Expression of Immune Checkpoints in Malignant Tumors: Therapy Targets and Biomarkers for the Gastric Cancer Prognosis
Abstract
:1. Introduction
2. Immune Checkpoint Expression in Connection with Clinical Features and the Therapeutic Efficacy of Their Inhibition
2.1. PD-L1 (B7-H1)
2.2. B7-H3 (CD276)
2.3. B7-H4 (VTCN1)
2.4. Galectin-3
2.5. Galectin-9
2.6. IDO1
2.7. CEACAM1
2.8. CD155
2.9. Siglec-15
2.10. ADAM17
3. Immune Checkpoints as Biomarkers of GC
3.1. PD-L1
3.2. B7-H3
3.3. B7-H4
3.4. Galectin-3
3.5. Galectin-9
3.6. IDO1
3.7. CEACAM1,CD155 and Siglec-15
3.8. ADAM17
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Doroshow, D.B.; Bhalla, S.; Beasley, M.B.; Sholl, L.M.; Kerr, K.M.; Gnjatic, S.; Wistuba, I.I.; Rimm, D.L.; Tsao, M.S.; Hirsch, F.R. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2021, 18, 345–362. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.P.; Kurzrock, R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol. Cancer Ther. 2015, 14, 847–856. [Google Scholar] [CrossRef] [Green Version]
- Bolandi, N.; Derakhshani, A.; Hemmat, N.; Baghbanzadeh, A.; Asadzadeh, Z.; Afrashteh Nour, M.; Brunetti, O.; Bernardini, R.; Silvestris, N.; Baradaran, B. The Positive and Negative Immunoregulatory Role of B7 Family: Promising Novel Targets in Gastric Cancer Treatment. Int. J. Mol. Sci. 2021, 22, 10719. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, X.; Jin, T.; Tian, Y.; Dai, C.; Widarma, C.; Song, R.; Xu, F. Immune checkpoint molecules in natural killer cells as potential targets for cancer immunotherapy. Signal Transduct. Target. Ther. 2020, 5, 250. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, J. Functions of Immune Checkpoint Molecules Beyond Immune Evasion. Adv. Exp. Med. Biol. 2020, 1248, 201–226. [Google Scholar] [CrossRef]
- Qin, S.; Xu, L.; Yi, M.; Yu, S.; Wu, K.; Luo, S. Novel immune checkpoint targets: Moving beyond PD-1 and CTLA-4. Mol. Cancer 2019, 18, 155. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Etemadi, A.; Safiri, S.; Sepanlou, S.G.; Ikuta, K.; Bisignano, C.; Shakeri, R.; Amani, M.; Fitzmaurice, C.; Nixon, M.; Abbasi, N.; et al. The global, regional, and national burden of stomach cancer in 195 countries, 1990–2017: A systematic analysis for the Global Burden of Disease study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 42–54. [Google Scholar] [CrossRef] [Green Version]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and Its Ligands in Tolerance and Immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Chen, Z.; Li, Y.; Zhao, W.; Wu, J.B.; Zhang, Z. PD-1/PD-L1 Checkpoint Inhibitors in Tumor Immunotherapy. Front. Pharmacol. 2021, 12, 731798. [Google Scholar] [CrossRef] [PubMed]
- Francisco, L.M.; Sage, P.T.; Sharpe, A.H. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev. 2010, 236, 219–242. [Google Scholar] [CrossRef] [PubMed]
- Hirano, F.; Kaneko, K.; Tamura, H.; Dong, H.; Wang, S.; Ichikawa, M.; Rietz, C.; Flies, D.B.; Lau, J.S.; Zhu, G.; et al. Blockade of B7-H1 and PD-1 by monoclonal antibodies potentiates cancer therapeutic immunity. Cancer Res. 2005, 65, 1089–1096. [Google Scholar]
- Böger, C.; Behrens, H.-M.; Mathiak, M.; Krüger, S.; Kalthoff, H.; Röcken, C. PD-L1 is an independent prognostic predictor in gastric cancer of Western patients. Oncotarget 2016, 7, 24269–24283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.; Wu, D.; Li, L.; Chai, Y.; Huang, J. PD-L1 and Survival in Solid Tumors: A Meta-Analysis. PLoS ONE 2015, 10, e0131403. [Google Scholar] [CrossRef]
- Xiang, X.; Yu, P.-C.; Long, D.; Liao, X.-L.; Zhang, S.; You, X.-M.; Zhong, J.-H.; Li, L.-Q. Prognostic value of PD-L1 expression in patients with primary solid tumors. Oncotarget 2018, 9, 5058–5072. [Google Scholar] [CrossRef] [Green Version]
- Dai, C.; Wang, M.; Lu, J.; Dai, Z.; Lin, S.; Yang, P.; Tian, T.; Liu, X.; Min, W.; Dai, Z. Prognostic and predictive values of PD-L1 expression in patients with digestive system cancer: A meta-analysis. Onco. Targets. Ther. 2017, 10, 3625–3634. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Z.; Du, Y. Clinicopathological and prognostic significance of programmed death ligant-1 expression in gastric cancer: A meta-analysis. J. Gastrointest. Oncol. 2021, 12, 112–120. [Google Scholar] [CrossRef]
- Gu, L.; Chen, M.; Guo, D.; Zhu, H.; Zhang, W.; Pan, J.; Zhong, X.; Li, X.; Qian, H.; Wang, X. PD-L1 and gastric cancer prognosis: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0182692. [Google Scholar] [CrossRef]
- Zhang, M.; Dong, Y.; Liu, H.; Wang, Y.; Zhao, S.; Xuan, Q.; Wang, Y.; Zhang, Q. The clinicopathological and prognostic significance of PD-L1 expression in gastric cancer: A meta-analysis of 10 studies with 1,901 patients. Sci. Rep. 2016, 6, 37933. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.-S.; Wang, X.-S.; Wang, Y.-F.; Hu, X.-C.; Yan, J.-Q.; Wang, W.; Yang, R.-J.; Feng, Y.-Y.; Gao, S.-G.; Liu, Y.-X.; et al. Prognostic significance of PD-L1 expression in patients with gastric cancer in East Asia: A meta-analysis. Onco. Targets. Ther. 2016, 9, 2649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.; Feng, G.; Zhao, H.; Liu, F.; Xu, L.; Wang, Q.; An, G. Clinicopathologic Significance and Prognostic Value of B7 Homolog 1 in Gastric Cancer: A Systematic Review and Meta-Analysis. Medicine 2015, 94, e1911. [Google Scholar] [CrossRef]
- Kim, D.H.; Bae, G.E.; Suh, K.S.; Ryuman, D.; Song, K.S.; Kim, J.S.; Lee, S.-I.; Yeo, M.-K. Clinical Significance of Tumor and Immune Cell PD-L1 Expression in Gastric Adenocarcinoma. In Vivo 2020, 34, 3171–3180. [Google Scholar] [CrossRef] [PubMed]
- Kawazoe, A.; Kuwata, T.; Kuboki, Y.; Shitara, K.; Nagatsuma, A.K.; Aizawa, M.; Yoshino, T.; Doi, T.; Ohtsu, A.; Ochiai, A. Clinicopathological features of programmed death ligand 1 expression with tumor-infiltrating lymphocyte, mismatch repair, and Epstein-Barr virus status in a large cohort of gastric cancer patients. Gastric Cancer 2017, 20, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Hu, W.; Zhu, Y.; Wu, Y.; Lin, H. The Prognostic Value of Circulating Soluble Programmed Death Ligand-1 in Cancers: A Meta-Analysis. Front. Oncol. 2021, 10, 626932. [Google Scholar] [CrossRef] [PubMed]
- Shigemori, T.; Toiyama, Y.; Okugawa, Y.; Yamamoto, A.; Yin, C.; Narumi, A.; Ichikawa, T.; Ide, S.; Shimura, T.; Fujikawa, H.; et al. Soluble PD-L1 Expression in Circulation as a Predictive Marker for Recurrence and Prognosis in Gastric Cancer: Direct Comparison of the Clinical Burden Between Tissue and Serum PD-L1 Expression. Ann. Surg. Oncol. 2019, 26, 876–883. [Google Scholar] [CrossRef]
- Ito, S.; Fukagawa, T.; Noda, M.; Hu, Q.; Nambara, S.; Shimizu, D.; Kuroda, Y.; Eguchi, H.; Masuda, T.; Sato, T.; et al. Prognostic Impact of Immune-Related Gene Expression in Preoperative Peripheral Blood from Gastric Cancer Patients. Ann. Surg. Oncol. 2018, 25, 3755–3763. [Google Scholar] [CrossRef]
- Roviello, G.; Corona, S.P.; D’Angelo, A.; Rosellini, P.; Nobili, S.; Mini, E. Immune Checkpoint Inhibitors in Pre-Treated Gastric Cancer Patients: Results from a Literature-Based Meta-Analysis. Int. J. Mol. Sci. 2020, 21, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapoval, A.I.; Ni, J.; Lau, J.S.; Wilcox, R.A.; Flies, D.B.; Liu, D.; Dong, H.; Sica, G.L.; Zhu, G.; Tamada, K.; et al. B7-H3: A costimulatory molecule for T cell activation and IFN-gamma production. Nat. Immunol. 2001, 2, 269–274. [Google Scholar] [CrossRef]
