts/rcr-position-statement-artificial-intelligence (accessed on 11 December 2020).
- Khemasuwan, D.; Sorensen, J.S.; Colt, H.G. Artificial intelligence in pulmonary medicine: Computer vision, predictive model and COVID-19. Eur. Respir. Rev. 2020, 29. [Google Scholar] [CrossRef] [PubMed]
- Weitzenblum, E.; Chaouat, A.; Canuet, M.; Kessler, R. Pulmonary hypertension in chronic obstructive pulmonary disease and interstitial lung diseases. Semin. Respir. Crit. Care Med. 2009, 30, 458–470. [Google Scholar] [CrossRef]
- Raghu, G.; Behr, J.; Brown, K.K.; Egan, J.J.; Kawut, S.M.; Flaherty, K.R.; Martinez, F.J.; Nathan, S.D.; Wells, A.U.; Collard, H.R.; et al. Treatment of idiopathic pulmonary fibrosis with ambrisentan: A parallel, randomized trial. Ann. Intern. Med. 2013, 158, 641–649. [Google Scholar] [CrossRef]
- Nathan, S.D.; Behr, J.; Collard, H.R.; Cottin, V.; Hoeper, M.M.; Martinez, F.J.; Corte, T.J.; Keogh, A.M.; Leuchte, H.; Mogulkoc, N.; et al. Riociguat for idiopathic interstitial pneumonia-associated pulmonary hypertension (RISE-IIP): A randomised, placebo-controlled phase 2b study. Lancet Respir. Med. 2019, 7, 780–790. [Google Scholar] [CrossRef]
- Waxman, A.; Restrepo-Jaramillo, R.; Thenappan, T.; Ravichandran, A.; Engel, P.; Bajwa, A.; Allen, R.; Feldman, J.; Argula, R.; Smith, P.; et al. Inhaled Treprostinil in Pulmonary Hypertension Due to Interstitial Lung Disease. N. Engl. J. Med. 2021, 384, 325–334. [Google Scholar] [CrossRef]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.S.; Darzi, A.; et al. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Grace, K.; Salvatier, J.; Dafoe, A.; Zhang, B.; Evans, O. When Will AI Exceed Human Performance? Evidence from AI Experts. arXiv 2017, arXiv:1705.08807. [Google Scholar] [CrossRef]
- Moravčík, M.; Schmid, M.; Burch, N.; Lisý, V.; Morrill, D.; Bard, N.; Davis, T.; Waugh, K.; Johanson, M.; Bowling, M. DeepStack: Expert-level artificial intelligence in heads-up no-limit poker. Science 2017, 356, 508–513. [Google Scholar] [CrossRef]
- Silver, D.; Huang, A.; Maddison, C.J.; Guez, A.; Sifre, L.; van den Driessche, G.; Schrittwieser, J.; Antonoglou, I.; Panneershelvam, V.; Lanctot, M.; et al. Mastering the game of Go with deep neural networks and tree search. Nature 2016, 529, 484–489. [Google Scholar] [CrossRef]
- Liu, X.; Faes, L.; Kale, A.U.; Wagner, S.K.; Fu, D.J.; Bruynseels, A.; Mahendiran, T.; Moraes, G.; Shamdas, M.; Kern, C.; et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digit. Health 2019, 1, e271–e297. [Google Scholar] [CrossRef]
- Augusto, J.B.; Davies, R.H.; Bhuva, A.N.; Knott, K.D.; Seraphim, A.; Alfarih, M.; Lau, C.; Hughes, R.K.; Lopes, L.R.; Shiwani, H.; et al. Diagnosis and risk stratification in hypertrophic cardiomyopathy using machine learning wall thickness measurement: A comparison with human test-retest performance. Lancet Digit. Health 2021, 3, e20–e28. [Google Scholar] [CrossRef]
- Yang, J.; Vetterli, T.; Balte, P.P.; Barr, R.G.; Laine, A.F.; Angelini, E.D. Unsupervised Domain Adaption with Adversarial Learning (UDAA) for Emphysema Subtyping on Cardiac CT Scans: The Mesa Study. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 289–293. [Google Scholar]
- Harvey, H. Why AI Will Not Replace Radiologists. Towards Data Science. Available online: https://towardsdatascience.com/why-ai-will-not-replace-radiologists-c7736f2c7d80 (accessed on 22 February 2021).
