Isolation of Extracellular Vesicles from Biological Fluids via the Aggregation–Precipitation Approach for Downstream miRNAs Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemical and Biochemical Reagents
2.2. Sample Collection
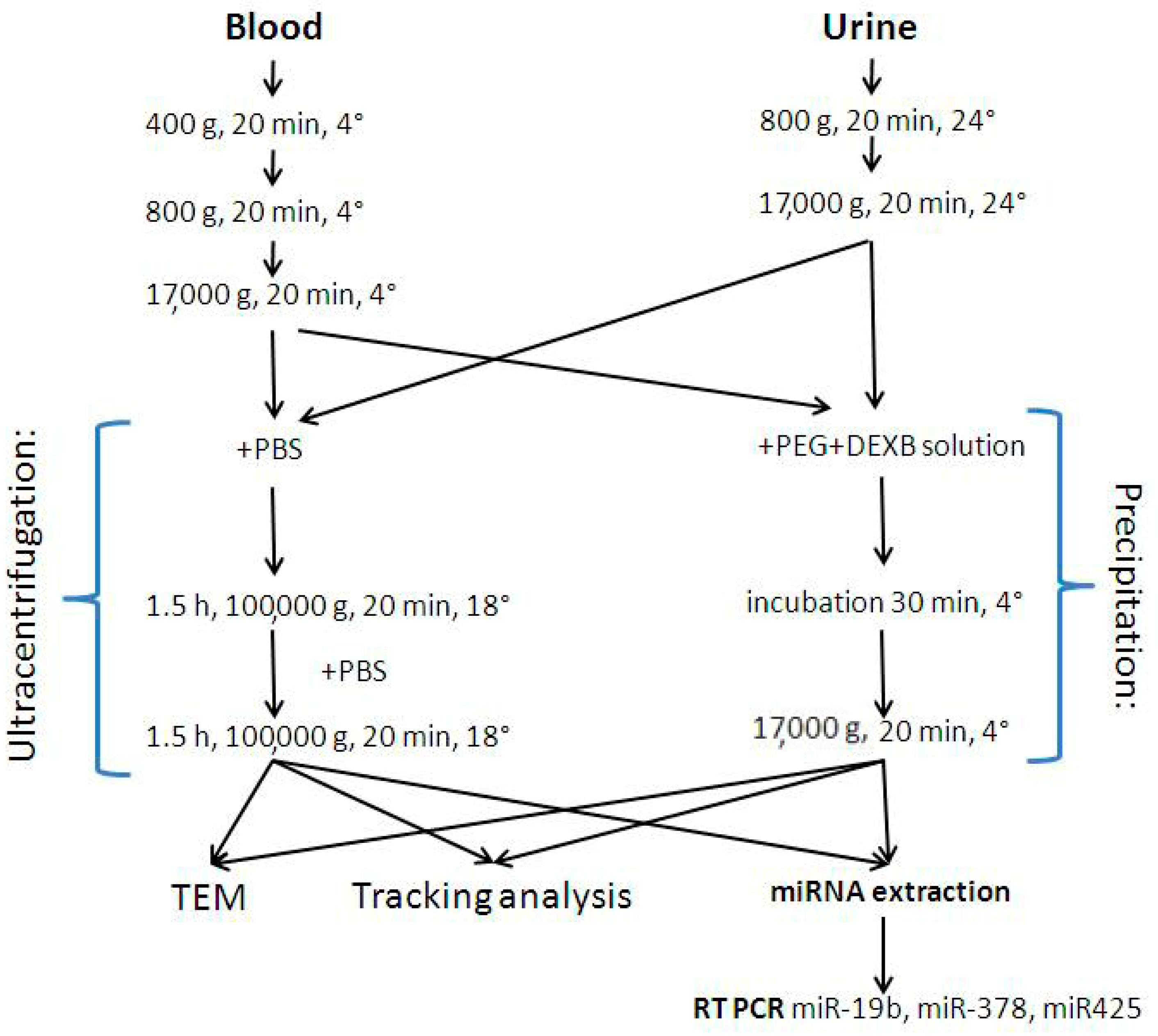
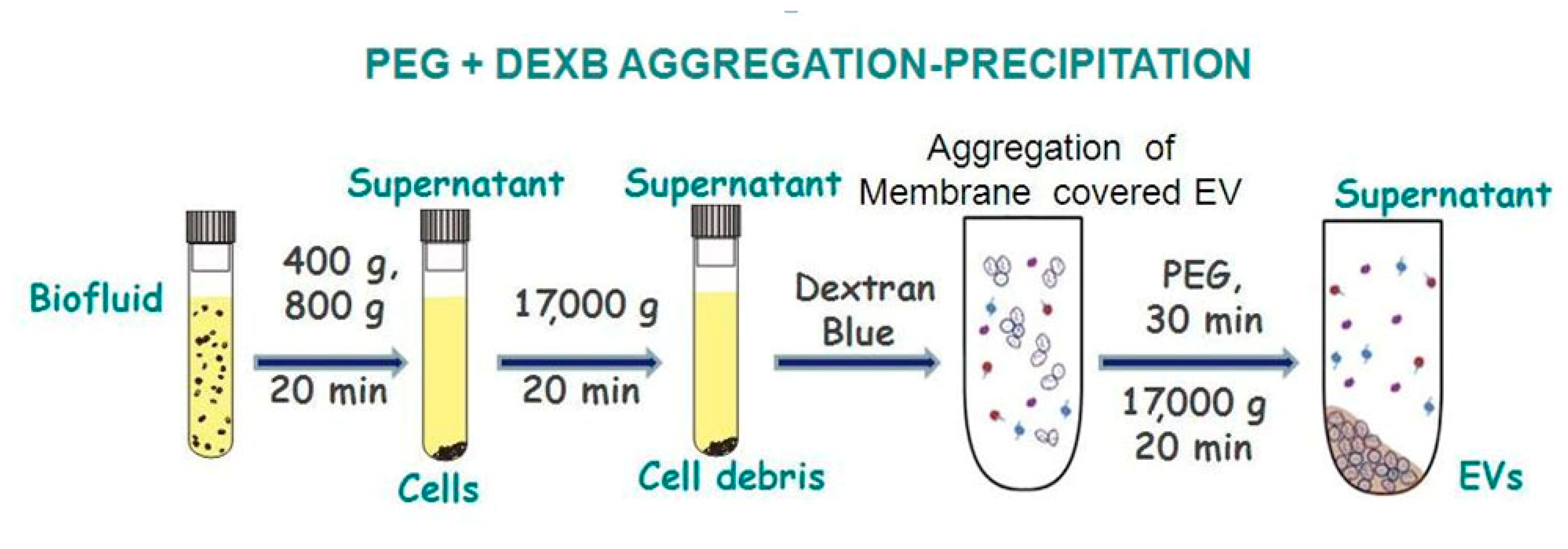
2.3. EV Enrichment Procedures
2.3.1. Ultracentrifugation
2.3.2. EV precipitation
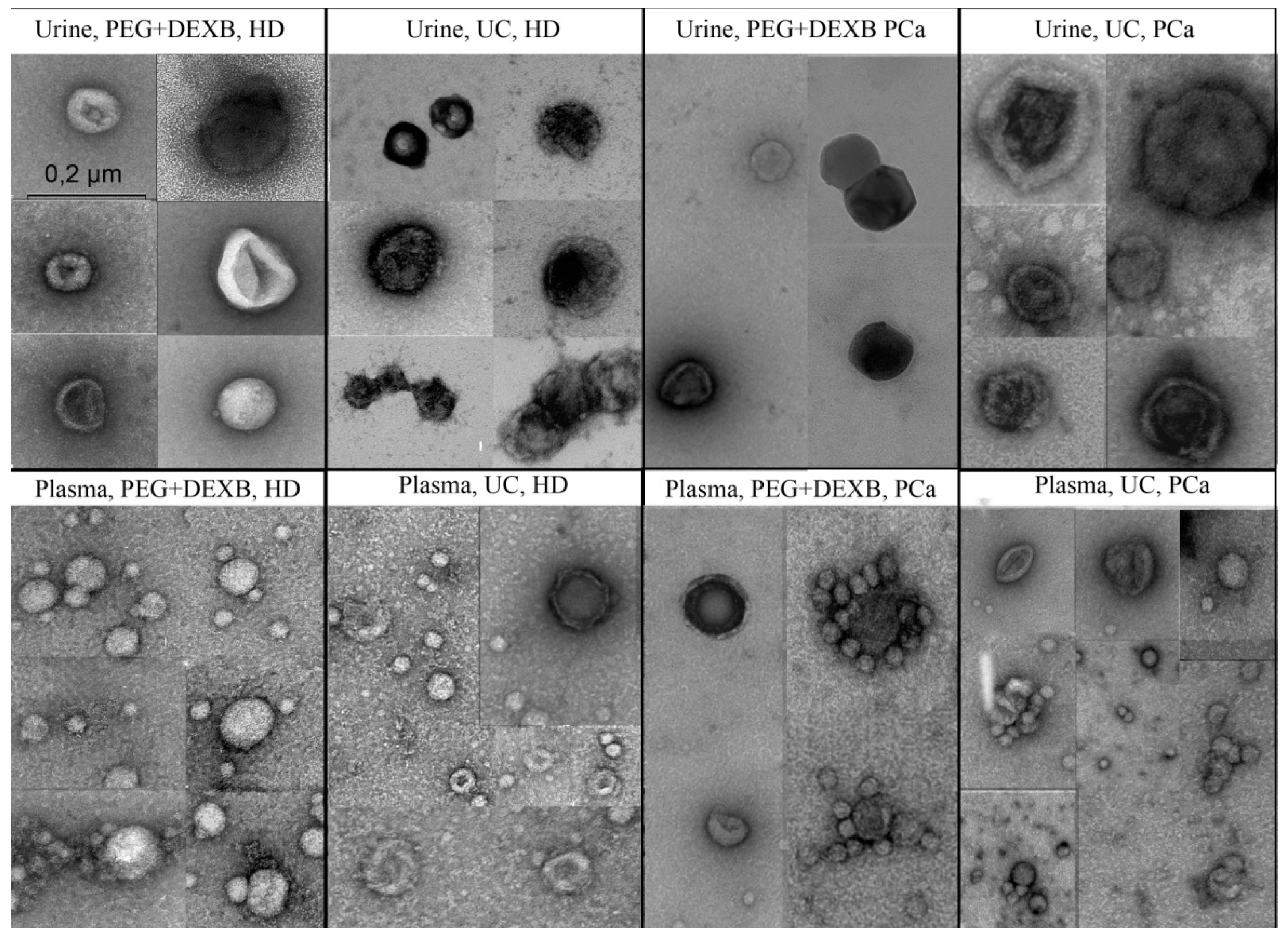
2.4. Transmission Electron Microscopy
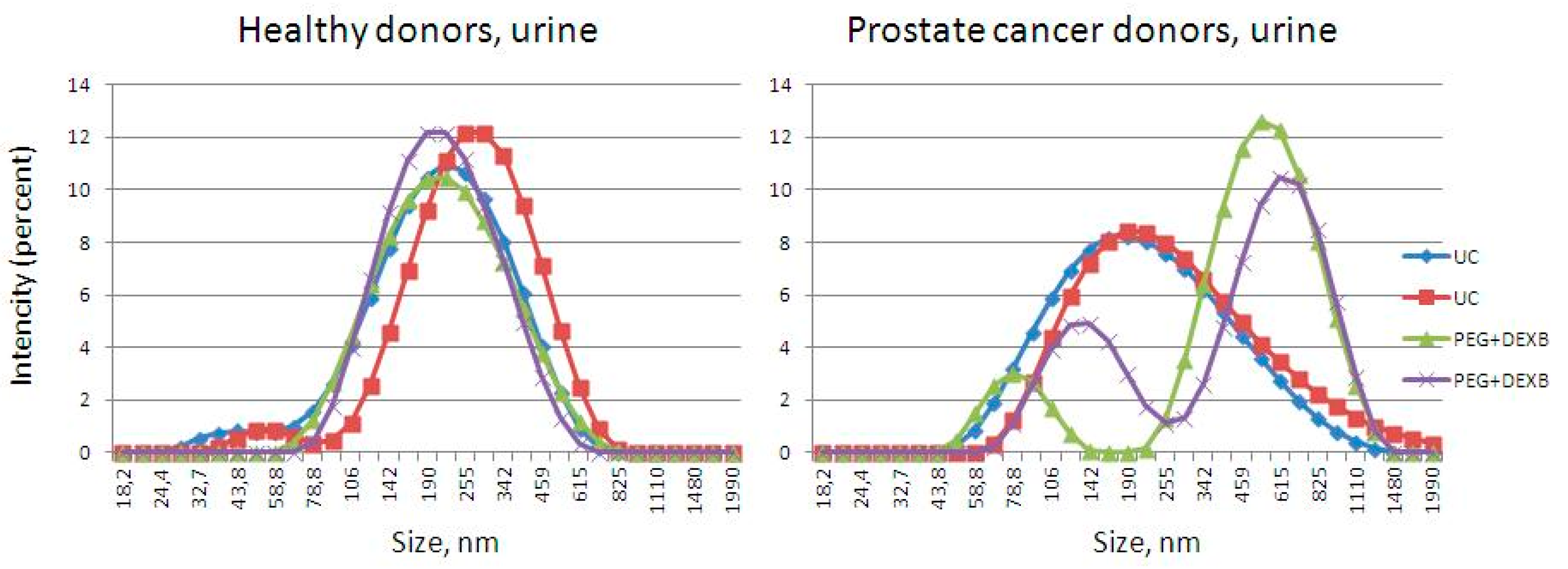
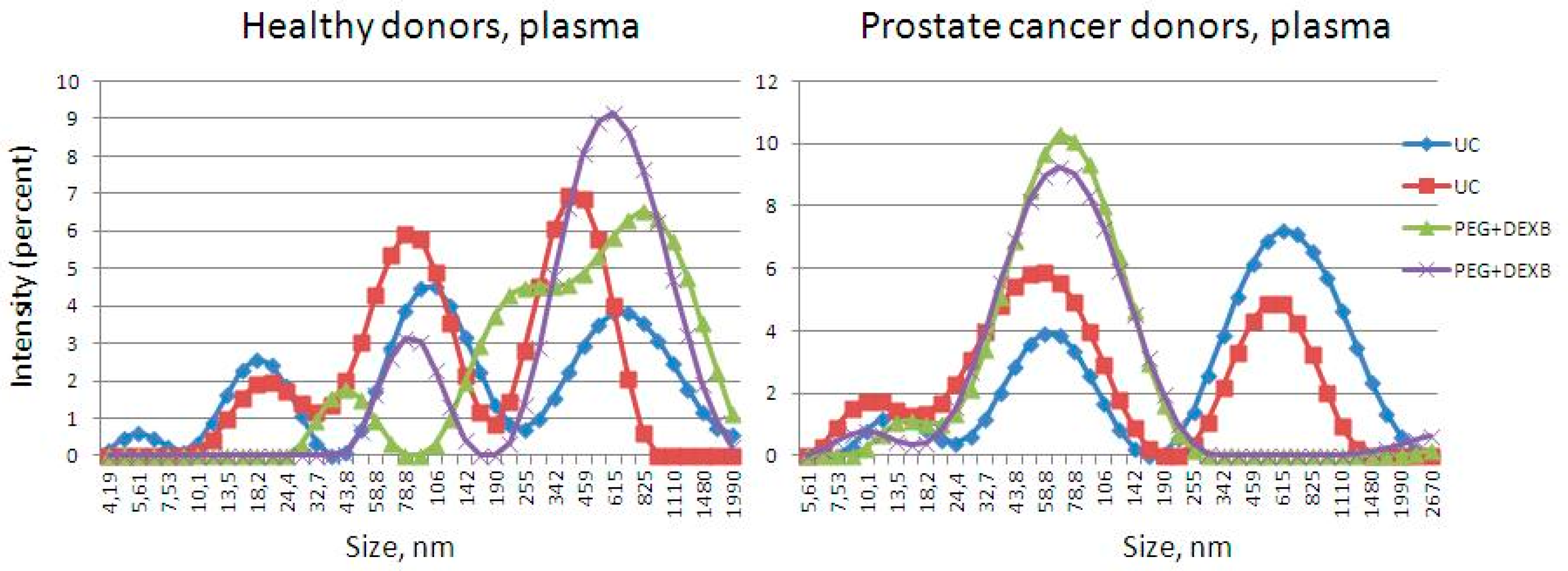
2.5. Characterization via Dynamic Light Scattering
2.6. miRNA Isolation by the Gu/OcA Protocol
2.7. RNA Precipitation
2.8. ReverseTranscription and Quantitative RT-PCR
2.9. Statistical Analysis
3. Results
3.1. Description of Extracellular Vesicles
3.2. Differential Efficiency of Isolation Methods for the Detection of miRNAs by RT-qPCR
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Denzer, K.; Kleijmeer, M.J.; Heijnen, H.F.; Stoorvoxgel, W.; Geuze, H.J. Exosome: From internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 2000, 113, 3365–3374. [Google Scholar]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. 2016, 6, 287–296. [Google Scholar] [CrossRef]
- Gould, S.J.; Raposo, G. As we wait: Coping with an imperfect nomenclature for extracellular vesicles. J. Extracell. Vesicles 2013, 15, 2. [Google Scholar] [CrossRef]
- Kourembanas, S. Exosomes: Vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu. Rev. Physiol. 2015, 77, 13–27. [Google Scholar] [CrossRef]
- Gonda, D.D.; Akers, J.C.; Kim, R.; Kalkanis, S.N.; Hochberg, F.H.; Chen, C.C.; Carter, B.S. Neuro-oncologic applications of exosomes, microvesicles, and other nano-sized extracellular particles. Neurosurgery 2013, 72, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Iraci, N.; Leonardi, T.; Gessler, F.; Vega, B.; Pluchino, S. Focuson Extracellular Vesicles: Physiological Role and Signaling Properties of Extracellular Membrane Vesicles. Int. J. Mol. Sci. 2016, 17, 171. [Google Scholar] [CrossRef]