- Hashiguchi, M.; Kobori, H.; Ritprajak, P.; Kamimura, Y.; Kozono, H.; Azuma, M. Triggering receptor expressed on myeloid cell-like transcript 2 (TLT-2) is a counter-receptor for B7-H3 and enhances T cell responses. Proc. Natl. Acad. Sci. USA 2008, 105, 10495–10500. [Google Scholar] [CrossRef] [Green Version]
- Suh, W.-K.; Gajewska, B.U.; Okada, H.; Gronski, M.A.; Bertram, E.M.; Dawicki, W.; Duncan, G.S.; Bukczynski, J.; Plyte, S.; Elia, A.; et al. The B7 family member B7-H3 preferentially down-regulates T helper type 1–mediated immune responses. Nat. Immunol. 2003, 4, 899–906. [Google Scholar] [CrossRef]
- Prasad, D.V.R.; Nguyen, T.; Li, Z.; Yang, Y.; Duong, J.; Wang, Y.; Dong, C. Murine B7-H3 Is a Negative Regulator of T Cells. J. Immunol. 2004, 173, 2500–2506. [Google Scholar] [CrossRef] [Green Version]
- Tekle, C.; Nygren, M.K.; Chen, Y.-W.; Dybsjord, I.; Nesland, J.M.; Maelandsmo, G.M.; Fodstad, O. B7-H3 contributes to the metastatic capacity of melanoma cells by modulation of known metastasis-associated genes. Int. J. cancer 2012, 130, 2282–2290. [Google Scholar] [CrossRef]
- Dai, W.; Shen, G.; Qiu, J.; Zhao, X.; Gao, Q. Aberrant expression of B7-H3 in gastric adenocarcinoma promotes cancer cell metastasis. Oncol. Rep. 2014, 32, 2086–2092. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Yang, X.; Wu, Y.; Zhao, K.; Ye, Z.; Zhu, J.; Xu, X.; Zhao, X.; Xing, C. B7-H3 promotes gastric cancer cell migration and invasion. Oncotarget 2017, 8, 71725–71735. [Google Scholar] [CrossRef] [Green Version]
- Ulase, D.; Behrens, H.-M.; Krüger, S.; Zeissig, S.; Röcken, C. Gastric Carcinomas with Stromal B7-H3 Expression Have Lower Intratumoural CD8+ T Cell Density. Int. J. Mol. Sci. 2021, 22, 2129. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, X.; Yao, P.; Shen, W.; Wu, Y.; Ye, Z.; Zhao, K.; Chen, H.; Cao, J.; Xing, C. B7-H3 increases the radioresistance of gastric cancer cells through regulating baseline levels of cell autophagy. Am. J. Transl. Res. 2019, 11, 4438–4449. [Google Scholar]
- Ye, Z.; Zheng, Z.; Li, X.; Zhu, Y.; Zhong, Z.; Peng, L.; Wu, Y. B7-H3 Overexpression Predicts Poor Survival of Cancer Patients: A Meta-Analysis. Cell. Physiol. Biochem. 2016, 39, 1568–1580. [Google Scholar] [CrossRef]
- Li, G.; Quan, Y.; Che, F.; Wang, L. B7-H3 in tumors: Friend or foe for tumor immunity? Cancer Chemother. Pharmacol. 2018, 81, 245–253. [Google Scholar] [CrossRef]
- Zhang, X.; Fang, C.; Zhang, G.; Jiang, F.; Wang, L.; Hou, J. Prognostic value of B7-H3 expression in patients with solid tumors: A meta-analysis. Oncotarget 2017, 8, 93156–93167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, S.; Liu, Z.; Zhang, M.; Guo, T.; Quan, Q.; Huang, L.; Guo, L.; Cao, L.; Zhang, X. Overexpression of B7-H3 in α-SMA-Positive Fibroblasts Is Associated with Cancer Progression and Survival in Gastric Adenocarcinomas. Front. Oncol. 2020, 9, 1466. [Google Scholar] [CrossRef] [PubMed]
- Arigami, T.; Uenosono, Y.; Hirata, M.; Yanagita, S.; Ishigami, S.; Natsugoe, S. B7-H3 expression in gastric cancer: A novel molecular blood marker for detecting circulating tumor cells. Cancer Sci. 2011, 102, 1019–1024. [Google Scholar] [CrossRef]
- Wu, C.-P.; Jiang, J.-T.; Tan, M.; Zhu, Y.-B.; Ji, M.; Xu, K.-F.; Zhao, J.-M.; Zhang, G.-B.; Zhang, X.-G. Relationship between co-stimulatory molecule B7-H3 expression and gastric carcinoma histology and prognosis. World J. Gastroenterol. 2006, 12, 457. [Google Scholar] [CrossRef]
- Loo, D.; Alderson, R.F.; Chen, F.Z.; Huang, L.; Zhang, W.; Gorlatov, S.; Burke, S.; Ciccarone, V.; Li, H.; Yang, Y.; et al. Development of an Fc-enhanced anti-B7-H3 monoclonal antibody with potent antitumor activity. Clin. Cancer Res. 2012, 18, 3834–3845. [Google Scholar] [CrossRef] [Green Version]
- Safety Study of MGA271 in Refractory Cancer. Available online: https://clinicaltrials.gov/ct2/show/NCT01391143 (accessed on 18 November 2021).
- Safety Study of Enoblituzumab (MGA271) in Combination with Pembrolizumab or MGA012 in Refractory Cancer. Available online: https://clinicaltrials.gov/ct2/show/NCT02475213 (accessed on 18 November 2021).
- Enoblituzumab Plus Retifanlimab or Tebotelimab in Head and Neck Cancer. Available online: https://clinicaltrials.gov/ct2/show/NCT04634825 (accessed on 18 November 2021).
- Aggarwal, C.; Joshua, A.; Ferris, R.; Antonia, S.; Rahma, O.; Tolcher, A.; Cohen, R.B.; Lou, Y.; Hauke, R.; Vogelzang, N.; et al. A phase 1, open-label, dose-escalation study of enoblituzumab in combination with pembrolizumab in patients with select solid tumors. J. Immunother. Cancer 2018, 6 (Suppl. 2), 114, abstract P24. [Google Scholar]
- Shenderov, E.; Demarzo, A.; Boudadi, K.; Allaf, M.; Wang, H.; Chapman, C.; Pavlovich, C.; Bivalacqua, T.; O’Neal, T.S.; Harb, R.; et al. Phase II neoadjuvant and immunologic study of B7-H3 targeting with enoblituzumab in localized intermediate- and high-risk prostate cancer. J. Clin. Oncol. 2018, 36, TPS5099. [Google Scholar] [CrossRef]
- Ohaegbulam, K.C.; Liu, W.; Jeon, H.; Almo, S.C.; Zang, X. Tumor-expressed immune checkpoint B7x promotes cancer progression and antigen-specific CD8 T cell exhaustion and suppressive innate immune cells. Oncotarget 2017, 8, 82740–82753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podojil, J.R.; Miller, S.D. Potential targeting of B7-H4 for the treatment of cancer. Immunol. Rev. 2017, 276, 40–51. [Google Scholar] [CrossRef]
- Choi, I.-H.; Zhu, G.; Sica, G.L.; Strome, S.E.; Cheville, J.C.; Lau, J.S.; Zhu, Y.; Flies, D.B.; Tamada, K.; Chen, L. Genomic Organization and Expression Analysis of B7-H4, an Immune Inhibitory Molecule of the B7 Family. J. Immunol. 2003, 171, 4650–4654. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, N.; Gavrieli, M.; Sedy, J.R.; Yang, J.; Fallarino, F.; Loftin, S.K.; Hurchla, M.A.; Zimmerman, N.; Sim, J.; Zang, X.; et al. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat. Immunol. 2003, 4, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Zang, X.; Loke, P.; Kim, J.; Murphy, K.; Waitz, R.; Allison, J.P. B7x: A widely expressed B7 family member that inhibits T cell activation. Proc. Natl. Acad. Sci. USA 2003, 100, 10388–10392. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.Y.; Wang, W.P. B7-H4, a promising target for immunotherapy. Cell. Immunol. 2020, 347, 104008. [Google Scholar] [CrossRef] [PubMed]
- Sica, G.L.; Choi, I.H.; Zhu, G.; Tamada, K.; Wang, S.D.; Tamura, H.; Chapoval, A.I.; Flies, D.B.; Bajorath, J.; Chen, L. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity 2003, 18, 849–861. [Google Scholar] [CrossRef] [Green Version]
- Ni, L.; Dong, C. New B7 Family Checkpoints in Human Cancers. Mol. Cancer Ther. 2017, 16, 1203–1211. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Wang, X.; Mo, N.; Zhang, L.; Yuan, X.; Lü, Z. B7-Homolog 4 Promotes Epithelial-Mesenchymal Transition and Invasion of Bladder Cancer Cells via Activation of Nuclear Factor-κB. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2018, 26, 1267–1274. [Google Scholar] [CrossRef]
- Chen, C.; Zhu, W.-D.; Xie, F.; Huang, J.-A. Nuclear localization of B7-H4 in pulmonary adenocarcinomas presenting as a solitary pulmonary nodule. Oncotarget 2016, 7, 58563–58568. [Google Scholar] [CrossRef] [Green Version]
- Shan, Z.; Yan, Z.; Peng, L.; Cheng, P.; Teng, Y.; Mao, F.; Fan, K.; Zhuang, Y.; Zhao, Y. Granulocyte-Macrophage Colony-Stimulating Factor-Activated Neutrophils Express B7-H4 That Correlates with Gastric Cancer Progression and Poor Patient Survival. J. Immunol. Res. 2021, 2021, 6613247. [Google Scholar] [CrossRef]