- Sathyakumar, K.; Munoz, M.; Singh, J.; Hussain, N.; Babu, B.A. Automated Lung Cancer Detection Using Artificial Intelligence (AI) Deep Convolutional Neural Networks: A Narrative Literature Review. Cureus 2020, 12, e10017. [Google Scholar] [CrossRef]
- Cui, S.; Ming, S.; Lin, Y.; Chen, F.; Shen, Q.; Li, H.; Chen, G.; Gong, X.; Wang, H. Development and clinical application of deep learning model for lung nodules screening on CT images. Sci. Rep. 2020, 10, 13657. [Google Scholar] [CrossRef]
- Svoboda, E. Artificial intelligence is improving the detection of lung cancer. Nature 2020, 587, S20–S22. [Google Scholar] [CrossRef] [PubMed]
- Armato, S.G., 3rd; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef]
- Van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging-“how-to” guide and critical reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Sollini, M.; Antunovic, L.; Chiti, A.; Kirienko, M. Towards clinical application of image mining: A systematic review on artificial intelligence and radiomics. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2656–2672. [Google Scholar] [CrossRef]
- Chen, A.; Karwoski, R.A.; Gierada, D.S.; Bartholmai, B.J.; Koo, C.W. Quantitative CT Analysis of Diffuse Lung Disease. Radiographics 2020, 40, 28–43. [Google Scholar] [CrossRef]
- Silva, M.; Milanese, G.; Seletti, V.; Ariani, A.; Sverzellati, N. Pulmonary quantitative CT imaging in focal and diffuse disease: Current research and clinical applications. Br. J. Radiol. 2018, 91, 20170644. [Google Scholar] [CrossRef]
- Lessmann, N.; Sánchez, C.I.; Beenen, L.; Boulogne, L.H.; Brink, M.; Calli, E.; Charbonnier, J.-P.; Dofferhoff, T.; van Everdingen, W.M.; Gerke, P.K.; et al. Automated Assessment of CO-RADS and Chest CT Severity Scores in Patients with Suspected COVID-19 Using Artificial Intelligence. Radiology 2020, 202439. [Google Scholar] [CrossRef]
- Salisbury, M.L.; Lynch, D.A.; van Beek, E.J.R.; Kazerooni, E.A.; Guo, J.; Xia, M.; Murray, S.; Anstrom, K.J.; Yow, E.; Martinez, F.J.; et al. Idiopathic Pulmonary Fibrosis: The Association between the Adaptive Multiple Features Method and Fibrosis Outcomes. Am. J. Respir. Crit. Care Med. 2017, 195, 921–929. [Google Scholar] [CrossRef]
- Watadani, T.; Sakai, F.; Johkoh, T.; Noma, S.; Akira, M.; Fujimoto, K.; Bankier, A.A.; Lee, K.S.; Müller, N.L.; Song, J.-W.; et al. Interobserver variability in the CT assessment of honeycombing in the lungs. Radiology 2013, 266, 936–944. [Google Scholar] [CrossRef]
- Bankier, A.A.; De Maertelaer, V.; Keyzer, C.; Gevenois, P.A. Pulmonary emphysema: Subjective visual grading versus objective quantification with macroscopic morphometry and thin-section CT densitometry. Radiology 1999, 211, 851–858. [Google Scholar] [CrossRef]
- Widell, J.; Lidén, M. Interobserver variability in high-resolution CT of the lungs. Eur. J. Radiol. Open 2020, 7, 100228. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, K.R.; Andrei, A.-C.; King, T.E., Jr.; Raghu, G.; Colby, T.V.; Wells, A.; Bassily, N.; Brown, K.; du Bois, R.; Flint, A.; et al. Idiopathic interstitial pneumonia: Do community and academic physicians agree on diagnosis? Am. J. Respir. Crit. Care Med. 2007, 175, 1054–1060. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.; Bartholmai, B.J.; Rajagopalan, S.; Kokosi, M.; Nair, A.; Karwoski, R.; Raghunath, S.M.; Walsh, S.L.F.; Wells, A.U.; Hansell, D.M. Automated Quantitative Computed Tomography Versus Visual Computed Tomography Scoring in Idiopathic Pulmonary Fibrosis. J. Thorac. Imaging 2016, 31, 304–311. [Google Scholar] [CrossRef]