- Bryzgunova, O.E.; Zaripov, M.M.; Skvortsova, T.E.; Lekchnov, E.A.; Grigor’eva, A.E.; Zaporozhchenko, I.A.; Morozkin, E.S.; Ryabchikova, E.I.; Yurchenko, Y.B.; Voitsitskiy, V.E.; et al. Comparative Study of Extracellular Vesicles from the Urine of Healthy Individuals and Prostate Cancer Patients. PLoS ONE 2016, 11, e0157566. [Google Scholar]
- Tamkovich, S.N.; Tutanov, O.S.; Laktionov, P.P. Exosomes: Generation, structure, transport, biological activity, and diagnostic application. Biol. Membr. 2016, 33, 163–175. (In Russian) [Google Scholar] [CrossRef]
- Campos, J.H.; Soares, R.P.; Ribeiro, K.; Andrade, A.C.; Batista, W.L.; Torrecilhas, A.C. Extracellular Vesicles: Role in inflammatory responses and potential uses in vaccination in cancer and infectious diseases. J. Immunol. Res. 2015, 2015, 832057. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Shen, Q.; Yang, X.; Qiu, Y.; Zhang, W. The Role of Extracellular Vesicles: An Epigenetic View of the Cancer Microenvironment. Biomed. Res. Int. 2015, 2015, 649161. [Google Scholar] [CrossRef] [PubMed]
- Urabe, F.; Kosaka, N.; Yoshioka, Y.; Egawa, S.; Ochiya, T. The small vesicular culprits: The investigation of extracellular vesicles as new targets for cancer treatment. Clin. Transl. Med. 2017, 6, 45. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, Y.; Katsuda, T.; Ochiya, T. Extracellular vesicles and encapsulated miRNAs as emerging cancer biomarkers for novel liquid biopsy. Jpn. J. Clin. Oncol. 2018, 48, 869–876. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Lekchnov, E.A.; Bryzgunova, O.E.; Zaporozhchenko, I.A.; Yarmoschuk, S.V.; Pashkovskaya, O.A.; Pak, S.V.; Laktionov, P.P. The Panel of 12 Cell-Free MicroRNAs as Potential Biomarkers in Prostate Neoplasms. Diagnostics 2020, 10, 38. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Bryzgunova, O.E.; Lekchnov, E.A.; Amelina, E.V.; Yarmoschuk, S.V.; Pak, S.V.; Laktionov, P.P. The Influence of Radical Prostatectomy on the Expression of Cell-Free MiRNA. Diagnostics 2020, 10, 600. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Wienholds, E.; Plasterk, R.H. MicroRNA function in animal development. FEBS Lett. 2005, 579, 5911–5922. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Luu, H.N.; Lin, H.Y.; Sørensen, K.D.; Ogunwobi, O.O.; Kumar, N.; Chornokur, G.; Phelan, C.; Jones, D.; Kidd, L.; Batra, J.; et al. miRNAs associated with prostate cancer risk and progression. BMC Urol. 2017, 17, 18. [Google Scholar] [CrossRef] [PubMed]
- Bryzgunova, O.E.; Konoshenko, M.Y.; Laktionov, P.P. MicroRNA-guided gene expression in prostate cancer: Literature and database overview. J. Gene Med. 2018, 20, e3016. [Google Scholar] [CrossRef]
- Fredsøe, J.; Rasmussen, A.K.I.; Thomsen, A.R.; Mouritzen, P.; Høyer, S.; Borre, M. Diagnostic and Prognostic MicroRNA Biomarkers for Prostate Cancerin Cell-free Urine. Eur. Urol. Focus 2018, 4, 825–833. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed. Res. Int. 2018, 2018, 8545347. [Google Scholar] [CrossRef]
- Gámez-Valero, A.; Monguió-Tortajada, M.; Carreras-Planella, L.; Franquesa, M.L.; Beyer, K.; Borràs, F.E. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci. Rep. 2016, 6, 33641. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Lekchnov, E.A.; Bryzgunova, O.E.; Laktionov, P.P. The Method for Isolation of Extracellular Vesicles from Biofluids. Patent 2678988, 5 February 2019. [Google Scholar]
- Lekchnov, E.A.; Zaporozhchenko, I.A.; Morozkin, E.S.; Bryzgunova, O.E.; Vlassov, V.V.; Laktionov, P.P. Protocol for miRNA isolation from biofluids. Anal. Biochem. 2016, 499, 78–84. [Google Scholar] [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 26, 789–804. [Google Scholar] [CrossRef]
