ARG1 mRNA Level Is a Promising Prognostic Marker in Head and Neck Squamous Cell Carcinomas
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Processing, and Characterization
2.2. Gene Expression Analysis
2.3. Tissue Slides Preparation and fIHC Validation
2.4. Multiplex fIHC Staining
2.5. Multiplex fIHC Data Analysis
2.6. Statistical Analysis
3. Results
3.1. Patients Characterization
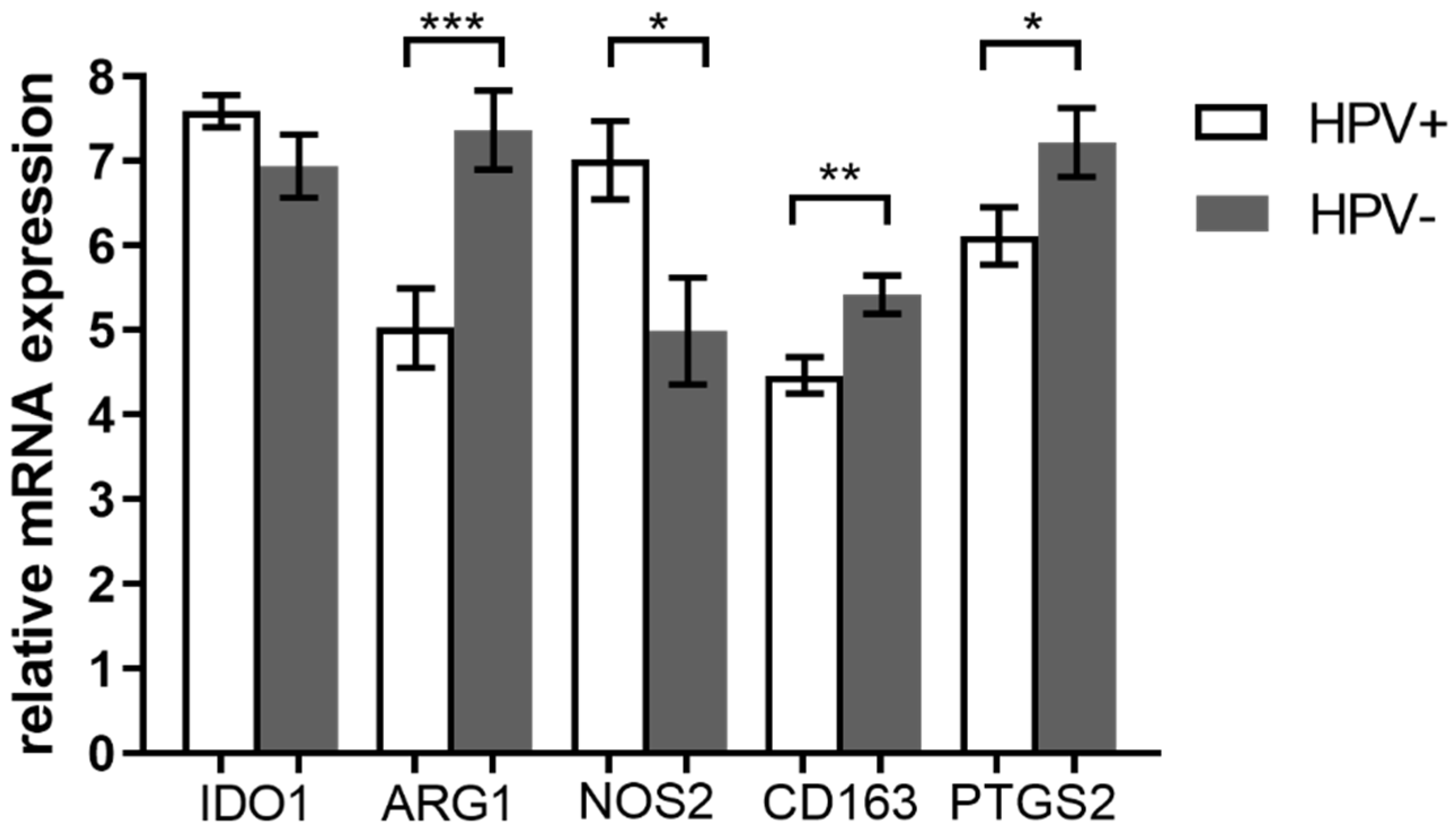
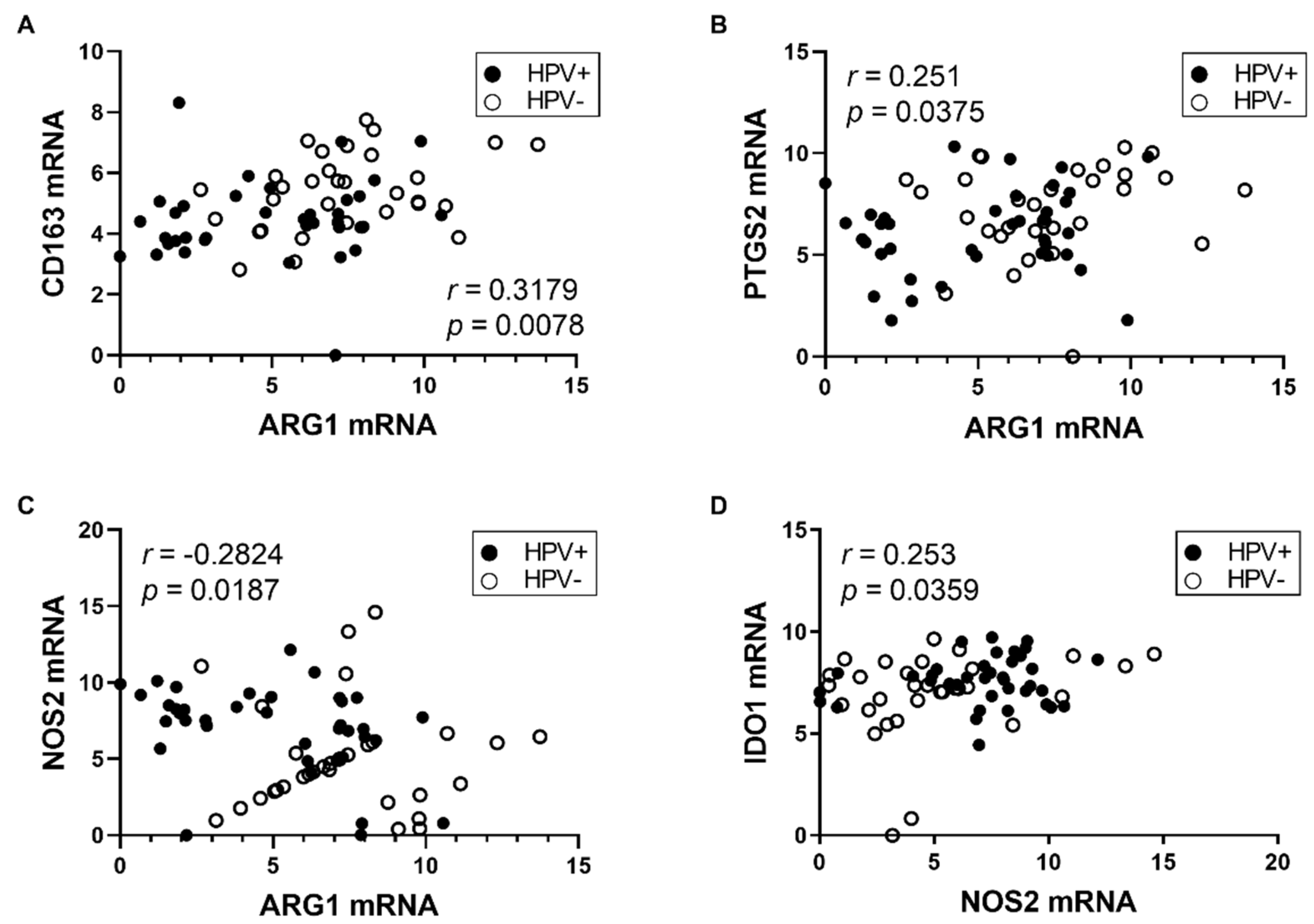
3.2. Expression of M2 TAM Markers Is Higher in HPV− Tumors
3.3. The Stroma of HNSCC Is More Infiltrated by TAMs Than the Tumor Parenchyma
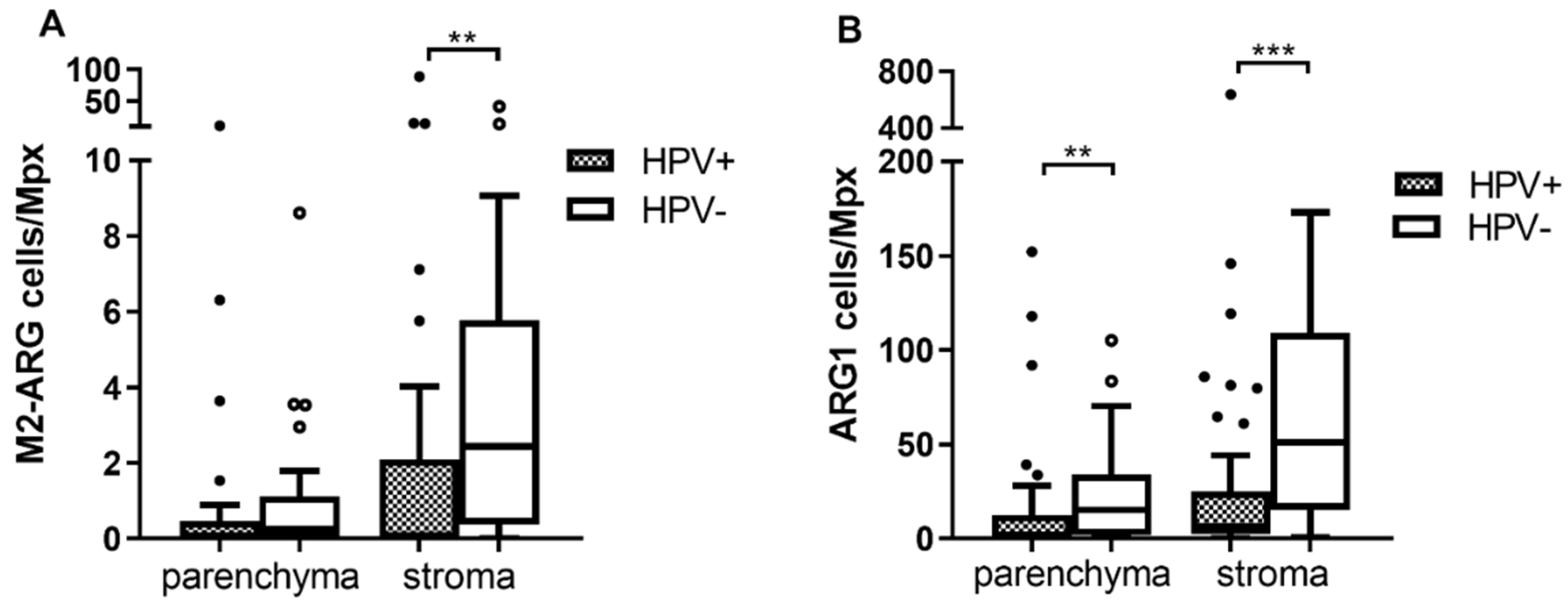
3.4. The Numbers of Arginase 1 Positive Cells Are Higher in Both Tumor Parenchyma and Stroma of HPV− Patients
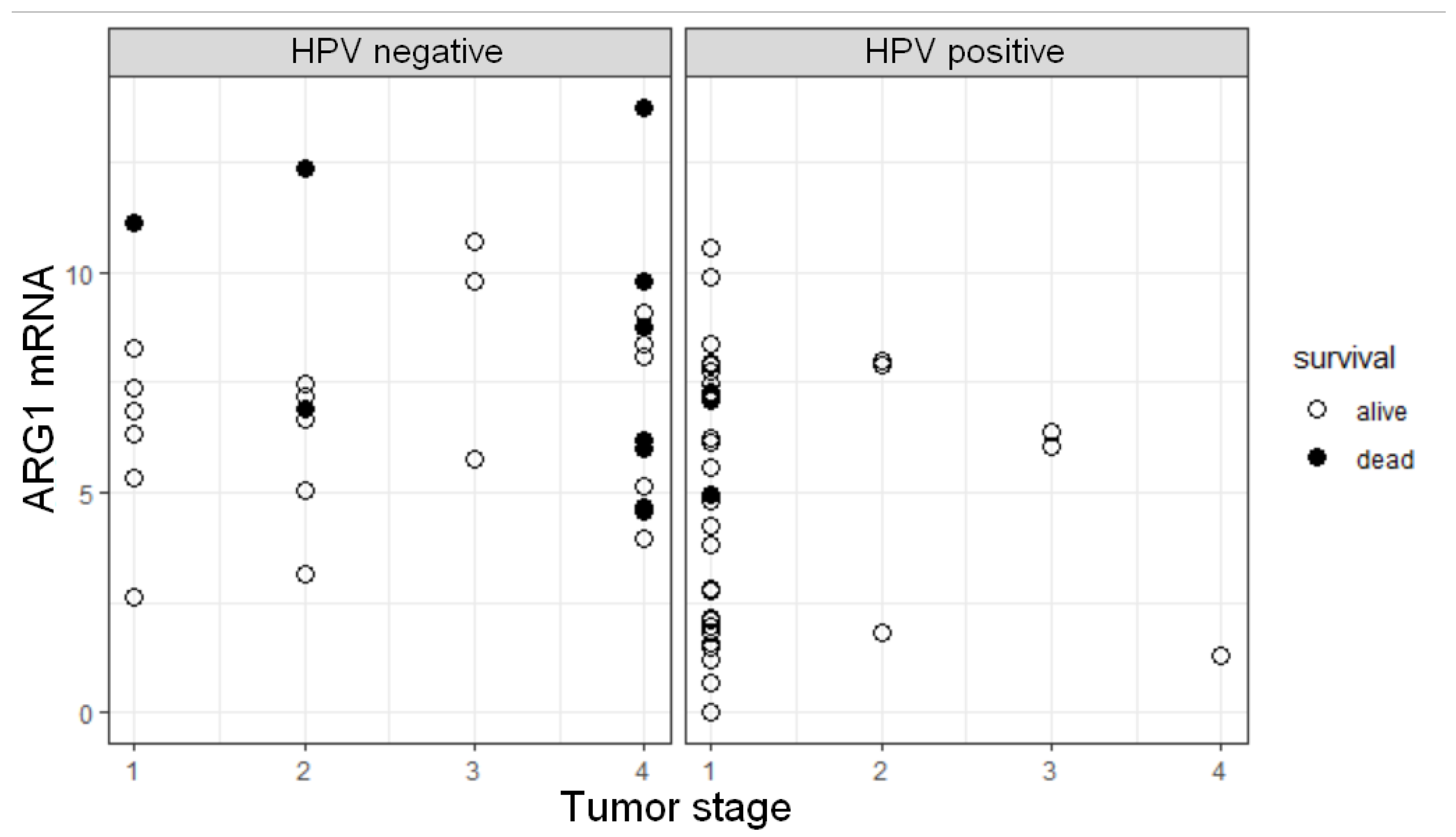
3.5. ARG1 Expression Level, HPV Status, and Tumor Stage Are Factors Influencing OS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Rettig, E.M.; D’Souza, G. Epidemiology of head and neck cancer. Surg. Oncol. Clin. N. Am. 2015, 24, 379–396. [Google Scholar] [CrossRef]
- Economopoulou, P.; Kotsantis, I.; Psyrri, A. Special Issue about Head and Neck Cancers: HPV Positive Cancers. Int. J. Mol. Sci. 2020, 21, 3388. [Google Scholar] [CrossRef]
- Ferris, R.L. Immunology and Immunotherapy of Head and Neck Cancer. J. Clin. Oncol. 2015, 33, 3293–3304. [Google Scholar] [CrossRef]
- Andersen, A.S.; Koldjaer Sølling, A.S.; Ovesen, T.; Rusan, M. The interplay between HPV and host immunity in head and neck squamous cell carcinoma. Int. J. Cancer 2014, 134, 2755–2763. [Google Scholar] [CrossRef] [PubMed]
- Balkwill, F.R.; Capasso, M.; Hagemann, T. The tumor microenvironment at a glance. J. Cell Sci. 2012, 125, 5591–5596. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, M.R.; Bonavita, E.; Barajon, I.; Garlanda, C.; Mantovani, A.; Jaillon, S. Tumor associated macrophages and neutrophils in cancer. Immunobiology 2013, 218, 1402–1410. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Marchesi, F.; Malesci, A.; Laghi, L.; Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017, 14, 399–416. [Google Scholar] [CrossRef]
- Lewis, C.; Murdoch, C. Macrophage responses to hypoxia: Implications for tumor progression and anti-cancer therapies. Am. J. Pathol. 2005, 167, 627–635. [Google Scholar] [CrossRef]
- Henze, A.T.; Mazzone, M. The impact of hypoxia on tumor-associated macrophages. J. Clin. Investig. 2016, 126, 3672–3679. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef] [PubMed]
