Exploring Machine Learning Techniques to Predict the Response to Omalizumab in Chronic Spontaneous Urticaria
Abstract
:1. Introduction
2. Methods
2.1. Participants
2.2. Machine Learning Approach
2.3. Data Preparation
2.4. Ethical Approval
3. Results
3.1. Concomitant Diseases
3.2. Treatment Efficacy
3.3. Response to Treatment Based on Patient’s Characteristics
3.4. Impact of Disease Duration
3.5. Association between Total IgE Levels and Response to Omalizumab
4. Variable Importance Analysis
5. Machine Learning Methods Classification
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CSU | Chronic spontaneous urticaria |
| UAS7 | Urticaria Activity Score over 7 days |
| ML | Machine learning |
| ER | Early responder |
| LR | Late responder |
| NR | Non-responder |
| GERD | Gastroesophageal reflux disease |
| IgE | Immunoglobulin E |
| IBS | Irritable bowel syndrome |
| SNAS | Systemic nickel allergy syndrome |
| BPH | Benign prostatic hyperplasia |
| TPO | Thyroid peroxidase antibody |
| CRP | C-reactive protein |
| ANA | Anti-nuclear antibody |
| Tg | Thyroglobulin |
| ANCA | Anti-neutrophil cytoplasmic antibodies |
| ESR | Erythrocyte sedimentation rate |
| SVM | Support Vector Machine |
| k-NN | k-nearest neighbors |
References
- Fok, J.S.; Kolkhir, P.; Church, M.K.; Maurer, M. Predictors of treatment response in chronic spontaneous urticaria. Allergy 2021, 76, 2965–2981. [Google Scholar] [CrossRef]
- O’donnell, B.F.; Lawlor, F.; Simpson, J.; Morgan, M.; Greaves, M. The impact of chronic urticaria on the quality of life. Br. J. Dermatol. 1997, 136, 197–201. [Google Scholar] [CrossRef]
- Staubach, P.; Eckhardt-Henn, A.; Dechene, M.; Vonend, A.; Metz, M.; Magerl, M.; Breuer, P.; Maurer, M. Quality of life in patients with chronic urticaria is differentially impaired and determined by psychiatric comorbidity. Br. J. Dermatol. 2006, 154, 294–298. [Google Scholar] [CrossRef]
- Zuberbier, T.; Aberer, W.; Asero, R.; Bindslev-Jensen, C.; Brzoza, Z.; Canonica, G.W.; Church, M.K.; Ensina, L.F.; Giménez-Arnau, A.; Godse, K.; et al. The EAACI/GA2LEN/EDF/WAO guideline for the definition, classification, diagnosis and management of urticaria. Allergy 2018, 73, 1393–1414. [Google Scholar] [CrossRef]
- Maurer, M.; Altrichter, S.; Bieber, T.; Biedermann, T.; Braeutigam, M.; Seyfried, S.; Brehler, R.; Grabbe, J.; Hunzelmann, N.; Jakob, T.; et al. Efficacy and safety of omalizumab in patients with chronic urticaria who exhibit IgE against thyroperoxidase. J. Allergy Clin. Immunol. 2011, 128, 202–209. [Google Scholar] [CrossRef]
- Zhao, Z.-T.; Ji, C.-M.; Yu, W.-J.; Meng, L.; Hawro, T.; Wei, J.F.; Maurer, M. Omalizumab for the treatment of chronic spontaneous urticaria: A meta-analysis of randomized clinical trials. J. Allergy Clin. Immunol. 2016, 137, 1742–1750. [Google Scholar] [CrossRef] [Green Version]