- The Royal College of Radiologists. Clinical Radiology UK Workforce Census 2018 Report; The Royal College of Radiologists: London, UK, 2019. [Google Scholar]
- Recht, M.; Bryan, R.N. Artificial Intelligence: Threat or Boon to Radiologists? J. Am. Coll. Radiol. 2017, 14, 1476–1480. [Google Scholar] [CrossRef]
- McDonald, R.J.; Schwartz, K.M.; Eckel, L.J.; Diehn, F.E.; Hunt, C.H.; Bartholmai, B.J.; Erickson, B.J.; Kallmes, D.F. The effects of changes in utilization and technological advancements of cross-sectional imaging on radiologist workload. Acad. Radiol. 2015, 22, 1191–1198. [Google Scholar] [CrossRef]
- Shiraishi, J.; Li, Q.; Appelbaum, D.; Doi, K. Computer-aided diagnosis and artificial intelligence in clinical imaging. Semin. Nucl. Med. 2011, 41, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Fazal, M.I.; Patel, M.E.; Tye, J.; Gupta, Y. The past, present and future role of artificial intelligence in imaging. Eur. J. Radiol. 2018, 105, 246–250. [Google Scholar] [CrossRef]
- Shiraishi, J.; Appelbaum, D.; Pu, Y.; Li, Q.; Pesce, L.; Doi, K. Usefulness of temporal subtraction images for identification of interval changes in successive whole-body bone scans: JAFROC analysis of radiologists’ performance. Acad. Radiol. 2007, 14, 959–966. [Google Scholar] [CrossRef]
- Annarumma, M.; Withey, S.J.; Bakewell, R.J.; Pesce, E.; Goh, V.; Montana, G. Automated Triaging of Adult Chest Radiographs with Deep Artificial Neural Networks. Radiology 2019, 291, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Kulik, T.J.; Clark, R.L.; Hasan, B.S.; Keane, J.F.; Springmuller, D.; Mullen, M.P. Pulmonary arterial hypertension: What the large pulmonary arteries tell us. Pediatr. Cardiol. 2011, 32, 759–765. [Google Scholar] [CrossRef]
- Lesage, D.; Angelini, E.D.; Bloch, I.; Funka-Lea, G. A review of 3D vessel lumen segmentation techniques: Models, features and extraction schemes. Med. Image Anal. 2009, 13, 819–845. [Google Scholar] [CrossRef] [PubMed]
- Nardelli, P.; Jimenez-Carretero, D.; Bermejo-Pelaez, D.; Washko, G.R.; Rahaghi, F.N.; Ledesma-Carbayo, M.J.; San Jose Estepar, R. Pulmonary Artery-Vein Classification in CT Images Using Deep Learning. IEEE Trans. Med. Imaging 2018, 37, 2428–2440. [Google Scholar] [CrossRef]
- Armstrong, I.; Billings, C.; Kiely, D.G.; Yorke, J.; Harries, C.; Clayton, S.; Gin-Sing, W. The patient experience of pulmonary hypertension: A large cross-sectional study of UK patients. BMC Pulm. Med. 2019, 19, 67. [Google Scholar] [CrossRef] [PubMed]
- Jairam, P.M.; van der Graaf, Y.; Lammers, J.-W.J.; Mali, W.P.T.M.; de Jong, P.A.; PROVIDI Study Group. Incidental findings on chest CT imaging are associated with increased COPD exacerbations and mortality. Thorax 2015, 70, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Binder, P.; Batmanghelich, N.K.; Estepar, R.S.J.; Golland, P. Unsupervised Discovery of Emphysema Subtypes in a Large Clinical Cohort. Mach. Learn. Med. Imaging 2016, 10019, 180–187. [Google Scholar] [CrossRef]
- Yang, J.; Angelini, E.D.; Balte, P.P.; Hoffman, E.A.; Austin, J.H.M.; Smith, B.M.; Song, J.; Barr, R.G.; Laine, A.F. Unsupervised Discovery of Spatially-Informed Lung Texture Patterns for Pulmonary Emphysema: The MESA COPD Study. Med. Image Comput. Comput. Assist. Interv. 2017, 10433, 116–124. [Google Scholar] [CrossRef]