- Medrek, C.; Pontén, F.; Jirström, K.; Leandersson, K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer 2012, 12, 306. [Google Scholar] [CrossRef]
- Park, J.Y.; Sung, J.Y.; Lee, J.; Park, Y.K.; Kim, Y.W.; Kim, G.Y.; Won, K.Y.; Lim, S.J. Polarized CD163+ tumor-associated macrophages are associated with increased angiogenesis and CXCL12 expression in gastric cancer. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 357–365. [Google Scholar] [CrossRef]
- Shabo, I.; Olsson, H.; Sun, X.F.; Svanvik, J. Expression of the macrophage antigen CD163 in rectal cancer cells is associated with early local recurrence and reduced survival time. Int. J. Cancer 2009, 125, 1826–1831. [Google Scholar] [CrossRef]
- Ino, Y.; Yamazaki-Itoh, R.; Shimada, K.; Iwasaki, M.; Kosuge, T.; Kanai, Y.; Hiraoka, N. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br. J. Cancer 2013, 108, 914–923. [Google Scholar] [CrossRef] [PubMed]
- Troiano, G.; Caponio, V.C.A.; Adipietro, I.; Tepedino, M.; Santoro, R.; Laino, L.; Lo Russo, L.; Cirillo, N.; Lo Muzio, L. Prognostic significance of CD68(+) and CD163(+) tumor associated macrophages in head and neck squamous cell carcinoma: A systematic review and meta-analysis. Oral Oncol. 2019, 93, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Heusinkveld, M.; van der Burg, S.H. Identification and manipulation of tumor associated macrophages in human cancers. J. Transl. Med. 2011, 9, 216. [Google Scholar] [CrossRef]
- Jayasingam, S.D.; Citartan, M.; Thang, T.H.; Mat Zin, A.A.; Ang, K.C.; Ch’ng, E.S. Evaluating the Polarization of Tumor-Associated Macrophages Into M1 and M2 Phenotypes in Human Cancer Tissue: Technicalities and Challenges in Routine Clinical Practice. Front. Oncol. 2019, 9, 1512. [Google Scholar] [CrossRef]
- Rath, M.; Müller, I.; Kropf, P.; Closs, E.I.; Munder, M. Metabolism via Arginase or Nitric Oxide Synthase: Two Competing Arginine Pathways in Macrophages. Front. Immunol. 2014, 5, 532. [Google Scholar] [CrossRef]
- Colegio, O.R.; Chu, N.Q.; Szabo, A.L.; Chu, T.; Rhebergen, A.M.; Jairam, V.; Cyrus, N.; Brokowski, C.E.; Eisenbarth, S.C.; Phillips, G.M.; et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014, 513, 559–563. [Google Scholar] [CrossRef]
- Yeung, A.W.; Terentis, A.C.; King, N.J.; Thomas, S.R. Role of indoleamine 2,3-dioxygenase in health and disease. Clin. Sci. (Lond.) 2015, 129, 601–672. [Google Scholar] [CrossRef]