- Maurer, M.; Rosén, K.; Hsieh, H.-J.; Saini, S.; Grattan, C.; Gimenéz-Arnau, A.; Agarwal, S.; Doyle, R.; Canvin, J.; Kaplan, A.; et al. Omalizumab for the Treatment of Chronic Spontaneous or Spontaneous Urticaria. N. Engl. J. Med. 2013, 368, 924–935. [Google Scholar] [CrossRef] [Green Version]
- Metz, M.; Ohanyan, T.; Church, M.K.; Maurer, M. Omalizumab is an effective and rapidly acting therapy in difficult-to-treat chronic urticaria: A retrospective clinical analysis. J. Dermatol. Sci. 2014, 73, 57–62. [Google Scholar] [CrossRef]
- Maurer, M.; Giménez-Arnau, A.M.; Sussman, G.; Metz, M.; Baker, D.R.; Bauer, A.; Bernstein, J.A.; Brehler, R.; Chu, C.-Y.; Hung, S.-I.; et al. Ligelizumab for Chronic Spontaneous Urticaria. N. Engl. J. Med. 2019, 381, 1321–1332. [Google Scholar] [CrossRef] [Green Version]
- Deo, R.C. Machine Learning in Medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef] [Green Version]
- Segura-Bedmar, I.; Colón-Ruiz, C.; Tejedor-Alonso, M.A.; Moro-Moro, M. Predicting of anaphylaxis in big data EMR by exploring machine learning approaches. J. Biomed. Inform. 2018, 87, 50–59. [Google Scholar] [CrossRef]
- Moreno-Indias, I.; Lahti, L.; Nedyalkova, M.; Elbere, I.; Roshchupkin, G.; Adilovic, M.; Aydemir, O.; Bakir-Gungor, B.; Pau, E.C.-D.S.; D’Elia, D.; et al. Statistical and Machine Learning Techniques in Human Microbiome Studies: Contemporary Challenges and Solutions. Front. Microbiol. 2021, 12, 635781. [Google Scholar] [CrossRef]
- Connor, C.W. Artificial Intelligence and Machine Learning in Anesthesiology. Anesthesiology 2019, 131, 1346–1359. [Google Scholar] [CrossRef] [Green Version]
- De Gregory, K.W.; Kuiper, P.; De Silvio, T.; Pleuss, J.D.; Miller, R.; Roginski, J.W.; Fisher, C.B.; Harness, D.; Viswanath, S.; Heymsfield, S.B.; et al. A review of machine learning in obesity. Obes. Rev. 2018, 19, 668–685. [Google Scholar] [CrossRef]
- Patel, L.; Shukla, T.; Huang, X.; Ussery, D.W.; Wang, S. Machine Learning Methods in Drug Discovery. Molecules 2020, 25, 5277. [Google Scholar] [CrossRef]
- Messinger, A.I.; Luo, G.; Deterding, R.R. The doctor will see you now: How machine learning and artificial intelligence can extend our understanding and treatment of asthma. J. Allergy Clin. Immunol. 2019, 145, 476–478. [Google Scholar] [CrossRef] [Green Version]
- Eckman, J.A.; Hamilton, R.G.; Gober, L.M.; Sterba, P.M.; Saini, S.S. Basophil phenotypes in chronic spontaneous urticaria in relation to disease activity and autoantibodies. J. Investig. Dermatol. 2008, 128, 1956–1963. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. (TIST) 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Lever, J.; Krzywinski, M.; Altman, N. Points of significance: Model selection and overfitting. Nat. Methods 2016, 13, 703–705. [Google Scholar] [CrossRef]