- Shin, K.E.; Chung, M.J.; Jung, M.P.; Choe, B.K.; Lee, K.S. Quantitative computed tomographic indexes in diffuse interstitial lung disease: Correlation with physiologic tests and computed tomography visual scores. J. Comput. Assist. Tomogr. 2011, 35, 266–271. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, S.M.; Song, J.W.; Lee, S.M.; Oh, S.Y.; Kim, N.; Seo, J.B. Texture-Based Automated Quantitative Assessment of Regional Patterns on Initial CT in Patients with Idiopathic Pulmonary Fibrosis: Relationship to Decline in Forced Vital Capacity. AJR Am. J. Roentgenol. 2016, 207, 976–983. [Google Scholar] [CrossRef]
- Condon, D.F.; Nickel, N.P.; Anderson, R.; Mirza, S.; de Jesus Perez, V.A. The 6th World Symposium on Pulmonary Hypertension: What’s old is new. F1000Research 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Hemnes, A.; PVDOMICS Study Group. PVDOMICS: Early Clinical Findings Across the Spectrum of Pulmonary Hypertension. In A105. GLORY DAYS: THE LATEST CLINICAL RESEARCH IN PAH; American Thoracic Society International Conference Abstracts; American Thoracic Society: Dallas, TX, USA, 2019; p. A2517. [Google Scholar]
- Zhu, N.; Pauciulo, M.W.; Welch, C.L.; Lutz, K.A.; Coleman, A.W.; Gonzaga-Jauregui, C.; Wang, J.; Grimes, J.M.; Martin, L.J.; He, H.; et al. Novel risk genes and mechanisms implicated by exome sequencing of 2572 individuals with pulmonary arterial hypertension. Genome Med. 2019, 11, 69. [Google Scholar] [CrossRef]
- National Cohort Study of Idiopathic and Heritable PAH. Available online: https://www.ipahcohort.com/ (accessed on 21 January 2021).
- Evans, J.D.W.; Girerd, B.; Montani, D.; Wang, X.-J.; Galiè, N.; Austin, E.D.; Elliott, G.; Asano, K.; Grünig, E.; Yan, Y.; et al. BMPR2 mutations and survival in pulmonary arterial hypertension: An individual participant data meta-analysis. Lancet Respir. Med. 2016, 4, 129–137. [Google Scholar] [CrossRef]
- Elliott, C.G.; Glissmeyer, E.W.; Havlena, G.T.; Carlquist, J.; McKinney, J.T.; Rich, S.; McGoon, M.D.; Scholand, M.B.; Kim, M.; Jensen, R.L.; et al. Relationship of BMPR2 mutations to vasoreactivity in pulmonary arterial hypertension. Circulation 2006, 113, 2509–2515. [Google Scholar] [CrossRef] [PubMed]
- Dunmore, B.J.; Jones, R.J.; Toshner, M.R.; Upton, P.D.; Morrell, N.W. Approaches to treat pulmonary arterial hypertension by targeting bmpr2—From cell membrane to nucleus. Cardiovasc. Res. 2021. [Google Scholar] [CrossRef]
- van der Bruggen, C.E.; Happé, C.M.; Dorfmüller, P.; Trip, P.; Spruijt, O.A.; Rol, N.; Hoevenaars, F.P.; Houweling, A.C.; Girerd, B.; Marcus, J.T.; et al. Bone Morphogenetic Protein Receptor Type 2 Mutation in Pulmonary Arterial Hypertension: A View on the Right Ventricle. Circulation 2016, 133, 1747–1760. [Google Scholar] [CrossRef]
- Jacob, J.; Bartholmai, B.J.; Rajagopalan, S.; Kokosi, M.; Nair, A.; Karwoski, R.; Walsh, S.L.F.; Wells, A.U.; Hansell, D.M. Mortality prediction in idiopathic pulmonary fibrosis: Evaluation of computer-based CT analysis with conventional severity measures. Eur. Respir. J. 2017, 49. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, F.; Moua, T.; Rajagopalan, S.; Karwoski, R.A.; Raghunath, S.; Decker, P.A.; Hartman, T.E.; Bartholmai, B.J.; Robb, R.A.; Ryu, J.H. Automated quantification of radiological patterns predicts survival in idiopathic pulmonary fibrosis. Eur. Respir. J. 2014, 43, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.-Y.; Zhou, Y.-Q.; Xing, Y.-F.; Li, C.-F.; Lv, Q.; Dong, J.; Qin, J.; Guo, Y.-F.; Jiang, N.; Huang, C.; et al. Selection of glucocorticoid-sensitive patients in interstitial lung disease secondary to connective tissue diseases population by radiomics. Ther. Clin. Risk Manag. 2018, 14, 1975–1986. [Google Scholar] [CrossRef]