- Hennequart, M.; Pilotte, L.; Cane, S.; Hoffmann, D.; Stroobant, V.; Plaen, E.; Eynde, B. Constitutive IDO1 Expression in Human Tumors Is Driven by Cyclooxygenase-2 and Mediates Intrinsic Immune Resistance. Cancer Immunol. Res. 2017, 5, 695–709. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, B.; Huang, J.; Lin, Y.; Xiang, T.; Wan, J.; Chouaib, S.; Ren, G. Cyclooxygenase-2 in tumor-associated macrophages promotes breast cancer cell survival by triggering a positive-feedback loop between macrophages and cancer cells. Oncotarget 2015, 6, 29637–29650. [Google Scholar] [CrossRef]
- Chen, I.C.; Lee, K.H.; Hsu, Y.H.; Wang, W.R.; Chen, C.M.; Cheng, Y.W. Expression Pattern and Clinicopathological Relevance of the Indoleamine 2,3-Dioxygenase 1/Tryptophan 2,3-Dioxygenase Protein in Colorectal Cancer. Dis. Markers 2016, 2016, 8169724. [Google Scholar] [CrossRef] [PubMed]
- Mei, J.; Li, M.Q.; Ding, D.; Li, D.J.; Jin, L.P.; Hu, W.G.; Zhu, X.Y. Indoleamine 2,3-dioxygenase-1 (IDO1) enhances survival and invasiveness of endometrial stromal cells via the activation of JNK signaling pathway. Int. J. Clin. Exp. Pathol. 2013, 6, 431–444. [Google Scholar] [PubMed]
- Inaba, T.; Ino, K.; Kajiyama, H.; Yamamoto, E.; Shibata, K.; Nawa, A.; Nagasaka, T.; Akimoto, H.; Takikawa, O.; Kikkawa, F. Role of the immunosuppressive enzyme indoleamine 2,3-dioxygenase in the progression of ovarian carcinoma. Gynecol. Oncol. 2009, 115, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.M.; Liu, J.; Liu, H.B.; Ye, M.; Zhang, Y.F.; Yang, D.S. Abnormal COX2 protein expression may be correlated with poor prognosis in oral cancer: A meta-analysis. BioMed Res. Int. 2014, 2014, 364207. [Google Scholar] [CrossRef] [PubMed]
- Frejborg, E.; Salo, T.; Salem, A. Role of Cyclooxygenase-2 in Head and Neck Tumorigenesis. Int. J. Mol. Sci. 2020, 21, 9246. [Google Scholar] [CrossRef] [PubMed]
- Polakova, I.; Pelak, O.; Thurner, D.; Pokryvkova, B.; Tachezy, R.; Kalina, T.; Smahel, M. Implementation of Mass Cytometry for Immunoprofiling of Patients with Solid Tumors. J. Immunol. Res. 2019, 2019, 6705949. [Google Scholar] [CrossRef]
- Koslabova, E.; Hamsikova, E.; Salakova, M.; Klozar, J.; Foltynova, E.; Salkova, E.; Rotnaglova, E.; Ludvikova, V.; Tachezy, R. Markers of HPV infection and survival in patients with head and neck tumors. Int. J. Cancer 2013, 133, 1832–1839. [Google Scholar] [CrossRef]
- Rotnáglová, E.; Tachezy, R.; Saláková, M.; Procházka, B.; Košl’abová, E.; Veselá, E.; Ludvíková, V.; Hamšíková, E.; Klozar, J. HPV involvement in tonsillar cancer: Prognostic significance and clinically relevant markers. Int. J. Cancer 2011, 129, 101–110. [Google Scholar] [CrossRef]