- Huang, C.; Clayton, E.A.; Matyunina, L.V.; McDonald, L.D.; Benigno, B.B.; Vannberg, F.; McDonald, J.F. Machine learning predicts individual cancer patient responses to therapeutic drugs with high accuracy. Sci. Rep. 2018, 8, 16444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, E.W.; Bhope, A.; Lim, J.; Sinha, S.; Emad, A. Tissue-guided LASSO for prediction of clinical drug response using preclinical samples. PLoS Comput. Biol. 2020, 16, e1007607. [Google Scholar] [CrossRef] [Green Version]
- Fukushima, A.; Sugimoto, M.; Hiwa, S.; Hiroyasu, T. Elastic net-based prediction of IFN-β treatment response of patients with multiple sclerosis using time series microarray gene expression profiles. Sci. Rep. 2019, 9, 1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, W.; Phung, Q.-D.; Tran, T.; Gupta, S.; Rana, S.; Karmakar, C.; Shilton, A.; Yearwood, J.L.; Dimitrova, N.; Ho, T.B.; et al. Guidelines for developing and reporting machine learning predictive models in biomedical research: A multidisciplinary view. J. Med. Internet Res. 2016, 18, e323. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, A.; Ferrer, M.; Bernstein, J.A.; Antonova, E.; Trzaskoma, B.; Raimundo, K.; Rosén, K.; Omachi, T.A.; Khalil, S.; Zazzali, J.L. Timing and duration of omalizumab response in patients with chronic spontaneous/spontaneous urticaria. J. Allergy Clin. Immunol. 2015, 137, 474–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bousquet, J.; Rabe, K.; Humbert, M.; Chung, K.F.; Berger, W.; Fox, H.; Ayre, G.; Chen, H.; Thomas, K.; Blogg, M.; et al. Predicting and evaluating response to omalizumab in patients with severe allergic asthma. Respir. Med. 2007, 101, 1483–1492. [Google Scholar] [CrossRef] [Green Version]
- Platts-Mills, T.A.E.; Perzanowski, M. The use of machine learning to understand the relationship between IgE to specific allergens and asthma. PLoS Med. 2018, 15, e1002696. [Google Scholar] [CrossRef]
- Anto, J.M.; Bousquet, J.; Akdis, M.; Auffray, C.; Keil, T.; Momas, I.; Postma, D.S.; Valenta, R.; Wickman, M.; Cambon-Thomsen, A.; et al. Mechanisms of the Development of Allergy (MeDALL): Introducing novel concepts in allergy phenotypes. J. Allergy Clin. Immunol. 2017, 139, 388–399. [Google Scholar] [CrossRef] [Green Version]
- Vidyasagar, M. Identifying Predictive Features in Drug Response Using Machine Learning: Opportunities and Challenges. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 15–34. [Google Scholar] [CrossRef]
- Gianfrancesco, M.A.; Tamang, S.; Yazdany, J.; Schmajuk, G. Potential Biases in Machine Learning Algorithms Using Electronic Health Record Data. JAMA Intern. Med. 2018, 178, 1544–1547. [Google Scholar] [CrossRef]

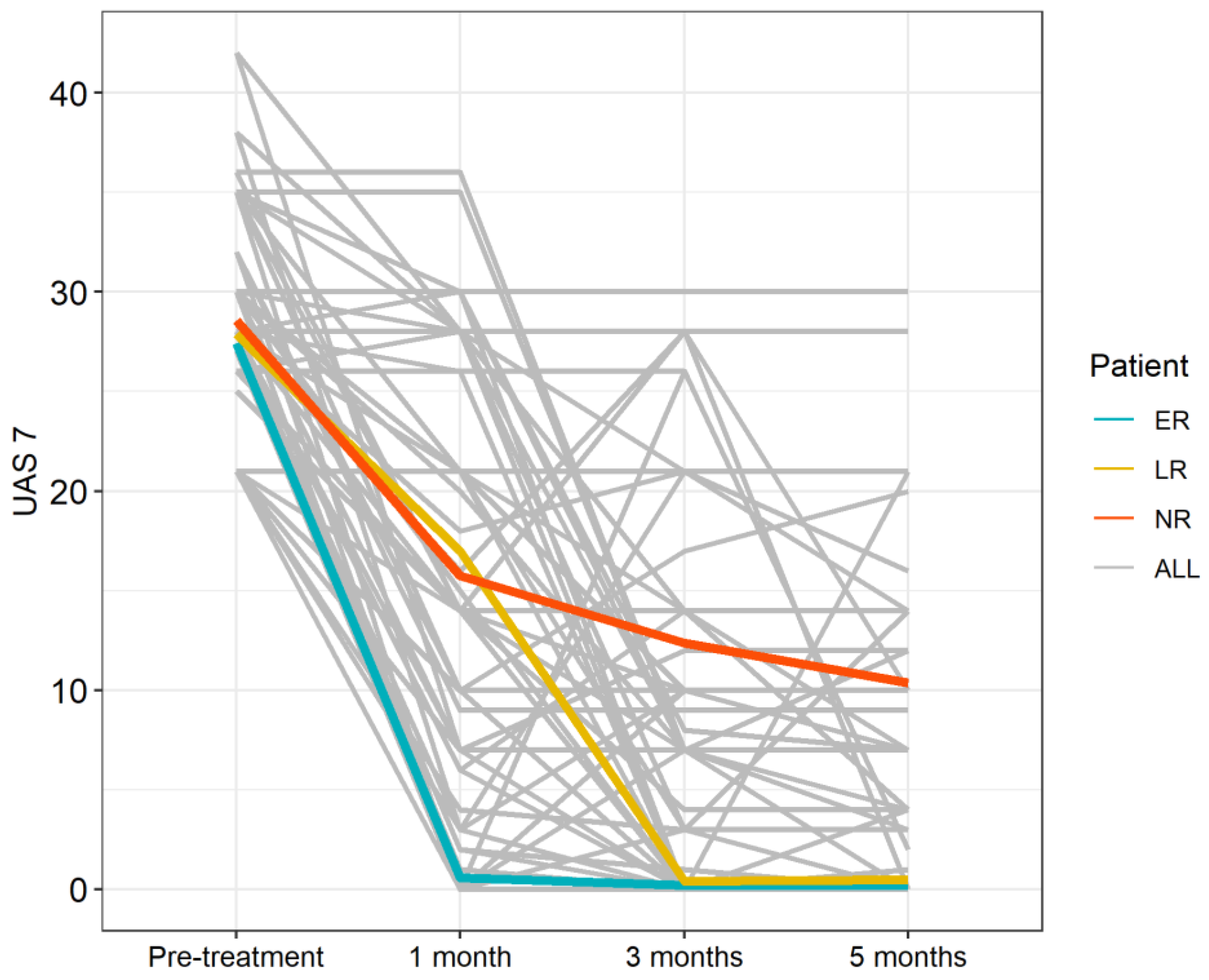





| ER | LR | NR | |
|---|---|---|---|