- Wachalska, M.; Koppers-Lalic, D.; van Eijndhoven, M.; Pegtel, M.; Geldof, A.A.; Lipinska, A.D.; van Moorselaar, R.; Bijnsdorp, I.V. Protein Complexes in Urine Interfere with Extracellular Vesicle Biomarker Studies. J.Circ. Biomark. 2016, 5, 4. [Google Scholar] [CrossRef]
- Nakai, W.; Yoshida, T.; Diez, D.; Miyatake, Y.; Nishibu, T.; Imawaka, N.; Naruse, K.; Sadamura, Y.; Hanayama, R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci. Rep. 2016, 6, 33935. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, D.; Kirchner, B.; Hermann, S.; Märte, M.; Wurmser, C.; Brandes, F.; Kotschote, S.; Bonin, M.; Steinlein, O.K.; Pfaffl, M.W.; et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles 2018, 7, 1481321. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.X.F.; Johansson, H.; Wood, M.J.A.; El Andaloussi, S. Considerations and Implications in the Purification of Extracellular Vesicles—A Cautionary Tale. Front. Neurosci. 2019, 13, 1067. [Google Scholar] [CrossRef] [PubMed]
- Nolte-’t Hoen, E.N.; Buermans, H.P.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.; ’tHoen, P.A. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef]
- Baglio, S.R.; Rooijers, K.; Koppers-Lalic, D.; Verweij, F.J.; Pérez Lanzón, M.; Zini, N.; Naaijkens, B.; Perut, F.; Niessen, H.W.; Baldini, N.; et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. 2015, 6, 127. [Google Scholar] [CrossRef]
- Andreu, Z.; Rivas, E.; Sanguino-Pascual, A.; Lamana, A.; Marazuela, M.; Gonzalez-Alvaro, I.; Sanchez-Madrid, F.; delaFuente, H.; Yanez-Mo, M. Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. J. Extracell. Vesicles 2016, 5, 31655. [Google Scholar] [CrossRef]
- VanDeun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; DeWever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3, 24858. [Google Scholar] [CrossRef]