- Gao, G.; Chernock, R.D.; Gay, H.A.; Thorstad, W.L.; Zhang, T.R.; Wang, H.; Ma, X.J.; Luo, Y.; Lewis, J.S., Jr.; Wang, X. A novel RT-PCR method for quantification of human papillomavirus transcripts in archived tissues and its application in oropharyngeal cancer prognosis. Int. J. Cancer 2013, 132, 882–890. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Vojtechova, Z.; Sabol, I.; Salakova, M.; Turek, L.; Grega, M.; Smahelova, J.; Vencalek, O.; Lukesova, E.; Klozar, J.; Tachezy, R. Analysis of the integration of human papillomaviruses in head and neck tumours in relation to patients’ prognosis. Int. J. Cancer 2016, 138, 386–395. [Google Scholar] [CrossRef]
- Romano, A.; Parrinello, N.L.; Vetro, C.; Tibullo, D.; Giallongo, C.; La Cava, P.; Chiarenza, A.; Motta, G.; Caruso, A.L.; Villari, L.; et al. The prognostic value of the myeloid-mediated immunosuppression marker Arginase-1 in classic Hodgkin lymphoma. Oncotarget 2016, 7, 67333–67346. [Google Scholar] [CrossRef]
- Ren, W.; Zhang, X.; Li, W.; Feng, Q.; Feng, H.; Tong, Y.; Rong, H.; Wang, W.; Zhang, D.; Zhang, Z.; et al. Circulating and tumor-infiltrating arginase 1-expressing cells in gastric adenocarcinoma patients were mainly immature and monocytic Myeloid-derived suppressor cells. Sci. Rep. 2020, 10, 8056. [Google Scholar] [CrossRef] [PubMed]
- Heuvers, M.E.; Muskens, F.; Bezemer, K.; Lambers, M.; Dingemans, A.C.; Groen, H.J.M.; Smit, E.F.; Hoogsteden, H.C.; Hegmans, J.; Aerts, J. Arginase-1 mRNA expression correlates with myeloid-derived suppressor cell levels in peripheral blood of NSCLC patients. Lung Cancer 2013, 81, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Khaled, Y.S.; Ammori, B.J.; Elkord, E. Increased levels of granulocytic myeloid-derived suppressor cells in peripheral blood and tumour tissue of pancreatic cancer patients. J. Immunol. Res. 2014, 2014, 879897. [Google Scholar] [CrossRef]
- Pagès, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- El-Naggar, A.K.; Westra, W.H. p16 expression as a surrogate marker for HPV-related oropharyngeal carcinoma: A guide for interpretative relevance and consistency. Head Neck 2012, 34, 459–461. [Google Scholar] [CrossRef] [PubMed]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tan, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Partlova, S.; Boucek, J.; Kloudova, K.; Lukesova, E.; Zabrodsky, M.; Grega, M.; Fucikova, J.; Truxova, I.; Tachezy, R.; Spisek, R.; et al. Distinct patterns of intratumoral immune cell infiltrates in patients with HPV-associated compared to non-virally induced head and neck squamous cell carcinoma. Oncoimmunology 2015, 4, e965570. [Google Scholar] [CrossRef]