- Song, X.; Shao, Y.; Gu, W.; Xu, C.; Mao, H.; Pei, H.; Jiang, J. Prognostic role of high B7-H4 expression in patients with solid tumors: A meta-analysis. Oncotarget 2016, 7, 76523–76533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Li, Z. B7-H4 is Predictive of Poor Prognosis in Patients with Gastric Cancer. Med. Sci. Monit. 2016, 22, 4233–4237. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Zhu, Y.; Wu, C.; Shen, Y.; Wei, W.; Chen, L.; Zheng, X.; Sun, J.; Lu, B.; Zhang, X. Tumor expression of B7-H4 predicts poor survival of patients suffering from gastric cancer. Cancer Immunol. Immunother. 2010, 59, 1707–1714. [Google Scholar] [CrossRef]
- Arigami, T.; Uenosono, Y.; Ishigami, S.; Hagihara, T.; Haraguchi, N.; Natsugoe, S. Clinical significance of the B7-H4 coregulatory molecule as a novel prognostic marker in gastric cancer. World J. Surg. 2011, 35, 2051–2057. [Google Scholar] [CrossRef]
- Geng, Y.; Wang, H.; Lu, C.; Li, Q.; Xu, B.; Jiang, J.; Wu, C. Expression of costimulatory molecules B7-H1, B7-H4 and Foxp3+ Tregs in gastric cancer and its clinical significance. Int. J. Clin. Oncol. 2015, 20, 273–281. [Google Scholar] [CrossRef]
- Maskey, N.; Li, K.; Hu, M.; Xu, Z.; Peng, C.; Yu, F.; Cao, H.; Chen, J.; Li, Y.; Yang, G. Impact of neoadjuvant chemotherapy on lymphocytes and co-inhibitory B7-H4 molecule in gastric cancer: Low B7-H4 expression associates with favorable prognosis. Tumor Biol. 2014, 35, 11837–11843. [Google Scholar] [CrossRef]
- Arigami, T.; Uenosono, Y.; Hirata, M.; Hagihara, T.; Yanagita, S.; Ishigami, S.; Natsugoe, S. Expression of B7-H4 in blood of patients with gastric cancer predicts tumor progression and prognosis. J. Surg. Oncol. 2010, 102, 748–752. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ji, M.; Wu, J.; Zhou, Q.; Li, X.; Li, Z.; Zheng, X.; Xu, B.; Zhao, W.; Wu, C.; et al. Serum B7-H4 expression is a significant prognostic indicator for patients with gastric cancer. World J. Surg. Oncol. 2014, 12, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sachdev, J.C.; Bauer, T.M.; Chawla, S.P.; Pant, S.; Patnaik, A.; Wainberg, Z.A.; Inamdar, S.P.; Marina, N.; Sun, S.; Schmidt, M.; et al. Phase 1a/1b study of first-in-class B7-H4 antibody, FPA150, as monotherapy in patients with advanced solid tumors. J. Clin. Oncol. 2019, 37, 2529. [Google Scholar] [CrossRef]
- Yang, R.-Y.; Rabinovich, G.A.; Liu, F.-T. Galectins: Structure, function and therapeutic potential. Expert Rev. Mol. Med. 2008, 10, e17. [Google Scholar] [CrossRef] [PubMed]
- Sciacchitano, S.; Lavra, L.; Morgante, A.; Ulivieri, A.; Magi, F.; De Francesco, G.; Bellotti, C.; Salehi, L.; Ricci, A. Galectin-3: One Molecule for an Alphabet of Diseases, from A to Z. Int. J. Mol. Sci. 2018, 19, 379. [Google Scholar] [CrossRef] [Green Version]
- Okada, K.; Shimura, T.; Suehiro, T.; Mochiki, E.; Kuwano, H. Reduced galectin-3 expression is an indicator of unfavorable prognosis in gastric cancer. Anticancer Res. 2006, 26, 1369–1376. [Google Scholar]
- Dong, R.; Zhang, M.; Hu, Q.; Zheng, S.; Soh, A.; Zheng, Y.; Yuan, H. Galectin-3 as a novel biomarker for disease diagnosis and a target for therapy (Review). Int. J. Mol. Med. 2017, 41, 599–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newlaczyl, A.U.; Yu, L.-G. Galectin-3—A jack-of-all-trades in cancer. Cancer Lett. 2011, 313, 123–128. [Google Scholar] [CrossRef]
- de Oliveira, J.T.; de Matos, A.J.; Gomes, J.; Vilanova, M.; Hespanhol, V.; Manninen, A.; Rutteman, G.; Chammas, R.; Gärtner, F.; Bernardes, E.S. Coordinated expression of galectin-3 and galectin-3-binding sites in malignant mammary tumors: Implications for tumor metastasis. Glycobiology 2010, 20, 1341–1352. [Google Scholar] [CrossRef] [Green Version]
- Chocarro, L.; Blanco, E.; Zuazo, M.; Arasanz, H.; Bocanegra, A.; Fernández-Rubio, L.; Morente, P.; Fernández-Hinojal, G.; Echaide, M.; Garnica, M.; et al. Understanding LAG-3 Signaling. Int. J. Mol. Sci. 2021, 22, 5282. [Google Scholar] [CrossRef]
- Farhad, M.; Rolig, A.S.; Redmond, W.L. The role of Galectin-3 in modulating tumor growth and immunosuppression within the tumor microenvironment. Oncoimmunology 2018, 7, e1434467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Compagno, D.; Tiraboschi, C.; Garcia, J.D.; Rondón, Y.; Corapi, E.; Velazquez, C.; Laderach, D.J. Galectins as Checkpoints of the Immune System in Cancers, Their Clinical Relevance, and Implication in Clinical Trials. Biomolecules 2020, 10, 750. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, S.; Raz, A. Regulation of cancer-related gene expression by galectin-3 and the molecular mechanism of its nuclear import pathway. Cancer Metastasis Rev. 2007, 26, 605–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elad-Sfadia, G.; Haklai, R.; Balan, E.; Kloog, Y. Galectin-3 Augments K-Ras Activation and Triggers a Ras Signal That Attenuates ERK but Not Phosphoinositide 3-Kinase Activity. J. Biol. Chem. 2004, 279, 34922–34930. [Google Scholar] [CrossRef] [Green Version]
- Thijssen, V.L.; Heusschen, R.; Caers, J.; Griffioen, A.W. Galectin expression in cancer diagnosis and prognosis: A systematic review. Biochim. Biophys. Acta-Rev. Cancer 2015, 1855, 235–247. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Tian, Y.; Wang, Y.; Zhang, Q.; Zhou, X.; Meng, X.; Song, N. Prognostic role of galectin-3 expression in patients with solid tumors: A meta-analysis of 36 eligible studies. Cancer Cell Int. 2018, 18, 172. [Google Scholar] [CrossRef]
- Wang, C.; Zhou, X.; Ma, L.; Zhuang, Y.; Wei, Y.; Zhang, L.; Jin, S.; Liang, W.; Shen, X.; Li, C.; et al. Galectin-3 may serve as a marker for poor prognosis in colorectal cancer: A meta-analysis. Pathol.-Res. Pract. 2019, 215, 152612. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; He, J.; Chen, Z.; Wu, C. Prognostic role of galectins expression in patients with hepatic cancer. Medicine 2020, 99, e19622. [Google Scholar] [CrossRef]
- Long, B.; Yu, Z.; Zhou, H.; Ma, Z.; Ren, Y.; Zhan, H.; Li, L.; Cao, H.; Jiao, Z. Clinical characteristics and prognostic significance of galectins for patients with gastric cancer: A meta-analysis. Int. J. Surg. 2018, 56, 242–249. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, D.C.; Kim, M.C.; Jung, G.J.; Kim, K.H.; Jang, J.S.; Kwon, H.C.; Kim, Y.M.; Jeong, J.S. Fascin expression is related to poor survival in gastric cancer. Pathol. Int. 2012, 62, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.-G.; Yu, Q.-F.; Xu, Y.; Fan, L.-F. Li-cadherin is Inversely Correlated with Galectin-3 Expression in Gastric Cancer. Dig. Dis. Sci. 2008, 53, 1811–1817. [Google Scholar] [CrossRef]
- Miyazaki, J.; Hokari, R.; Kato, S.; Tsuzuki, Y.; Kawaguchi, A.; Nagao, S.; Itoh, K.; Miura, S. Increased expression of galectin-3 in primary gastric cancer and the metastatic lymph nodes. Oncol. Rep. 2002, 9, 1307–1312. [Google Scholar] [CrossRef] [PubMed]
- Li, Y. Serum Galectin-3 as a Potential Marker for Gastric Cancer. Med. Sci. Monit. 2015, 21, 755–760. [Google Scholar] [CrossRef] [Green Version]
- Chou, F.-C.; Chen, H.-Y.; Kuo, C.-C.; Sytwu, H.-K. Role of Galectins in Tumors and in Clinical Immunotherapy. Int. J. Mol. Sci. 2018, 19, 430. [Google Scholar] [CrossRef] [Green Version]
- Heusschen, R.; Griffioen, A.W.; Thijssen, V.L. Galectin-9 in tumor biology: A jack of multiple trades. Biochim. Biophys. Acta-Rev. Cancer 2013, 1836, 177–185. [Google Scholar] [CrossRef]