- Greenfield, E.A. Polyethylene Glycol Fusion for Hybridoma Production. Cold Spring Harb. Protoc. 2018, 3, pdb.prot103176. [Google Scholar] [CrossRef]
- Leary, H.L.; Halsey, J.F. An assay to measure antigen-specific immune complexes in food-allergy patients. J. Allergy Clin. Immunol. 1984, 74, 190–195. [Google Scholar] [CrossRef]
- Mooney, N.A.; Hay, F.C.; Poulton, T.A. A comparative study of complement components in polyethylene glycol precipitated immune complexes from patients with ovarian cancer and patients with rheumatoid arthritis. Clin. Exp. Immunol. 1983, 52, 561–568. [Google Scholar]
- Ray, U.; Roy, S.S. Aberrant lipid metabolism in cancer cells—The role of oncolipid-activated signaling. FEBSJ. 2018, 285, 432–443. [Google Scholar] [CrossRef]
- Parrales, A.; Iwakuma, T. p53 as a Regulator of Lipid Metabolism in Cancer. Int. J. Mol. Sci. 2016, 17, 2074. [Google Scholar] [CrossRef] [PubMed]
- Foj, L.; Ferrer, F.; Serra, M.; Arévalo, A.; Gavagnach, M.; Giménez, N.; Filella, X. Exosomal and Non-Exosomal Urinary miRNAs in Prostate Cancer Detectionand Prognosis. Prostate 2017, 77, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Balacescu, O.; Petrut, B.; Tudoran, O.; Feflea, D.; Balacescu, L.; Anghel, A.; Sirbu, I.O.; Seclaman, E.; Marian, C. Urinary microRNAs for prostate cancer diagnosis, prognosis, and treatment response: Are we there yet? Wiley Interdiscip. Rev. RNA 2017, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Duong, P.; Chung, A.; Bouchareychas, L.; Raffai, R.L. Cushioned-Density Gradient Ultracentrifugation (C-DGUC) improves the isolation efficiency of extracellular vesicles. PLoS ONE 2019, 14, e0215324. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Skog, J.; Nordstrand, A.; Baranov, V.; Mincheva-Nilsson, L.; Breakefield, X.O.; Widmark, A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Welton, J.L.; Khanna, S.; Giles, P.J.; Brennan, P.; Brewis, I.A.; Staffurth, J.; Mason, M.D.; Clayton, A. Proteomics analysis of bladder cancer exosomes. Mol. Cell. Proteom. 2010, 9, 1324–1338. [Google Scholar] [CrossRef]