- Russell, S.; Angell, T.; Lechner, M.; Liebertz, D.; Correa, A.; Sinha, U.; Kokot, N.; Epstein, A. Immune cell infiltration patterns and survival in head and neck squamous cell carcinoma. Head Neck Oncol. 2013, 5, 24. [Google Scholar]
- Fang, J.; Li, X.; Ma, D.; Liu, X.; Chen, Y.; Wang, Y.; Lui, V.W.Y.; Xia, J.; Cheng, B.; Wang, Z. Prognostic significance of tumor infiltrating immune cells in oral squamous cell carcinoma. BMC Cancer 2017, 17, 375. [Google Scholar] [CrossRef]
- Oguejiofor, K.; Galletta-Williams, H.; Dovedi, S.J.; Roberts, D.L.; Stern, P.L.; West, C.M. Distinct patterns of infiltrating CD8+ T cells in HPV+ and CD68 macrophages in HPV- oropharyngeal squamous cell carcinomas are associated with better clinical outcome but PD-L1 expression is not prognostic. Oncotarget 2017, 8, 14416–14427. [Google Scholar] [CrossRef]
- Seminerio, I.; Kindt, N.; Descamps, G.; Bellier, J.; Lechien, J.R.; Mat, Q.; Pottier, C.; Journé, F.; Saussez, S. High infiltration of CD68+ macrophages is associated with poor prognoses of head and neck squamous cell carcinoma patients and is influenced by human papillomavirus. Oncotarget 2018, 9, 11046–11059. [Google Scholar] [CrossRef]
- Ni, Y.H.; Ding, L.; Huang, X.F.; Dong, Y.C.; Hu, Q.G.; Hou, Y.Y. Microlocalization of CD68+ tumor-associated macrophages in tumor stroma correlated with poor clinical outcomes in oral squamous cell carcinoma patients. Tumour Biol. 2015, 36, 5291–5298. [Google Scholar] [CrossRef]
- Ou, D.; Adam, J.; Garberis, I.; Blanchard, P.; Nguyen, F.; Levy, A.; Casiraghi, O.; Gorphe, P.; Breuskin, I.; Janot, F.; et al. Influence of tumor-associated macrophages and HLA class I expression according to HPV status in head and neck cancer patients receiving chemo/bioradiotherapy. Radiother. Oncol. 2019, 130, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Edin, S.; Wikberg, M.L.; Dahlin, A.M.; Rutegård, J.; Öberg, Å.; Oldenborg, P.A.; Palmqvist, R. The distribution of macrophages with a M1 or M2 phenotype in relation to prognosis and the molecular characteristics of colorectal cancer. PLoS ONE 2012, 7, e47045. [Google Scholar] [CrossRef]
- Krijgsman, D.; De Vries, N.L.; Andersen, M.N.; Skovbo, A.; Tollenaar, R.; Møller, H.J.; Hokland, M.; Kuppen, P.J.K. CD163 as a Biomarker in Colorectal Cancer: The Expression on Circulating Monocytes and Tumor-Associated Macrophages, and the Soluble Form in the Blood. Int. J. Mol. Sci. 2020, 21, 5925. [Google Scholar] [CrossRef]