- Moar, P.; Tandon, R. Galectin-9 as a biomarker of disease severity. Cell. Immunol. 2021, 361, 104287. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Sun, L.; Li, C.-F.; Wang, Y.-H.; Yao, J.; Li, H.; Yan, M.; Chang, W.-C.; Hsu, J.-M.; Cha, J.-H.; et al. Galectin-9 interacts with PD-1 and TIM-3 to regulate T cell death and is a target for cancer immunotherapy. Nat. Commun. 2021, 12, 832. [Google Scholar] [CrossRef]
- Oomizu, S.; Arikawa, T.; Niki, T.; Kadowaki, T.; Ueno, M.; Nishi, N.; Yamauchi, A.; Hattori, T.; Masaki, T.; Hirashima, M. Cell surface galectin-9 expressing Th cells regulate Th17 and Foxp3+ Treg development by galectin-9 secretion. PLoS ONE 2012, 7, e48574. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhao, E.; Zhang, Z.; Zhao, G.; Cao, H. Association between Tim-3 and Gal-9 expression and gastric cancer prognosis. Oncol. Rep. 2018, 40, 2115–2126. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-H.; Park, Y.; Kim, J.-H.; Kang, K.-W.; Lee, S.-J.; Kim, S.J.; Kim, B.S. Prognostic Value of Galectin-9 Relates to Programmed Death-Ligand 1 in Patients with Multiple Myeloma. Front. Oncol. 2021, 11, 669817. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Z.; Fu, Q.; Wang, Z.; Fu, H.; Liu, W.; Wang, Y.; Xu, J. Galectin-9 as a prognostic and predictive biomarker in bladder urothelial carcinoma. Urol. Oncol. Semin. Orig. Investig. 2017, 35, 349–355. [Google Scholar] [CrossRef]
- Chen, P.; He, Y.; Zhou, C. P47.13 Galectin-9, A Novel Prognostic Factor in Small Cell Lung Cancer. J. Thorac. Oncol. 2021, 16, S498. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, L.; Jing, D.; Xu, G.; Zhang, J.; Lin, L.; Zhao, J.; Yao, Z.; Lin, H. Galectin-9 Expression Predicts Favorable Clinical Outcome in Solid Tumors: A Systematic Review and Meta-Analysis. Front. Physiol. 2018, 9, 452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Chen, Z.; Wu, R.; Yin, J.; Fan, M.; Xu, X. Prognostic Role of High Gal-9 Expression in Solid Tumours: A Meta-Analysis. Cell. Physiol. Biochem. 2018, 45, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Jin, M.-S.; Kong, F.; Cao, D.; Ma, H.-X.; Jia, Z.; Wang, Y.-P.; Suo, J.; Cao, X. Decreased galectin-9 and increased Tim-3 expression are related to poor prognosis in gastric cancer. PLoS ONE 2013, 8, e81799. [Google Scholar] [CrossRef]
- Choi, S.I.; Seo, K.W.; Kook, M.C.; Kim, C.G.; Kim, Y.W.; Cho, S.J. Prognostic value of tumoral expression of galectin-9 in gastric cancer. Turkish J. Gastroenterol. 2017, 28, 166–170. [Google Scholar] [CrossRef] [Green Version]
- Schulz, H.; Kuhn, C.; Hofmann, S.; Mayr, D.; Mahner, S.; Jeschke, U.; Schmoeckel, E. Overall survival of ovarian cancer patients is determined by expression of galectins-8 and -9. Int. J. Mol. Sci. 2018, 19, 323. [Google Scholar] [CrossRef] [Green Version]
- Ye, Z.; Yue, L.; Shi, J.; Shao, M.; Wu, T. Role of IDO and TDO in Cancers and Related Diseases and the Therapeutic Implications. J. Cancer 2019, 10, 2771–2782. [Google Scholar] [CrossRef] [Green Version]
- Zhai, L.; Ladomersky, E.; Lenzen, A.; Nguyen, B.; Patel, R.; Lauing, K.L.; Wu, M.; Wainwright, D.A. IDO1 in cancer: A Gemini of immune checkpoints. Cell. Mol. Immunol. 2018, 15, 447–457. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.-P.; Fu, S.-F.; Chen, X.; Ye, J.; Ye, Y.; Kong, L.-D.; Zhu, Z. The Clinicopathological and Prognostic Significance of IDO1 Expression in Human Solid Tumors: Evidence from a Systematic Review and Meta-Analysis. Cell. Physiol. Biochem. 2018, 49, 134–143. [Google Scholar] [CrossRef]
- Prendergast, G.C.; Malachowski, W.J.; Mondal, A.; Scherle, P.; Muller, A.J. Indoleamine 2,3-Dioxygenase and Its Therapeutic Inhibition in Cancer. Int. Rev. Cell Mol. Biol. 2018, 336, 175–203. [Google Scholar] [CrossRef]
- Opitz, C.A.; Somarribas Patterson, L.F.; Mohapatra, S.R.; Dewi, D.L.; Sadik, A.; Platten, M.; Trump, S. The therapeutic potential of targeting tryptophan catabolism in cancer. Br. J. Cancer 2020, 122, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, X.; Wang, L.; Ma, X.; Gong, Z.; Zhang, S.; Li, Y. Targeting the IDO1 pathway in cancer: From bench to bedside. J. Hematol. Oncol. 2018, 11, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prendergast, G.C.; Mondal, A.; Dey, S.; Laury-Kleintop, L.D.; Muller, A.J. Inflammatory Reprogramming with IDO1 Inhibitors: Turning Immunologically Unresponsive ‘Cold’ Tumors ‘Hot’. Trends in Cancer 2018, 4, 38–58. [Google Scholar] [CrossRef]
- Wang, S.; Wu, J.; Shen, H.; Wang, J. The prognostic value of IDO expression in solid tumors: A systematic review and meta-analysis. BMC Cancer 2020, 20, 471. [Google Scholar] [CrossRef]
- Liu, H.; Shen, Z.; Wang, Z.; Wang, X.; Zhang, H.; Qin, J.; Qin, X.; Xu, J.; Sun, Y. Increased expression of IDO associates with poor postoperative clinical outcome of patients with gastric adenocarcinoma. Sci. Rep. 2016, 6, 21319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Wang, L.J.; Lombardo, K.; Kwak, Y.; Kim, W.H.; Resnick, M.B. Expression of Indoleamine 2, 3-dioxygenase 1 (IDO1) and Tryptophanyl-tRNA Synthetase (WARS) in Gastric Cancer Molecular Subtypes. Appl. Immunohistochem. Mol. Morphol. 2020, 28, 360–368. [Google Scholar] [CrossRef]
- Nishi, M.; Yoshikawa, K.; Higashijima, J.; Tokunaga, T.; Kashihara, H.; Takasu, C.; Ishikawa, D.; Wada, Y.; Shimada, M. The Impact of Indoleamine 2,3-dioxygenase (IDO) Expression on Stage III Gastric Cancer. Anticancer Res. 2018, 38, 3387–3392. [Google Scholar] [CrossRef]
- Li, F.; Sun, Y.; Huang, J.; Xu, W.; Liu, J.; Yuan, Z. CD4/CD8 + T cells, DC subsets, Foxp3, and IDO expression are predictive indictors of gastric cancer prognosis. Cancer Med. 2019, 8, 7330–7344. [Google Scholar] [CrossRef]
- Patil, P.A.; Blakely, A.M.; Lombardo, K.A.; Machan, J.T.; Miner, T.J.; Wang, L.-J.; Marwaha, A.S.; Matoso, A. Expression of PD-L1, indoleamine 2,3-dioxygenase and the immune microenvironment in gastric adenocarcinoma. Histopathology 2018, 73, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Nam, K.H.; Ahn, S.-H.; Park, D.J.; Kim, H.-H.; Kim, S.H.; Chang, H.; Lee, J.-O.; Kim, Y.J.; Lee, H.S.; et al. Prognostic implications of immunosuppressive protein expression in tumors as well as immune cell infiltration within the tumor microenvironment in gastric cancer. Gastric Cancer 2016, 19, 42–52. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Liu, H.; Li, F.; Li, H.; Yu, J.; Ren, X. The Correlation Between the Subsets of Tumor Infiltrating Memory T Cells and the Expression of Indoleamine 2,3-Dioxygenase in Gastric Cancer. Dig. Dis. Sci. 2013, 58, 3494–3502. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Huang, J.; Li, S.; Li, H.; Yu, J.; Ren, X.; Liu, J. The subsets of dendritic cells and memory T cells correspond to indoleamine 2,3-dioxygenase in stomach tumor microenvironment. Tumor Biol. 2014, 35, 8691–8698. [Google Scholar] [CrossRef] [PubMed]
- Meireson, A.; Devos, M.; Brochez, L. IDO Expression in Cancer: Different Compartment, Different Functionality? Front. Immunol. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Schalper, K.A.; Carvajal-Hausdorf, D.; McLaughlin, J.; Altan, M.; Velcheti, V.; Gaule, P.; Sanmamed, M.F.; Chen, L.; Herbst, R.S.; Rimm, D.L. Differential Expression and Significance of PD-L1, IDO-1, and B7-H4 in Human Lung Cancer. Clin. Cancer Res. 2017, 23, 370–378. [Google Scholar] [CrossRef] [Green Version]