- Wood, S.L.; Knowles, M.A.; Thompson, D.; Selby, P.J.; Banks, R.E. Proteomic studies of urinary biomarkers for prostate, bladder and kidney cancers. Nat. Rev. Urol. 2013, 4, 206–218. [Google Scholar] [CrossRef]
- Fernández-Llama, P.; Khositseth, S.; Gonzales, P.A.; Star, R.A.; Pisitkun, T.; Knepper, M.A. Tamm-Horsfall protein and urinary exosome isolation. Kidney Int. 2010, 77, 736–742. [Google Scholar] [CrossRef]
- Alvarez, M.L.; Khosroheidari, M.; Ravi, R.K.; DiStefano, J.K. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012, 82, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Musante, L.; Tataruch, D.; Gu, D.; Benito-Martin, A.; Calzaferri, G.; Aherne, S.; Holthofer, H. A simplified method to recover urinary vesicles for clinical applications, and sample banking. Sci. Rep. 2014, 4, 7532. [Google Scholar] [CrossRef] [PubMed]
- Daveloose, D. Isolation and Characterization of Urinary Exosomes in Metastatic Castration—Refractory Prostate Cancer. Master’s Thesis, French National Centre for Scientific Research, Paris, France, 2016. [Google Scholar]
- Livshits, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 2015, 5, 17319. [Google Scholar] [CrossRef]
- Zeringer, E.; Barta, T.; Li, M.; Vlassov, A.V. Strategies for isolation of exosomes. Cold Spring Harb. Protoc. 2015, 4, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Deregibus, M.C.; Figliolini, F.; D’Antico, S.; Manzini, P.M.; Pasquino, C.; DeLena, M.; Tetta, C.; Brizzi, M.F.; Camussi, G. Charge-based precipitation of extracellular vesicles. Int. J. Mol. Med. 2016, 38, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, Z.; Lynn, K.D.; Thorpe, P.E.; Schroit, A.J. Anovel “salting-out” procedure for the isolation of tumor-derived exosomes. J. Immunol. Methods 2014, 407, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Zacharias, W.; Gercel-Taylor, C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol 2011, 728, 235–246. [Google Scholar] [PubMed]