- Mooney, J.J.; Elicker, B.M.; Urbania, T.H.; Agarwal, M.R.; Ryerson, C.J.; Nguyen, M.L.T.; Woodruff, P.G.; Jones, K.D.; Collard, H.R.; King, T.E., Jr.; et al. Radiographic fibrosis score predicts survival in hypersensitivity pneumonitis. Chest 2013, 144, 586–592. [Google Scholar] [CrossRef]
- Walsh, S.L.F.; Sverzellati, N.; Devaraj, A.; Wells, A.U.; Hansell, D.M. Chronic hypersensitivity pneumonitis: High resolution computed tomography patterns and pulmonary function indices as prognostic determinants. Eur. Radiol. 2012, 22, 1672–1679. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.; Aksman, L.; Mogulkoc, N.; Procter, A.J.; Gholipour, B.; Cross, G.; Barnett, J.; Brereton, C.J.; Jones, M.G.; van Moorsel, C.H.; et al. Serial CT analysis in idiopathic pulmonary fibrosis: Comparison of visual features that determine patient outcome. Thorax 2020, 75, 648–654. [Google Scholar] [CrossRef]
- Regan, E.A.; Hokanson, J.E.; Murphy, J.R.; Make, B.; Lynch, D.A.; Beaty, T.H.; Curran-Everett, D.; Silverman, E.K.; Crapo, J.D. Genetic epidemiology of COPD (COPDGene) study design. COPD J. Chronic Obstr. Pulm. Dis. 2010, 7, 32–43. [Google Scholar] [CrossRef]
- Sieren, J.P.; Newell, J.D., Jr.; Barr, R.G.; Bleecker, E.R.; Burnette, N.; Carretta, E.E.; Couper, D.; Goldin, J.; Guo, J.; Han, M.K.; et al. SPIROMICS Protocol for Multicenter Quantitative Computed Tomography to Phenotype the Lungs. Am. J. Respir. Crit. Care Med. 2016, 194, 794–806. [Google Scholar] [CrossRef] [PubMed]
- Han, M.K.; Quibrera, P.M.; Carretta, E.E.; Barr, R.G.; Bleecker, E.R.; Bowler, R.P.; Cooper, C.B.; Comellas, A.; Couper, D.J.; Curtis, J.L.; et al. Frequency of exacerbations in patients with chronic obstructive pulmonary disease: An analysis of the SPIROMICS cohort. Lancet Respir. Med. 2017, 5, 619–626. [Google Scholar] [CrossRef]
- Han, M.K.; Kazerooni, E.A.; Lynch, D.A.; Liu, L.X.; Murray, S.; Curtis, J.L.; Criner, G.J.; Kim, V.; Bowler, R.P.; Hanania, N.A.; et al. Chronic obstructive pulmonary disease exacerbations in the COPDGene study: Associated radiologic phenotypes. Radiology 2011, 261, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Milanese, G.; Silva, M.; Sverzellati, N. Lung volume reduction of pulmonary emphysema: The radiologist task. Curr. Opin. Pulm. Med. 2016, 22, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Sverzellati, N.; Chetta, A.; Calabrò, E.; Carbognani, P.; Internullo, E.; Olivieri, D.; Zompatori, M. Reliability of quantitative computed tomography to predict postoperative lung function in patients with chronic obstructive pulmonary disease having a lobectomy. J. Comput. Assist. Tomogr. 2005, 29, 819–824. [Google Scholar] [CrossRef] [PubMed]
- Iannuzzi, G.L.; D’Alto, M.; Formisano, R.; Maniscalco, M. Biomarkers in clinical management of pulmonary hypertension: Has the emperor no clothes? A call for action. Biomark. Med. 2019, 13, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Röhrich, S.; Hofmanninger, J.; Prayer, F.; Müller, H.; Prosch, H.; Langs, G. Prospects and Challenges of Radiomics by Using Nononcologic Routine Chest CT. Radiol. Cardiothorac. Imaging 2020, 2, e190190. [Google Scholar] [CrossRef]