- Öztürk, S.; Kalter, V.; Roessner, P.M.; Sunbul, M.; Seiffert, M. IDO1-Targeted Therapy Does Not Control Disease Development in the Eµ-TCL1 Mouse Model of Chronic Lymphocytic Leukemia. Cancers 2021, 13, 1899. [Google Scholar] [CrossRef]
- Beauchemin, N.; Draber, P.; Dveksler, G.; Gold, P.; Gray-Owen, S.; Grunert, F.; Hammarstrom, S.; Holmes, K.V.; Karlsson, A.; Kuroki, M.; et al. Redefined Nomenclature for Members of the Carcinoembryonic Antigen Family. Exp. Cell Res. 1999, 252, 243–249. [Google Scholar] [CrossRef]
- Dankner, M.; Gray-Owen, S.D.; Huang, Y.-H.; Blumberg, R.S.; Beauchemin, N. CEACAM1 as a Multi-Purpose Target for Cancer Immunotherapy. Oncoimmunology 2017, 6, e1328336. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.-W.; Lyv, Z.-W.; Cui, B.; Wang, Y.-Y.; Cheng, J.-T.; Zhang, Y.; Cai, W.-Q.; Zhou, Y.; Ma, Z.-W.; Wang, X.-W.; et al. The old CEACAMs find their new role in tumor immunotherapy. Investig. New Drugs 2020, 38, 1888–1898. [Google Scholar] [CrossRef]
- Huang, Y.H.; Zhu, C.; Kondo, Y.; Anderson, A.C.; Gandhi, A.; Russell, A.; Dougan, S.K.; Petersen, B.S.; Melum, E.; Pertel, T.; et al. CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature 2015, 517, 386–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calinescu, A.; Turcu, G.; Nedelcu, R.I.; Brinzea, A.; Hodorogea, A.; Antohe, M.; Diaconu, C.; Bleotu, C.; Pirici, D.; Jilaveanu, L.B.; et al. On the Dual Role of Carcinoembryonic Antigen-Related Cell Adhesion Molecule 1 (CEACAM1) in Human Malignancies. J. Immunol. Res. 2018, 2018, 7169081. [Google Scholar] [CrossRef] [Green Version]
- Ergün, S.; Kilik, N.; Ziegeler, G.; Hansen, A.; Nollau, P.; Götze, J.; Wurmbach, J.H.; Horst, A.; Weil, J.; Fernando, M.; et al. CEA-related cell adhesion molecule 1: A potent angiogenic factor and a major effector of vascular endothelial growth factor. Mol. Cell 2000, 5, 311–320. [Google Scholar] [CrossRef]
- Yang, F.; Zeng, Z.; Li, J.; Ren, X.; Wei, F. TIM-3 and CEACAM1 are Prognostic Factors in Head and Neck Squamous Cell Carcinoma. Front. Mol. Biosci. 2021, 8, 619765. [Google Scholar] [CrossRef] [PubMed]
- Thies, A.; Moll, I.; Berger, J.; Wagener, C.; Brümmer, J.; Schulze, H.-J.; Brunner, G.; Schumacher, U. CEACAM1 expression in cutaneous malignant melanoma predicts the development of metastatic disease. J. Clin. Oncol. 2002, 20, 2530–2536. [Google Scholar] [CrossRef]
- Thöm, I.; Schult-Kronefeld, O.; Burkholder, I.; Schuch, G.; Andritzky, B.; Kastendieck, H.; Edler, L.; Wagener, C.; Bokemeyer, C.; Schumacher, U.; et al. Expression of CEACAM-1 in pulmonary adenocarcinomas and their metastases. Anticancer Res. 2009, 29, 249–254. [Google Scholar]
- Wang, J.-B.; Li, P.; Liu, X.-L.; Zheng, Q.-L.; Ma, Y.-B.; Zhao, Y.-J.; Xie, J.-W.; Lin, J.-X.; Lu, J.; Chen, Q.-Y.; et al. An immune checkpoint score system for prognostic evaluation and adjuvant chemotherapy selection in gastric cancer. Nat. Commun. 2020, 11, 6352. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, A.; Yokoyama, S.; Nakamori, M.; Nakamura, M.; Ojima, T.; Yamaguchi, S.; Mitani, Y.; Shively, J.E.; Yamaue, H. Loss of CEACAM1 is associated with poor prognosis and peritoneal dissemination of patients with gastric cancer. Sci. Rep. 2019, 9, 12702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.-F.; Xu, S.-X.; He, P.; Xi, Z.-H. Expression of carcinoembryonic antigen-related cell adhesion molecule 1(CEACAM1) and its correlation with angiogenesis in gastric cancer. Pathol.-Res. Pract. 2014, 210, 473–476. [Google Scholar] [CrossRef]
- Zhou, C.-J.; Liu, B.; Zhu, K.-X.; Zhang, Q.-H.; Zhang, T.-G.; Xu, W.-H.; Wang, H.-B.; Yu, W.-H.; Qu, Y.-D.; Wang, H.-J.; et al. The different expression of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) and possible roles in gastric carcinomas. Pathol.-Res. Pract. 2009, 205, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.H.; Lee, J.; Hur, M.; Park, H.-Y.; Yum, H.I.; Nam, H.; Oh, M.-Y.; Choi, H.; Kim, J.; Cho, B.C.; et al. MG1124, a novel CEACAM1-targeted monoclonal antibody, has therapeutic potential as a combination partner of PD-1 inhibitors in NSCLC patients. Ann. Oncol. 2019, 30, v490. [Google Scholar] [CrossRef]
- Cho, H.J.; Lee, J.-C.; Park, H.-Y.; Yang, W.S.; Nam, H.-M.; Ryu, J.H.; Oh, Y.; Hur, M. 1064P Efficacy of a novel anti-CEACAM1 monoclonal antibody and CEACAM1 up-regulation in tumour-infiltrating lymphocytes (TILs) of cancer patients. Ann. Oncol. 2020, 31, S725. [Google Scholar] [CrossRef]
- Molfetta, R.; Zitti, B.; Lecce, M.; Milito, N.D.; Stabile, H.; Fionda, C.; Cippitelli, M.; Gismondi, A.; Santoni, A.; Paolini, R. CD155: A Multi-Functional Molecule in Tumor Progression. Int. J. Mol. Sci. 2020, 21, 922. [Google Scholar] [CrossRef] [Green Version]
- Maier, M.K.; Seth, S.; Czeloth, N.; Qiu, Q.; Ravens, I.; Kremmer, E.; Ebel, M.; Müller, W.; Pabst, O.; Förster, R.; et al. The adhesion receptor CD155 determines the magnitude of humoral immune responses against orally ingested antigens. Eur. J. Immunol. 2007, 37, 2214–2225. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.J.; Andrews, D.M.; Smyth, M.J. Receptors that interact with nectin and nectin-like proteins in the immunosurveillance and immunotherapy of cancer. Curr. Opin. Immunol. 2012, 24, 246–251. [Google Scholar] [CrossRef]
- de Andrade, L.F.; Smyth, M.J.; Martinet, L. DNAM-1 control of natural killer cells functions through nectin and nectin-like proteins. Immunol. Cell Biol. 2014, 92, 237–244. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Madore, J.; Li, X.-Y.; Smyth, M.J. Tumor intrinsic and extrinsic immune functions of CD155. Semin. Cancer Biol. 2020, 65, 189–196. [Google Scholar] [CrossRef]
- Deuss, F.A.; Watson, G.M.; Fu, Z.; Rossjohn, J.; Berry, R. Structural Basis for CD96 Immune Receptor Recognition of Nectin-like Protein-5, CD155. Structure 2019, 27, 219–228.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Bi, J.; Zheng, X.; Chen, Y.; Wang, H.; Wu, W.; Wang, Z.; Wu, Q.; Peng, H.; Wei, H.; et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat. Immunol. 2018, 19, 723–732. [Google Scholar] [CrossRef]
- Chandramohan, V.; Bryant, J.D.; Piao, H.; Keir, S.T.; Lipp, E.S.; Lefaivre, M.; Perkinson, K.; Bigner, D.D.; Gromeier, M.; McLendon, R.E. Validation of an Immunohistochemistry Assay for Detection of CD155, the Poliovirus Receptor, in Malignant Gliomas. Arch. Pathol. Lab. Med. 2017, 141, 1697–1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.-Y.; Das, I.; Lepletier, A.; Addala, V.; Bald, T.; Stannard, K.; Barkauskas, D.; Liu, J.; Aguilera, A.R.; Takeda, K.; et al. CD155 loss enhances tumor suppression via combined host and tumor-intrinsic mechanisms. J. Clin. Investig. 2018, 128, 2613–2625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, W.; Zhang, H.; Han, F.; Chen, X.; Lin, R.; Wang, W.; Qiu, H.; Zhuang, Z.; Liao, Q.; Zhang, W.; et al. CD155T/TIGIT Signaling Regulates CD8+ T-cell Metabolism and Promotes Tumor Progression in Human Gastric Cancer. Cancer Res. 2017, 77, 6375–6388. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Ma, L.; Feng, L.; Huang, Z.; Meng, X.; Yu, J. CD155 Overexpression Correlates with Poor Prognosis in Primary Small Cell Carcinoma of the Esophagus. Front. Mol. Biosci. 2021, 7, 608404. [Google Scholar] [CrossRef]