- Huang, Y.K.; Wang, M.; Sun, Y.; Di Costanzo, N.; Mitchell, C.; Achuthan, A.; Hamilton, J.A.; Busuttil, R.A.; Boussioutas, A. Macrophage spatial heterogeneity in gastric cancer defined by multiplex immunohistochemistry. Nat. Commun. 2019, 10, 3928. [Google Scholar] [CrossRef]
- Hu, J.M.; Liu, K.; Liu, J.H.; Jiang, X.L.; Wang, X.L.; Chen, Y.Z.; Li, S.G.; Zou, H.; Pang, L.J.; Liu, C.X.; et al. CD163 as a marker of M2 macrophage, contribute to predicte aggressiveness and prognosis of Kazakh esophageal squamous cell carcinoma. Oncotarget 2017, 8, 21526–21538. [Google Scholar] [CrossRef] [PubMed]
- Balermpas, P.; Rödel, F.; Liberz, R.; Oppermann, J.; Wagenblast, J.; Ghanaati, S.; Harter, P.N.; Mittelbronn, M.; Weiss, C.; Rödel, C.; et al. Head and neck cancer relapse after chemoradiotherapy correlates with CD163+ macrophages in primary tumour and CD11b+ myeloid cells in recurrences. Br. J. Cancer 2014, 111, 1509–1518. [Google Scholar] [CrossRef]
- Mori, K.; Hiroi, M.; Shimada, J.; Ohmori, Y. Infiltration of m2 tumor-associated macrophages in oral squamous cell carcinoma correlates with tumor malignancy. Cancers 2011, 3, 3726–3739. [Google Scholar] [CrossRef]
- Ohashi, T.; Aoki, M.; Tomita, H.; Akazawa, T.; Sato, K.; Kuze, B.; Mizuta, K.; Hara, A.; Nagaoka, H.; Inoue, N.; et al. M2-like macrophage polarization in high lactic acid-producing head and neck cancer. Cancer Sci. 2017, 108, 1128–1134. [Google Scholar] [CrossRef]
- Li, L.; Wang, X.L.; Lei, Q.; Sun, C.Z.; Xi, Y.; Chen, R.; He, Y.W. Comprehensive immunogenomic landscape analysis of prognosis-related genes in head and neck cancer. Sci. Rep. 2020, 10, 6395. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, W.; Yang, S.; He, P.; Wang, J.; Gaedcke, J.; Ströbel, P.; Azizian, A.; Ried, T.; Gaida, M.M.; et al. NO(•) /RUNX3/kynurenine metabolic signaling enhances disease aggressiveness in pancreatic cancer. Int. J. Cancer 2020, 146, 3160–3169. [Google Scholar] [CrossRef] [PubMed]
- Soliman, H.; Rawal, B.; Fulp, J.; Lee, J.H.; Lopez, A.; Bui, M.M.; Khalil, F.; Antonia, S.; Yfantis, H.G.; Lee, D.H.; et al. Analysis of indoleamine 2-3 dioxygenase (IDO1) expression in breast cancer tissue by immunohistochemistry. Cancer Immunol. Immunother. 2013, 62, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Hurt, B.; Schulick, R.; Edil, B.; El Kasmi, K.C.; Barnett, C., Jr. Cancer-promoting mechanisms of tumor-associated neutrophils. Am. J. Surg. 2017, 214, 938–944. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Lian, J.; Yang, M.; Wuyang, J.; Zhao, C.; Chen, W.; Liu, C.; Zhao, Q.; Lou, C.; Han, J.; et al. Overexpression of Arginase-1 is an indicator of poor prognosis in patients with colorectal cancer. Pathol. Res. Pract. 