| Female/Male (%) | 57/43 | 65/35 | 67/33 |
| Age (ys) | 45.8 | 50.5 | 50.4 |
| Disease duration (ys) | 5.7 | 5.1 | 6.2 |
| Concomitant Diseases | Patients n = 132 |
|---|---|
| Respiratory | 37 |
| Thyroid | 24 |
| Hypertension | 23 |
| Dislipidemia | 13 |
| Autoimmune | 12 |
| Gastrointestinal | 12 |
| Allergies | 7 |
| Psychiatric | 9 |
| Miscellaneous | 16 |
| Blood Test | Total Patients (%) |
|---|---|
| Total IgE | 18.9 |
| Anti-TPO | 12.9 |
| CRP | 10.6 |
| D-dimer | 9.8 |
| ANA | 6.1 |
| Anti-Tg | 6.1 |
| Eosinophilia | 5.3 |
| ESR | 3.8 |
| ANCA | 2.3 |
| Glycemia | 2.2 |
| H. pylori | 2.3 |
| Specific IgE (cat) | 1.5 |
| Others | 6 |
| Months | Accuracy | Sensitivity | Specificity | Precision | Method |
|---|---|---|---|---|---|
| 1 | 0.631647 | 0.325238 | 0.801691 | 0.464167 | Elastic net |
| 3 | 0.483874 | 0.552738 | 0.473431 | 0.44404 | Elastic net |
| 5 | 0.493051 | 0.689466 | 0.279524 | 0.542756 | Elastic net |
| 1 | 0.71 | 0.171429 | 1 | 1 | k-NN |
| 3 | 0.473684 | 0.8 | 0.236364 | 0.433333 | k-NN |
| 5 | 0.5 | 1 | 0 | 0.5 | k-NN |
| 1 | 0.631647 | 0.325238 | 0.801691 | 0.464167 | Lasso |
| 3 | 0.483874 | 0.552738 | 0.473431 | 0.44404 | Lasso |
| 5 | 0.493051 | 0.689466 | 0.279524 | 0.542756 | Lasso |
| 1 | 0.362471 | 0.658095 | 0.198309 | 0.318586 | Logistic |
| 3 | 0.516126 | 0.447262 | 0.526569 | 0.419524 | Logistic |
| 5 | 0.506949 | 0.310534 | 0.720476 | NA | Logistic |
| 1 | 0.61672 | 0.26 | 0.818475 | 0.386191 | Ridge |
| 3 | 0.48104 | 0.563691 | 0.46434 | 0.439895 | Ridge |
| 5 | 0.495892 | 0.700577 | 0.267024 | 0.539624 | Ridge |
| 1 | 0.6022222 | 0.375 | 0.7533333 | 0.3541667 | SVM |
| 3 | 0.7666667 | 0.6875 | 0.8133333 | 0.5925926 | SVM |
| 5 | 0.315 | 0.5208333 | 0.3703704 | 0.4351852 | SVM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sardina, D.S.; Valenti, G.; Papia, F.; Uasuf, C.G. Exploring Machine Learning Techniques to Predict the Response to Omalizumab in Chronic Spontaneous Urticaria. Diagnostics 2021, 11, 2150. https://doi.org/10.3390/diagnostics11112150
Sardina DS, Valenti G, Papia F, Uasuf CG. Exploring Machine Learning Techniques to Predict the Response to Omalizumab in Chronic Spontaneous Urticaria. Diagnostics. 2021; 11(11):2150. https://doi.org/10.3390/diagnostics11112150
Chicago/Turabian StyleSardina, Davide Stefano, Giuseppe Valenti, Francesco Papia, and Carina Gabriela Uasuf. 2021. "Exploring Machine Learning Techniques to Predict the Response to Omalizumab in Chronic Spontaneous Urticaria" Diagnostics 11, no. 11: 2150. https://doi.org/10.3390/diagnostics11112150
APA StyleSardina, D. S., Valenti, G., Papia, F., & Uasuf, C. G. (2021). Exploring Machine Learning Techniques to Predict the Response to Omalizumab in Chronic Spontaneous Urticaria. Diagnostics, 11(11), 2150. https://doi.org/10.3390/diagnostics11112150