- Foroni, C.; Zarovni, N.; Bianciardi, L.; Bernardi, S.; Triggiani, L.; Zocco, D.; Venturella, M.; Chiesi, A.; Valcamonico, F.; Berruti, A. When Less Is More: Specific Capture and Analysis of Tumor Exosomes in Plasma Increases the Sensitivity of Liquid Biopsy for Comprehensive Detection of Multiple Androgen Receptor Phenotypes in Advanced Prostate Cancer Patients. Biomedicines 2020, 22, 131. [Google Scholar] [CrossRef]


| miRNA | Label | Sequence |
|---|---|---|
| Universal Reverse Primer | 5′-GTGCAGGGTCCGAGGT-3′ | |
| hsa-miR-19b-3p | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAGTT-3′ |
| (miR-19b) | Forward | 5′-CGCTGTGCAAATCCATGCAA-3′ |
| Probe | 5′-(FAM)-GCACTGGATACGACTCAGTT-(FQ)-3′ | |
| hsa-miR-378a-3p | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCCTTC-3′ |
| (miR-378) | Forward | 5′-GACTGGACTTGGAGTCA-3′ |
| Probe | 5′-(FAM)-CGCACTGGATACGACAAAGTC-(BHQ)-3′ | |
| hsa-miR-425-5p | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAACG-3′ |
| (miR-425) | Forward | 5′-TAATGACACGATCACTCC-3′ |
| Probe | 5′-(FAM)-CGCACTGGATACGACTCAACG-(BHQ)-3′ | |
| cel-miR-39-3p | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAAGCT-3′ |
| (miR-39) | Forward | 5′-ATTCACCGGGTGTAAATC-3′ |
| Probe | 5′-(FAM)-CACTGGATACGACCAAGCTGA-(BHQ)-3′ |
| HD | nm | 31–250 | 31–50 | 51–100 | 101–150 | 151–200 | 201–250 |
|---|---|---|---|---|---|---|---|
| PlasmaUC | % | 71 | 82 | 16 | 2 | 0 | 0 |
| median, nm | 45 | 41 | 55 | 133 | |||
| PlasmaPEG+DEXB | % | 86 | 67 | 32 | 1 | 0 | 0 |
| median, nm | 47 | 45 | 56 | 119 | |||
| UrineUC | % | 100 | 5 | 67 | 28 | 0 | 0 |
| median, nm | 89 | 43 | 80 | 119 | |||