2019, 215, 152383. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Ghosh, S.K. Modulation of L-Arginine-Arginase Metabolic Pathway Enzymes: Immunocytochemistry and mRNA Expression in Peripheral Blood and Tissue Levels in Head and Neck Squamous Cell Carcinomas in North East India. Asian Pac. J. Cancer Prev. 2015, 16, 7031–7038. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Gene | Sequence 5′–3′ | Length [bp] | |
|---|---|---|---|
| IDO1 | F | AAGAAACTGGAACTGCCTCCT | 121 |
| R | CACGAAATGAGAACAAAACGTCC | ||
| ARG1 | F | GGCAGAAGTCAAGAAGAACGGA | 127 |
| R | GTGAGCATCCACCCAGATGA | ||
| CD163 | F | GCAATGGGGTGGACTTACCT | 120 |
| R | TGCTTCACTTCAACACGTCC | ||
| NOS2 | F | GCTGTGCTCCATAGTTTCCAG | 137 |
| R | GGGACCAGCCAAATCCAGTC | ||
| PTGS2 | F | GCATTCTTTGCCCAGCACTT | 142 |
| R | GGCGCAGTTTACGCTGTCT | ||
| GUS | F | GAAAATATGTGGTTGGAGAGCTCATT | 101 |
| R | CCGAGTGAAGATCCCCTTTTTA | ||
| ACTB | F | CCACGAAACTACCTTCAACTCCA | 132 |
| R | GTGATCTCCTTCTGCATCCTGTC |
| # | AR | Antibody | Dilution | Incubation | Secondary | OPAL | Dilution |
|---|---|---|---|---|---|---|---|
| 1 | 6 | CD68 | 1:200 | 1 h/RT | Ms + Rb Opal HRP polymer | 540 | 1:200 |
| 2 | 6 | CD163 | 1:100 | OVN/4 °C | 620 | 1:100 | |
| 3 | 9 | ARG1 | 1:200 | 1 h/RT | 650 | 1:200 | |
| 4 | 9 | CD80 | 1:50 | OVN/4 °C | 520 | 1:50 | |
| 5 | 6 | Cytokeratin | 1:800 | 1 h/RT | 690 | 1:200 | |
| 6 | 6 | DAPI | 1:15 | 5 min/RT | – | – | – |
| Characteristics | Total | HPV+ | HPV− | |
|---|---|---|---|---|
| No. (%) | No. (%) | No. (%) | ||
| No. of patients | 73 (100%) | 38 (52%) | 35 (48%) | |
| Age (years) | Mean age | 61.86 | 61.66 | 62.14 |
| Median age | 61.50 | 60.00 | 62.50 | |
| Gender | female | 16 (22%) | 3 (8%) | 13 (37%) |
| male | 57 (78%) | 35 (92%) | 22 (63%) | |
| Localization | oropharynx | 50 (68%) | 38 (100%) | 12 (34%) |
| oral cavity | 23 (32%) | 0 (0%) | 23 (66%) | |
| Education | >12 years | 31 (50%) | 19 (54%) | 12 (44%) |
| ≤12 years | 31 (50%) | 16 (46%) | 15 (56%) | |
| Smoking | never | 24 (33%) | 19 (50%) | 5 (14%) |
| past | 23 (31%) | 12 (32%) | 11 (32%) | |
| current | 26 (36%) | 7 (18%) | 19 (54%) | |
| Alcohol consumption | never | 20 (27%) | 14 (37%) | 6 (17%) |
| past | 11 (15%) | 2 (5%) | 9 (26%) | |
| current | 42 (58%) | 22 (58%) | 20 (57%) | |
| Tumor extension (pT) | T1 | 17 (23%) | 7 (18%) | 10 (29%) |
| T2 | 48 (66%) | 29 (76%) | 19 (54%) | |
| T3 | 4 (5.5%) | 1 (3%) | 3 (8.5%) | |
| T4 | 4 (5.5%) | 1 (3%) | 3 (8.5%) | |
| Nodal status (pN) | N0 | 30 (41%) | 10 (29%) | 20 (57%) |