- Tan, R.T.; Kuzo, R.; Goodman, L.R.; Siegel, R.; Haasler, G.B.; Presberg, K.W. Utility of CT scan evaluation for predicting pulmonary hypertension in patients with parenchymal lung disease. Medical College of Wisconsin Lung Transplant Group. Chest 1998, 113, 1250–1256. [Google Scholar] [CrossRef]
- Ng, C.S.; Wells, A.U.; Padley, S.P. A CT sign of chronic pulmonary arterial hypertension: The ratio of main pulmonary artery to aortic diameter. J. Thorac. Imaging 1999, 14, 270–278. [Google Scholar] [CrossRef]
- Hansell, D.M.; Goldin, J.G.; King, T.E., Jr.; Lynch, D.A.; Richeldi, L.; Wells, A.U. CT staging and monitoring of fibrotic interstitial lung diseases in clinical practice and treatment trials: A position paper from the Fleischner Society. Lancet Respir. Med. 2015, 3, 483–496. [Google Scholar] [CrossRef]
- Labaki, W.W.; Martinez, C.H.; Martinez, F.J.; Galbán, C.J.; Ross, B.D.; Washko, G.R.; Barr, R.G.; Regan, E.A.; Coxson, H.O.; Hoffman, E.A.; et al. The Role of Chest Computed Tomography in the Evaluation and Management of the Patient with Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2017, 196, 1372–1379. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Hagiwara, Y.; Sudarshan, V.K.; Chan, W.Y.; Ng, K.H. Towards precision medicine: From quantitative imaging to radiomics. J. Zhejiang Univ. Sci. B 2018, 19, 6–24. [Google Scholar] [CrossRef]
- Herold, C.J.; Lewin, J.S.; Wibmer, A.G.; Thrall, J.H.; Krestin, G.P.; Dixon, A.K.; Schoenberg, S.O.; Geckle, R.J.; Muellner, A.; Hricak, H. Imaging in the Age of Precision Medicine: Summary of the Proceedings of the 10th Biannual Symposium of the International Society for Strategic Studies in Radiology. Radiology 2016, 279, 226–238. [Google Scholar] [CrossRef]
- Abramson, R.G.; Burton, K.R.; Yu, J.-P.J.; Scalzetti, E.M.; Yankeelov, T.E.; Rosenkrantz, A.B.; Mendiratta-Lala, M.; Bartholmai, B.J.; Ganeshan, D.; Lenchik, L.; et al. Methods and challenges in quantitative imaging biomarker development. Acad. Radiol. 2015, 22, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Quantitative Imaging Biomarkers Alliance. Available online: https://www.rsna.org/research/quantitative-imaging-biomarkers-alliance (accessed on 22 January 2021).
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Mackin, D.; Fave, X.; Zhang, L.; Fried, D.; Yang, J.; Taylor, B.; Rodriguez-Rivera, E.; Dodge, C.; Jones, A.K.; Court, L. Measuring Computed Tomography Scanner Variability of Radiomics Features. Investig. Radiol. 2015, 50, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Andrearczyk, V.; Depeursinge, A.; Müller, H. Learning cross-protocol radiomics and deep feature standardization from CT images of texture phantoms. In Proceedings of the Medical Imaging 2019: Imaging Informatics for Healthcare, Research, and Applications; International Society for Optics and Photonics: Bellingham, WA, USA, 2019; Volume 10954, p. 109540. [Google Scholar]
- Shafiq-Ul-Hassan, M.; Latifi, K.; Zhang, G.; Ullah, G.; Gillies, R.; Moros, E. Voxel size and gray level normalization of CT radiomic features in lung cancer. Sci. Rep. 2018, 8, 10545. [Google Scholar] [CrossRef] [PubMed]
- Shafiq-Ul-Hassan, M.; Zhang, G.G.; Latifi, K.; Ullah, G.; Hunt, D.C.; Balagurunathan, Y.; Abdalah, M.A.; Schabath, M.B.; Goldgof, D.G.; Mackin, D.; et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med. Phys. 2017, 44, 1050–1062. [Google Scholar] [CrossRef]
- Ligero, M.; Jordi-Ollero, O.; Bernatowicz, K.; Garcia-Ruiz, A.; Delgado-Muñoz, E.; Leiva, D.; Mast, R.; Suarez, C.; Sala-Llonch, R.; Calvo, N.; et al. Minimizing acquisition-related radiomics variability by image resampling and batch effect correction to allow for large-scale data analysis. Eur. Radiol. 2020. [Google Scholar] [CrossRef]