- Luo, C.; Ye, W.; Hu, J.; Othmane, B.; Li, H.; Chen, J.; Zu, X. A Poliovirus Receptor (CD155)-Related Risk Signature Predicts the Prognosis of Bladder Cancer. Front. Oncol. 2021, 11, 660273. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, Z.; Du, G.; Cao, L.; Tan, B. CD155-Prognostic and Immunotherapeutic Implications Based on Multiple Analyses of Databases Across 33 Human Cancers. Technol. Cancer Res. Treat. 2021, 20, 1533033820980088. [Google Scholar] [CrossRef]
- Li, Y.-C.; Zhou, Q.; Song, Q.-K.; Wang, R.-B.; Lyu, S.; Guan, X.; Zhao, Y.-J.; Wu, J.-P. Overexpression of an Immune Checkpoint (CD155) in Breast Cancer Associated with Prognostic Significance and Exhausted Tumor-Infiltrating Lymphocytes: A Cohort Study. J. Immunol. Res. 2020, 2020, 3948928. [Google Scholar] [CrossRef]
- Xu, D.; Zhao, E.; Zhu, C.; Zhao, W.; Wang, C.; Zhang, Z.; Zhao, G. TIGIT and PD-1 may serve as potential prognostic biomarkers for gastric cancer. Immunobiology 2020, 225, 151915. [Google Scholar] [CrossRef]
- Yong, H.; Cheng, R.; Li, X.; Gao, G.; Jiang, X.; Cheng, H.; Zhou, X.; Zhao, W. CD155 expression and its prognostic value in postoperative patients with breast cancer. Biomed. Pharmacother. 2019, 115, 108884. [Google Scholar] [CrossRef]
- Huang, D.-W.; Huang, M.; Lin, X.-S.; Huang, Q. CD155 expression and its correlation with clinicopathologic characteristics, angiogenesis, and prognosis in human cholangiocarcinoma. Onco. Targets. Ther. 2017, 10, 3817–3825. [Google Scholar] [CrossRef] [Green Version]
- Iguchi-Manaka, A.; Okumura, G.; Kojima, H.; Cho, Y.; Hirochika, R.; Bando, H.; Sato, T.; Yoshikawa, H.; Hara, H.; Shibuya, A.; et al. Increased Soluble CD155 in the Serum of Cancer Patients. PLoS ONE 2016, 11, e0152982. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Ma, J.; Lei, T.; Zhao, M.; Zhang, M. Targeting immunotherapy for bladder cancer by using anti-CD3 × CD155 bispecific antibody. J. Cancer 2019, 10, 5153–5161. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Mao, L.; Liu, J.-F.; Chen, L.; Yu, G.-T.; Yang, L.-L.; Wu, H.; Bu, L.-L.; Kulkarni, A.B.; Zhang, W.-F.; et al. Blockade of TIGIT/CD155 Signaling Reverses T-cell Exhaustion and Enhances Antitumor Capability in Head and Neck Squamous Cell Carcinoma. Cancer Immunol. Res. 2019, 7, 1700–1713. [Google Scholar] [CrossRef]
- Sun, J.; Lu, Q.; Sanmamed, M.F.; Wang, J. Siglec-15 as an Emerging Target for Next-generation Cancer Immunotherapy. Clin. Cancer Res. 2021, 27, 680–688. [Google Scholar] [CrossRef]
- Angata, T.; Tabuchi, Y.; Nakamura, K.; Nakamura, M. Siglec-15: An immune system Siglec conserved throughout vertebrate evolution. Glycobiology 2007, 17, 838–846. [Google Scholar] [CrossRef]
- Angata, T. Siglec-15: A potential regulator of osteoporosis, cancer, and infectious diseases. J. Biomed. Sci. 2020, 27, 10. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Huang, Z.; Chen, Y.; Yao, H.; Ke, Z.; He, X.; Qiu, M.; Wang, M.; Xiong, Z.; Yang, S. Integrative Analysis of Siglec-15 mRNA in Human Cancers Based on Data Mining. J. Cancer 2020, 11, 2453–2464. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, J.; Liu, L.N.; Flies, D.B.; Nie, X.; Toki, M.; Zhang, J.; Song, C.; Zarr, M.; Zhou, X.; et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat. Med. 2019, 25, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Sari-Ak, D.; Bagga, T. Siglecs as Therapeutic Targets in Cancer. Biology 2021, 10, 1178. [Google Scholar] [CrossRef]
- Li, B.; Zhang, B.; Wang, X.; Zeng, Z.; Huang, Z.; Zhang, L.; Wei, F.; Ren, X.; Yang, L. Expression signature, prognosis value, and immune characteristics of Siglec-15 identified by pan-cancer analysis. Oncoimmunology 2020, 9, 1807291. [Google Scholar] [CrossRef]
- Quirino, M.W.L.; Pereira, M.C.; de Souza, M.D.F.D.; da Rocha Pitta, I.; da Silva Filho, A.F.; de Souza Albuquerque, M.S.; de Barros Albuquerque, A.P.; Martins, M.R.; da Rocha Pitta, M.G.; de Melo Rêgo, M.J.B. Immunopositivity for Siglec-15 in gastric cancer and its association with clinical and pathological parameters. Eur. J. Histochem. 2021, 65. [Google Scholar] [CrossRef]
- A Safety and Tolerability Study of NC318 in Subjects with Advanced or Metastatic Solid Tumors. Available online: https://clinicaltrials.gov/ct2/show/NCT03665285 (accessed on 12 December 2021).
- Edwards, D.R.; Handsley, M.M.; Pennington, C.J. The ADAM metalloproteinases. Mol. Aspects Med. 2008, 29, 258–289. [Google Scholar] [CrossRef]
- Hassemer, E.L.; Endres, B.; Toonen, J.A.; Ronchetti, A.; Dubielzig, R.; Sidjanin, D.J. ADAM17 transactivates EGFR signaling during embryonic eyelid closure. Invest. Ophthalmol. Vis. Sci. 2013, 54, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Gooz, M. ADAM-17: The enzyme that does it all. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 146–169. [Google Scholar] [CrossRef] [Green Version]
- Blobel, C.P. ADAMs: Key components in EGFR signalling and development. Nat. Rev. Mol. Cell Biol. 2005, 6, 32–43. [Google Scholar] [CrossRef]
- Shen, H.; Li, L.; Zhou, S.; Yu, D.; Yang, S.; Chen, X.; Wang, D.; Zhong, S.; Zhao, J.; Tang, J. The role of ADAM17 in tumorigenesis and progression of breast cancer. Tumor Biol. 2016, 37, 15359–15370. [Google Scholar] [CrossRef] [PubMed]
- Göoz, P.; Göoz, M.; Baldys, A.; Hoffman, S. ADAM-17 regulates endothelial cell morphology, proliferation, and in vitro angiogenesis. Biochem. Biophys. Res. Commun. 2009, 380, 33–38. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Zhou, H.; Zhang, C.; He, J.; Wei, H.; Zhou, M.; Lu, Y.; Sun, Y.; Ding, J.W.; Zeng, J.; et al. ADAM17 promotes epithelial-mesenchymal transition via TGF-α/Smad pathway in gastric carcinoma cells. Int. J. Oncol. 2016, 49, 2520–2528. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, D.; Sun, X.; Zhang, Y.; Wang, L.; Suo, J. ADAM17 promotes lymph node metastasis in gastric cancer via activation of the Notch and Wnt signaling pathways. Int. J. Mol. Med. 2019, 43, 914–926. [Google Scholar] [CrossRef]
- Möller-Hackbarth, K.; Dewitz, C.; Schweigert, O.; Trad, A.; Garbers, C.; Rose-John, S.; Scheller, J. A Disintegrin and Metalloprotease (ADAM) 10 and ADAM17 Are Major Sheddases of T Cell Immunoglobulin and Mucin Domain 3 (Tim-3). J. Biol. Chem. 2013, 288, 34529–34544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orme, J.J.; Jazieh, K.A.; Xie, T.; Harrington, S.; Liu, X.; Ball, M.; Madden, B.; Charlesworth, M.C.; Azam, T.U.; Lucien, F.; et al. ADAM10 and ADAM17 cleave PD-L1 to mediate PD-(L)1 inhibitor resistance. Oncoimmunology 2020, 9, 1744980. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, T.; Tomita, T.; Dixon, M.F.; Axon, A.T.R.; Robinson, P.A.; Crabtree, J.E. ADAMs (a disintegrin and metalloproteinase) messenger RNA expression in Helicobacter pylori-infected, normal, and neoplastic gastric mucosa. J. Infect. Dis. 2002, 185, 332–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, X.; Hao, B.; Cao, N.; Wang, J.; Lv, X.; Zhang, X. ADAM17 overexpression is associated with poorer clinical outcomes in cancer patients: A systematic review and meta-analysis. Oncotarget 2018, 5. [Google Scholar] [CrossRef] [Green Version]
- Ni, P.; Yu, M.; Zhang, R.; He, M.; Wang, H.; Chen, S.; Duan, G. Prognostic Significance of ADAM17 for Gastric Cancer Survival: A Meta-Analysis. Medicina 2020, 56, 322. [Google Scholar] [CrossRef]
- Sun, X.; Wang, Y.; Ma, R.; Li, W. Clinical significance of ADAM10 and ADAM17 in gastric and colorectal cancers: A systematic review and meta-analysis. Int. J. Clin. Exp. Med. 2017, 10, 5941–5948. [Google Scholar]
- Moss, M.L.; Minond, D. Recent Advances in ADAM17 Research: A Promising Target for Cancer and Inflammation. Mediators Inflamm. 2017, 2017, 9673537. [Google Scholar] [CrossRef] [Green Version]