| UrinePEG+DEXB | % | 100 | 11 | 65 | 18 | 4 | 2 |
| median, nm | 70 | 41 | 68 | 117 | 174 | 240 |
| PCa | Range, nm | 31–250 | 31–50 | 51–100 | 101–150 | 151–200 | 201–250 |
|---|---|---|---|---|---|---|---|
| PlasmaUC | % | 53 | 91 | 7 | 2 | 0 | 0 |
| median, nm | 40 | 39 | 56 | 109 | |||
| Plasma PEG+DEXB | % | 56 | 86 | 14 | 0 | 0 | 0 |
| median, nm | 40 | 38 | 59 | ||||
| UrineUC | % | 96 | 13 | 47 | 23 | 10 | 7 |
| median, nm | 93 | 47 | 81 | 126 | 178 | 222 | |
| Urine PEG+DEXB | % | 86 | 9 | 64 | 21 | 5 | 1 |
| median, nm | 78 | 34 | 71 | 124 | 171 | 211 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konoshenko, M.Y.; Lekchnov, E.A.; Bryzgunova, O.E.; Kiseleva, E.; Pyshnaya, I.A.; Laktionov, P.P. Isolation of Extracellular Vesicles from Biological Fluids via the Aggregation–Precipitation Approach for Downstream miRNAs Detection. Diagnostics 2021, 11, 384. https://doi.org/10.3390/diagnostics11030384
Konoshenko MY, Lekchnov EA, Bryzgunova OE, Kiseleva E, Pyshnaya IA, Laktionov PP. Isolation of Extracellular Vesicles from Biological Fluids via the Aggregation–Precipitation Approach for Downstream miRNAs Detection. Diagnostics. 2021; 11(3):384. https://doi.org/10.3390/diagnostics11030384
Chicago/Turabian StyleKonoshenko, Maria Y., Evgeniy A. Lekchnov, Olga E. Bryzgunova, Elena Kiseleva, Inna A. Pyshnaya, and Pavel P. Laktionov. 2021. "Isolation of Extracellular Vesicles from Biological Fluids via the Aggregation–Precipitation Approach for Downstream miRNAs Detection" Diagnostics 11, no. 3: 384. https://doi.org/10.3390/diagnostics11030384
APA StyleKonoshenko, M. Y., Lekchnov, E. A., Bryzgunova, O. E., Kiseleva, E., Pyshnaya, I. A., & Laktionov, P. P. (2021). Isolation of Extracellular Vesicles from Biological Fluids via the Aggregation–Precipitation Approach for Downstream miRNAs Detection. Diagnostics, 11(3), 384. https://doi.org/10.3390/diagnostics11030384