- Mühlberg, A.; Katzmann, A.; Heinemann, V.; Kärgel, R.; Wels, M.; Taubmann, O.; Lades, F.; Huber, T.; Maurus, S.; Holch, J.; et al. The Technome—A Predictive Internal Calibration Approach for Quantitative Imaging Biomarker Research. Sci. Rep. 2020, 10, 1103. [Google Scholar] [CrossRef] [PubMed]
- Parmar, C.; Rios Velazquez, E.; Leijenaar, R.; Jermoumi, M.; Carvalho, S.; Mak, R.H.; Mitra, S.; Shankar, B.U.; Kikinis, R.; Haibe-Kains, B.; et al. Robust Radiomics feature quantification using semiautomatic volumetric segmentation. PLoS ONE 2014, 9, e102107. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Baumgartner, C.; Ledig, C.; Newcombe, V.F.J.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Nori, A.; Criminisi, A.; Rueckert, D.; et al. Unsupervised domain adaptation in brain lesion segmentation with adversarial networks. arXiv 2016, arXiv:1612.08894. [Google Scholar]
- Zhang, S.; Sun, F.; Wang, N.; Zhang, C.; Yu, Q.; Zhang, M.; Babyn, P.; Zhong, H. Computer-Aided Diagnosis (CAD) of Pulmonary Nodule of Thoracic CT Image Using Transfer Learning. J. Digit. Imaging 2019, 32, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Park, C.M. Can Artificial Intelligence Fix the Reproducibility Problem of Radiomics? Radiology 2019, 292, 374–375. [Google Scholar] [CrossRef]
- Madani, A.; Van Muylem, A.; Gevenois, P.A. Pulmonary Emphysema: Effect of Lung Volume on Objective Quantification at Thin-Section CT. Radiology 2010, 257, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Galbán, C.J.; Han, M.K.; Boes, J.L.; Chughtai, K.A.; Meyer, C.R.; Johnson, T.D.; Galbán, S.; Rehemtulla, A.; Kazerooni, E.A.; Martinez, F.J.; et al. Computed tomography-based biomarker provides unique signature for diagnosis of COPD phenotypes and disease progression. Nat. Med. 2012, 18, 1711–1715. [Google Scholar] [CrossRef]
- Sverzellati, N.; Kuhnigk, J.-M.; Furia, S.; Diciotti, S.; Scanagatta, P.; Marchianò, A.; Molinari, F.; Stoecker, C.; Pastorino, U. CT-based weight assessment of lung lobes: Comparison with ex vivo measurements. Diagn. Interv. Radiol. 2013, 19, 355–359. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Huang, Y.; Ma, Z.; Liang, C.; Liang, C.; Liu, Z. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci. Rep. 2016, 6, 34921. [Google Scholar] [CrossRef] [PubMed]
- Duman, I.E.; Cimsit, C.; Yildizeli, S.O.; Cimsit, N.C. Parenchymal density changes in acute pulmonary embolism: Can quantitative CT be a diagnostic tool? A preliminary study. Clin. Imaging 2017, 41, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Shrivastava, A.; Singh, S.; Gupta, A. Revisiting Unreasonable Effectiveness of Data in Deep Learning Era. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 843–852. [Google Scholar]
- Halevy, A.; Norvig, P.; Pereira, F. The Unreasonable Effectiveness of Data. IEEE Intell. Syst. 2009, 24, 8–12. [Google Scholar] [CrossRef]
- Harvey, H.; Glocker, B. A standardised approach for preparing imaging data for machine learning tasks in radiology. Artif. Intell. Med. Imaging 2019. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Kohli, M.D.; Summers, R.M.; Geis, J.R. Medical Image Data and Datasets in the Era of Machine Learning-Whitepaper from the 2016 C-MIMI Meeting Dataset Session. J. Digit. Imaging 2017, 30, 392–399. [Google Scholar] [CrossRef]