- Saad, M.I.; Rose-John, S.; Jenkins, B.J. Jenkins ADAM17: An Emerging Therapeutic Target for Lung Cancer. Cancers 2019, 11, 1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Yuen, S.M.; Murphy, G.; Xie, R.; Kwok, H.F. Anti-tumor effects of a ‘human & mouse cross-reactive’ anti-ADAM17 antibody in a pancreatic cancer model in vivo. Eur. J. Pharm. Sci. 2017, 110, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Mishra, H.K.; Dixon, K.J.; Pore, N.; Felices, M.; Miller, J.S.; Walcheck, B. Activation of ADAM17 by IL-15 Limits Human NK Cell Proliferation. Front. Immunol. 2021, 12, 711621. [Google Scholar] [CrossRef]
- Shou, Z.-X.; Jin, X.; Zhao, Z.-S. Upregulated Expression of ADAM17 Is a Prognostic Marker for Patients with Gastric Cancer. Ann. Surg. 2012, 256, 1014–1022. [Google Scholar] [CrossRef]
- Zhang, T.; Zhu, W.; Huang, M.; Fan, R.; Chen, X. Prognostic value of ADAM17 in human gastric cancer. Med. Oncol. 2012, 29, 2684–2690. [Google Scholar] [CrossRef] [PubMed]
- Aydin, D.; Bilici, A.; Yavuzer, D.; Kefeli, U.; Tan, A.; Ercelep, O.; Mert, A.; Yuksel, S.; Ozcelik, M.; Isik, D.; et al. Prognostic significance of ADAM17 expression in patients with gastric cancer who underwent curative gastrectomy. Clin. Transl. Oncol. 2015, 17, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Jiang, J.; Lu, K.; Chen, Q.; Tao, D.; Chen, Z. Therapeutic potential of ADAM17 modulation in gastric cancer through regulation of the EGFR and TNF-α signalling pathways. Mol. Cell. Biochem. 2017, 426, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Qian, J.; Wu, Q.; Chen, Y.; Yu, G. ADAM-17 expression is enhanced by FoxM1 and is a poor prognostic sign in gastric carcinoma. J. Surg. Res. 2017, 220, 223–233. [Google Scholar] [CrossRef]
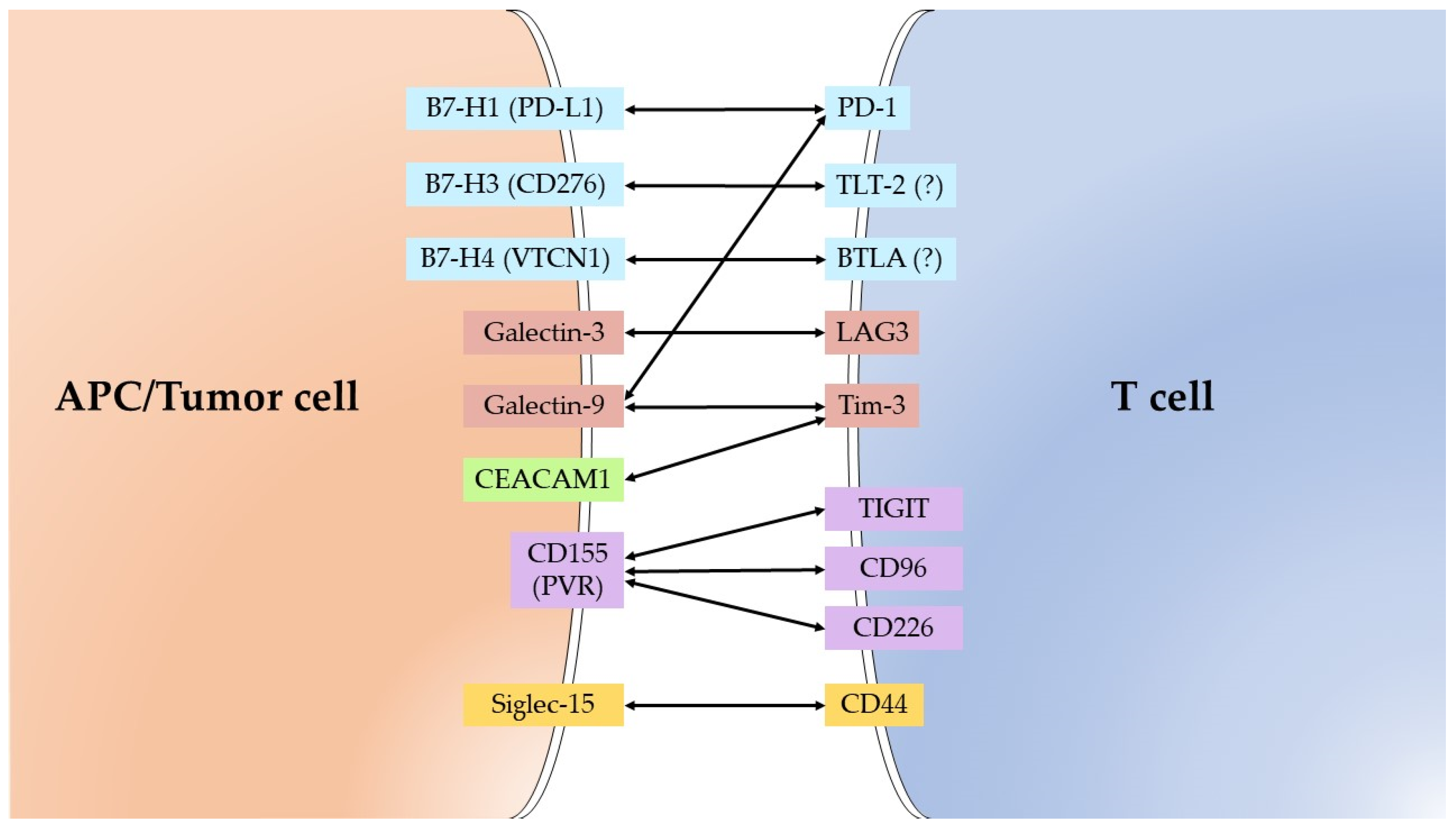
| Molecule | Alternative Name | Receptor/Substrate |
|---|---|---|
| B7-H1 | CD274/PD-L1/PDCD1L1 | PD-1 |
| B7-H3 | CD276/B7RP-2 | TLT-2 (?) |
| B7-H4 | VTCN1/B7S1/B7x | BTLA (?) |
| Galectin-3 | MAC-2 | LAG3/LGALS3BP |
| Galectin-9 | Ecalectin | Tim-3/PD-1/CD44 |
| IDO1 | INDO | * Tryptophan |
| CEACAM1 | CD66a/BGP | Tim-3 |
| CD155 | PVR/Necl-5 | TIGIT/CD96/CD226 |
| Siglec-15 | CD33L3/HsT1361 | CD44/Sialyl-Tn |
| ADAM17 | CD156b/TACE | * Pro-TNF-a/Pro-TGF- a/Notch1/Pro-Amphiregulin/Pro-HB-EGF/Pro-epiregulin/Neuregulin 1/IL6 Receptor/etc. |
| Checkpoint | Characteristics of Checkpoints as Candidates for GC Markers | Materials/ Method | Reference |
|---|---|---|---|
| B7-H1 | Increased expression is associated with poor 3-year OS (HR = 1.23, 95% CI = 1.02–1.49, p = 0.028), poor 5-year OS (HR = 1.39, 95% CI = 1.14–1.69, p = 0.001), lymph node metastasis (OR = 1.73, 95% CI = 1.18–2.54, p < 0.01) | Meta-analysis GC tissue/IHC | [17] |
| Increased expression is associated with poor OS (HR = 1.46, 95% CI = 1.08–1.98, p = 0.01), greater depth of infiltration (p = 0.03), lymph node metastasis (p = 0.03), venous invasion (p = 0.0003) | Meta-analysis GC tissue/IHC | [18] | |
| Increased expression is associated with poor OS (HR = 1.64, 95% CI = 1.11–2.43, p = 0.01), large tumor size (OR = 1.87, 95% CI = 1.25–2.78, p = 0.002), lymph node metastasis (OR = 2.17, 95% CI = 1.04–4.52, p = 0.04). | Meta-analysis GC tissue/IHC | [19] | |
| Increased expression is associated with poor OS (HR = 1.60, 95% CI = 1.09–2.36, p = 0.012) | Meta-analysis GC tissue/IHC | [20] | |
| Increased expression is associated with poor OS (HR = 1.74, 95% CI = 1.40–2.17, p = 0.146), lymph node metastasis (OR = 2.61, 95% CI = 1.78–3.84, p = 0.004), higher TNM stage (OR = 2.28, 95% CI = 1.39–3.74, p = 0.006) | Meta-analysis GC tissue, Serum/ IHC, ELISA | [21] | |
| Better OS in patients with PD-L1-positive tumors treated with ICIs (HR = 0.82, 95% CI = 0.67–0.99, p = 0.04) | Meta-analysis GC tissue/IHC | [27] | |
| Increased expression is associated with better OS (HR = 0.753, 95% CI = 0.584–0.971, p = 0.029), less advanced depth of infiltration (p = 0.001), absence of distant metastasis (p = 0.029), lower TNM stage (p = 0.01) | GC tissue/IHC | [13] | |
| Increased expression is associated with poor OS (HR = 1.81, 95% CI = 1.15–2.78, p < 0.05) | Blood/ qRT-PCR | [26] | |
| Increased expression in GC tissue is associated with poor OS (HR = 4.28, 95% CI = 1.43–12.8, p = 0.0094), depth of infiltration (p = 0.0003), vessel involvement (p < 0.0001), lymphatic vessel involvement (p = 0.0005), lymph node metastasis (p = 0.023), peritoneal metastasis (p = 0.0098) Increased expression in serum is associated with poor OS (HR = 11.2, 95% CI = 3.44–36.7, p = 0.0001) | GC tissue, Serum/ IHC, ELISA | [25] | |
| B7-H3 | Increased expression is associated with poor OS (p = 0.003), higher TNM stage (p = 0.000), greater depth of infiltration (p = 0.001), lymph node metastasis (p = 0.020) | GC tissue/IHC | [40] |
| Increased expression is associated with better OS (RR = 2.803, 95% CI = 1.051–7.477, p = 0.040) | GC tissue/IHC | [42] | |
| Increased expression is associated with greater depth of infiltration (p = 0.005) | GC tissue/IHC | [34] | |
| Increased stromal expression is associated with greater depth of infiltration (p = 0.013) | GC tissue/IHC | [35] | |
| Increased expression is associated with poor OS (HR = 1.56, 95% CI = 1.01–2.54, p = 0.046), higher TNM stage (p = 0.013) | Blood/qRT-PCR | [41] | |
| B7-H4 | Increased expression is associated with poor prognosis (OR = 1.63, 95% CI = 1.30–2.03) | Meta-analysis GC tissue, blood, serum/IHC, qRT-PCR, ELISA | [61] |
| Increased expression is associated with poor prognosis (RR = 1.85, 95% CI = 1.15–2.96, p = 0.0087), myometrial invasion (p = 0.004), lymph node metastasis (p < 0.0001), recurrence (p = 0.003) | GC tissue/IHC | [62] | |
| Increased expression is associated with poor prognosis (HR = 1.49, 95% CI = 1.03–2.17, p = 0.035), higher TNM stage (p = 0.04) | GC tissue/IHC | [63] | |
| Increased expression is associated with poor prognosis (HR = 1.41, 95% CI = 1.01–1.98, p = 0.049), lymph node metastasis (p = 0.007), higher TNM stage (p = 0.004), greater depth of infiltration (p = 0.011) | GC tissue/IHC | [64] | |
| Decreased expression is associated with better OS in NACT (neoadjuvant chemotherapy) group (p = 0.031) | GC tissue/IHC | [65] | |
| Increased expression is associated with poor prognosis (HR = 2.01, 95% CI = 1.08–5.03, p = 0.024), greater depth of infiltration (p = 0.006), lymph node metastasis (p = 0.001), higher TNM stage (p < 0.001), lymphatic invasion (p < 0.001), venous invasion (p = 0.010) | Blood/RT-PCR | [66] | |
| Increased expression is associated with poor prognosis (HR = 1.925, 95% CI = 1.033–3.857, p = 0.039), large tumor size (p = 0.002), lymph node metastasis (p = 0.001), greater depth of infiltration (p = 0.041) higher TNM stage (p < 0.001) | Serum/ ELISA | [67] | |
| Galectin-3 | Decreased expression is associated with poor prognosis (HR = 0.49, 95% CI = 0.36–0.67, p < 0.001), lymphatic vessel invasion (OR = 0.48, 95% CI = 0.26–0.89, p = 0.018), higher TNM stage (OR = 0.47, 95% CI = 0.32–0.40, p < 0.001), greater depth of infiltration (OR = 0.33, 95% CI = 0.21–0.51, p < 0.001), poorer differentiation grade (OR = 0.10, 95% CI = 0.04–0.25, p < 0.001) | Meta-analysis GC tissue, serum/IHC, ELISA | [84] |
| Increased expression is associated with better OS (p = 0.006), less advanced depth of infiltration (p < 0.001), absence of lymph node metastasis (p = 0.001), absence of distant metastasis (p = 0.004), lower TNM stage (p < 0.001), absence of lymphovascular invasion (p = 0.035) | GC tissue/IHC | [85] | |