| N1 | 32 (44%) | 26 (68%) | 6 (17%) | |
| N2 | 5 (7%) | 1 (1.5%) | 4 (12%) | |
| N3 | 6 (8%) | 1 (1.5%) | 5 (14%) | |
| Metastasis presence (M) | 0 | 69 (95%) | 38 (100%) | 31(89%) |
| 1 | 4 (5%) | 0 (0%) | 4 (11%) | |
| Tumor stage (S) | I | 40 (55%) | 32 (84%) | 8 (23%) |
| II | 12 (16%) | 3 (8%) | 9 (26%) | |
| III | 6 (8%) | 2 (5%) | 4 (11%) | |
| IV | 15 (21%) | 1 (3%) | 14 (40%) | |
| Extracapsular spreading | 0 | 57 (78%) | 28 (74%) | 29 (83%) |
| 1 | 16 (22%) | 10 (26%) | 6 (17%) | |
| Charlson comorbidity index | 0 | 2 (3%) | 2 (5%) | 0 (0%) |
| 1 | 19 (26%) | 13 (34%) | 6 (17%) | |
| 2 | 16 (22%) | 7 (18.5%) | 9 (26%) | |
| 3 | 16 (22%) | 7 (18.5%) | 9 (26%) | |
| 4 | 10 (13%) | 6 (16%) | 4 (11%) | |
| 5 | 5 (7%) | 1 (3%) | 4 (11%) | |
| 6 | 2 (3%) | 0 (0%) | 2 (6%) | |
| 7 | 3 (4%) | 2 (5%) | 1 (3%) | |
| HPV+ | HPV− | All Patients | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | Parenchyma | Stroma | p | Parenchyma | Stroma | p | Parenchyma | Stroma | p |
| M1 | 1.635 | 29.68 | <0.0001 | 4.693 | 22.3 | 0.0001 | 2.361 | 26.9 | <0.0001 |
| M2 | 0.982 | 17.6 | <0.0001 | 0.472 | 11.87 | <0.0001 | 0.789 | 12.8 | <0.0001 |
| M2-ARG | 0 | 0 | 0.0347 | 0.255 | 2.442 | <0.0001 | 0.203 | 1.007 | <0.0001 |
| ARG | 1.462 | 6.314 | <0.0001 | 15.1 | 51.12 | <0.0001 | 4.023 | 18.13 | <0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pokrývková, B.; Šmahelová, J.; Dalewská, N.; Grega, M.; Vencálek, O.; Šmahel, M.; Nunvář, J.; Klozar, J.; Tachezy, R. ARG1 mRNA Level Is a Promising Prognostic Marker in Head and Neck Squamous Cell Carcinomas. Diagnostics 2021, 11, 628. https://doi.org/10.3390/diagnostics11040628
Pokrývková B, Šmahelová J, Dalewská N, Grega M, Vencálek O, Šmahel M, Nunvář J, Klozar J, Tachezy R. ARG1 mRNA Level Is a Promising Prognostic Marker in Head and Neck Squamous Cell Carcinomas. Diagnostics. 2021; 11(4):628. https://doi.org/10.3390/diagnostics11040628
Chicago/Turabian StylePokrývková, Barbora, Jana Šmahelová, Natálie Dalewská, Marek Grega, Ondřej Vencálek, Michal Šmahel, Jaroslav Nunvář, Jan Klozar, and Ruth Tachezy. 2021. "ARG1 mRNA Level Is a Promising Prognostic Marker in Head and Neck Squamous Cell Carcinomas" Diagnostics 11, no. 4: 628. https://doi.org/10.3390/diagnostics11040628
APA StylePokrývková, B., Šmahelová, J., Dalewská, N., Grega, M., Vencálek, O., Šmahel, M., Nunvář, J., Klozar, J., & Tachezy, R. (2021). ARG1 mRNA Level Is a Promising Prognostic Marker in Head and Neck Squamous Cell Carcinomas. Diagnostics, 11(4), 628. https://doi.org/10.3390/diagnostics11040628