- Mutasa, S.; Sun, S.; Ha, R. Understanding artificial intelligence based radiology studies: What is overfitting? Clin. Imaging 2020, 65, 96–99. [Google Scholar] [CrossRef]
- Nagendran, M.; Chen, Y.; Lovejoy, C.A.; Gordon, A.C.; Komorowski, M.; Harvey, H.; Topol, E.J.; Ioannidis, J.P.A.; Collins, G.S.; Maruthappu, M. Artificial intelligence versus clinicians: Systematic review of design, reporting standards, and claims of deep learning studies. BMJ 2020, 368, m689. [Google Scholar] [CrossRef]
- Zech, J.R.; Badgeley, M.A.; Liu, M.; Costa, A.B.; Titano, J.J.; Oermann, E.K. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med. 2018, 15, e1002683. [Google Scholar] [CrossRef] [PubMed]
- Balki, I.; Amirabadi, A.; Levman, J.; Martel, A.L.; Emersic, Z.; Meden, B.; Garcia-Pedrero, A.; Ramirez, S.C.; Kong, D.; Moody, A.R.; et al. Sample-Size Determination Methodologies for Machine Learning in Medical Imaging Research: A Systematic Review. Can. Assoc. Radiol. J. 2019, 70, 344–353. [Google Scholar] [CrossRef]
- Cruz Rivera, S.; Liu, X.; Chan, A.-W.; Denniston, A.K.; Calvert, M.J.; SPIRIT-AI and CONSORT-AI Working Group. Guidelines for clinical trial protocols for interventions involving artificial intelligence: The SPIRIT-AI extension. Lancet Digit. Health 2020, 2, e549–e560. [Google Scholar] [CrossRef]
- Liu, X.; Cruz Rivera, S.; Moher, D.; Calvert, M.J.; Denniston, A.K.; SPIRIT-AI and CONSORT-AI Working Group. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: The CONSORT-AI extension. Lancet Digit. Health 2020, 2, e537–e548. [Google Scholar] [CrossRef]
- Reyes, M.; Meier, R.; Pereira, S.; Silva, C.A.; Dahlweid, F.-M.; von Tengg-Kobligk, H.; Summers, R.M.; Wiest, R. On the Interpretability of Artificial Intelligence in Radiology: Challenges and Opportunities. Radiol. Artif. Intell. 2020, 2, e190043. [Google Scholar] [CrossRef] [PubMed]
- The Lancet Respiratory Medicine Opening the black box of machine learning. Lancet Respir. Med. 2018, 6, 801. [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dwivedi, K.; Sharkey, M.; Condliffe, R.; Uthoff, J.M.; Alabed, S.; Metherall, P.; Lu, H.; Wild, J.M.; Hoffman, E.A.; Swift, A.J.; et al. Pulmonary Hypertension in Association with Lung Disease: Quantitative CT and Artificial Intelligence to the Rescue? State-of-the-Art Review. Diagnostics 2021, 11, 679. https://doi.org/10.3390/diagnostics11040679
Dwivedi K, Sharkey M, Condliffe R, Uthoff JM, Alabed S, Metherall P, Lu H, Wild JM, Hoffman EA, Swift AJ, et al. Pulmonary Hypertension in Association with Lung Disease: Quantitative CT and Artificial Intelligence to the Rescue? State-of-the-Art Review. Diagnostics. 2021; 11(4):679. https://doi.org/10.3390/diagnostics11040679
Chicago/Turabian StyleDwivedi, Krit, Michael Sharkey, Robin Condliffe, Johanna M. Uthoff, Samer Alabed, Peter Metherall, Haiping Lu, Jim M. Wild, Eric A. Hoffman, Andrew J. Swift, and et al. 2021. "Pulmonary Hypertension in Association with Lung Disease: Quantitative CT and Artificial Intelligence to the Rescue? State-of-the-Art Review" Diagnostics 11, no. 4: 679. https://doi.org/10.3390/diagnostics11040679
APA StyleDwivedi, K., Sharkey, M., Condliffe, R., Uthoff, J. M., Alabed, S., Metherall, P., Lu, H., Wild, J. M., Hoffman, E. A., Swift, A. J., & Kiely, D. G. (2021). Pulmonary Hypertension in Association with Lung Disease: Quantitative CT and Artificial Intelligence to the Rescue? State-of-the-Art Review. Diagnostics, 11(4), 679. https://doi.org/10.3390/diagnostics11040679