| Decreased expression is associated with poor prognosis (RR = 3.831, 95% CI = 1.574–9.329, p = 0.0031), diffuse type (p < 0.0001), poor tumor grade (p < 0.0001), lymph node metastasis (p = 0.0495), lymphatic invasion (p = 0.0086), higher TNM stage (p = 0.0433) | GC tissue/IHC | [71] | |
| Increased expression is associated with higher TNM stage (p = 0.038), poor differentiation (p = 0.001), lymph node metastasis (p = 0.022) | GC tissue/RT-PCR | [86] | |
| Increased expression is associated with higher TNM stage (p = 0.0019) | GC tissue/IHC | [87] | |
| Increased expression is associated with lymph node metastasis (p = 0.001), distant metastasis (p < 0.001) | Serum/ ELISA | [88] | |
| Galectin-9 | Increased expression is associated with better OS (HR = 0.51, 95% CI = 0.35–0.76, p = 0.001) Gal-9 negativity is associated with greater depth of infiltration (p < 0.001), lymph node metastasis (p < 0.001), higher TNM stage (p < 0.001) | GC tissue/IHC | [101] |
| Increased expression is associated with poor OS (p = 0.0028), lymph node metastasis (p = 0.0060), higher TNM stage (p = 0.0292), blood vessel invasion (p = 0.0410) | GC tissue/IHC | [94] | |
| Increased expression is associated with better OS (p = 0.002) Decreased expression is associated with lymph-vascular invasion (p = 0.034), lymph node metastasis (p = 0.009), distant metastasis (p = 0.002), higher TNM staging (p = 0.043) | GC tissue/IHC | [100] | |
| IDO1 | Increased expression is associated with poor OS (HR = 1.596, 95% CI = 1.156–2.204, p = 0.005), greater depth of infiltration (p = 0.045), lymph node metastasis (p < 0.001) | GC tissue/IHC | [111] |
| Increased expression is associated with poor OS (p = 0.0059) | GC tissue/IHC | [112] | |
| Increased expression is associated with poor OS (HR = 2.75, 95% CI = 1.01–7.58, p < 0.05) | GC tissue/IHC | [113] | |
| Increased expression is associated with greater depth of infiltration (p = 0.016), lymph node metastasis (p = 0.038) | GC tissue/IHC | [117] | |
| Increased expression is associated with poor OS (p = 0.043), large tumor size (p = 0.044), greater depth of infiltration (p = 0.027) | GC tissue/IHC | [114] | |
| Increased expression is associated with greater depth of infiltration (p = 0.016), lymph node metastasis (p = 0.046) | GC tissue/IHC | [118] | |
| Increased expression is associated with well/moderately differentiated histology (p < 0.001), intestinal type (p < 0.001), absence of vascular invasion (p = 0.012), lower TNM stage (p = 0.007) | GC tissue/IHC | [116] | |
| Increased expression is associated with lower rate of metastasis (p = 0.032), lower rate of recurrence (p = 0.010) | GC tissue/IHC | [115] | |
| CEACAM1 | Increased expression is associated with poor OS (p = 0.001) | GC tissue/IHC | [131] |
| Loss of CEACAM1 is associated with poor OS (HR = 3.472, 95% CI = 1.508–8.00, p = 0.03), peritoneal dissemination after gastrectomy (HR = 3.711, 95% CI = 1.253–10.995, p = 0.018) | GC tissue/IHC | [132] | |
| Increased expression is associated with lymph node metastasis (p < 0.05), higher TNM stage (p < 0.05) | GC tissue/IHC | [133] | |
| Increased expression is associated with lymph node metastasis (p < 0.05) | GC tissue/IHC | [134] | |
| CD155 | Increased expression is associated with poor OS (p = 0.001) | GC tissue/IHC | [131] |
| Increased TIGIT and CD155 expression were associated with poor OS (p = 0.011) | KM Plotter GC database | [151] | |
| Increased expression is associated with higher TNM stage (p < 0.05) | Serum/ ELISA | [154] | |
| Siglec-15 | Increased expression is associated with poor OS (HR = 4.87, 95% CI = 1.42–15.4, p = 0.006) | TCGA and GEO databases | [163] |
| Increased expression is associated with TNM stage (p = 0.01), histological grade (p = 0.0022), angiolymphatic invasion (p = 0.041) | GC tissue/IHC | [164] | |
| ADAM17 | Increased expression is associated with poor OS (HR = 2.04, 95% CI = 1.66–2.50, p = 0.299), higher TNM stage (OR = 4.09, 95% CI = 1.85–9.04, p = 0.000), lymph node metastasis (OR = 3.08, 95% CI = 1.13–8.36, p = 0.007) | Meta-analysis GC tissue/IHC | [178] |
| Increased ADAM10 and ADAM17 expression were associated with greater depth of infiltration (OR = 0.29, 95% CI = 0.21 to 0.40, p < 0.0001), lymph node metastasis (OR = 4.36, 95% CI = 2.25 to 8.45, p < 0.0001), distant metastasis (OR = 0.09, 95% CI = 0.02 to 0.37, p = 0.0008) | Meta-analysis GC tissue/IHC | [179] | |
| Increased expression is associated with poor prognosis (HR = 2.067, 95% CI = 1.475–2.883, p = 0.000), large tumor size (p = 0.000), greater depth of invasion (p = 0.000), higher TNM stage (p = 0.000), diffuse type (p = 0.000), vessel invasion (p = 0.000), lymph node metastasis (p = 0.000), distant metastasis (p = 0.000) | GC tissue/IHC | [184] | |
| Increased expression is associated with poor prognosis (HR = 5.87, 95% CI = 1.59–20.52, p = 0.008), poor differentiation (p = 0.006), greater depth of invasion (p < 0.0001), lymph node metastasis (p = 0.02), distant metastasis (p = 0.02), higher TNM stage (p = 0.03) | GC tissue/IHC | [185] | |
| Increased expression is associated with poor OS (p = 0.019), poor DFS (HR = 1.61, 95% CI = 0.93–2.79, p = 0.038), large tumor size (p = 0.04), lymph node metastasis (p = 0.003), vascular invasion (p = 0.015), recurrence (p = 0.032) | GC tissue/IHC | [186] | |
| Increased expression is associated with poor prognosis (HR = 2.239, 95% CI = 1.516–3.305, p < 0.001) (AUC = 0.618, p = 0.006), lymph node metastasis (OR = 2.161, 95% CI = 1.115–4.190, p = 0.022) | GC tissue/IHC | [173] | |
| Increased expression is associated with greater depth of invasion (p = 0.007), distant metastasis (p = 0.047), higher TNM stage (p = 0.001) | GC tissue/IHC | [187] | |
| Increased expression is associated with poor prognosis (p = 0.007), lymph node metastasis (p = 0.005), higher TNM stage (p = 0.002) | GC tissue/IHC | [188] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mansorunov, D.; Apanovich, N.; Apanovich, P.; Kipkeeva, F.; Muzaffarova, T.; Kuzevanova, A.; Nikulin, M.; Malikhova, O.; Karpukhin, A. Expression of Immune Checkpoints in Malignant Tumors: Therapy Targets and Biomarkers for the Gastric Cancer Prognosis. Diagnostics 2021, 11, 2370. https://doi.org/10.3390/diagnostics11122370
Mansorunov D, Apanovich N, Apanovich P, Kipkeeva F, Muzaffarova T, Kuzevanova A, Nikulin M, Malikhova O, Karpukhin A. Expression of Immune Checkpoints in Malignant Tumors: Therapy Targets and Biomarkers for the Gastric Cancer Prognosis. Diagnostics. 2021; 11(12):2370. https://doi.org/10.3390/diagnostics11122370
Chicago/Turabian StyleMansorunov, Danzan, Natalya Apanovich, Pavel Apanovich, Fatimat Kipkeeva, Tatyana Muzaffarova, Anna Kuzevanova, Maxim Nikulin, Olga Malikhova, and Alexander Karpukhin. 2021. "Expression of Immune Checkpoints in Malignant Tumors: Therapy Targets and Biomarkers for the Gastric Cancer Prognosis" Diagnostics 11, no. 12: 2370. https://doi.org/10.3390/diagnostics11122370
APA StyleMansorunov, D., Apanovich, N., Apanovich, P., Kipkeeva, F., Muzaffarova, T., Kuzevanova, A., Nikulin, M., Malikhova, O., & Karpukhin, A. (2021). Expression of Immune Checkpoints in Malignant Tumors: Therapy Targets and Biomarkers for the Gastric Cancer Prognosis. Diagnostics, 11(12), 2370. https://doi.org/10.3390/diagnostics11122370






