Can Autism Be Diagnosed with Artificial Intelligence? A Narrative Review
Abstract
:1. Introduction
2. Related Works
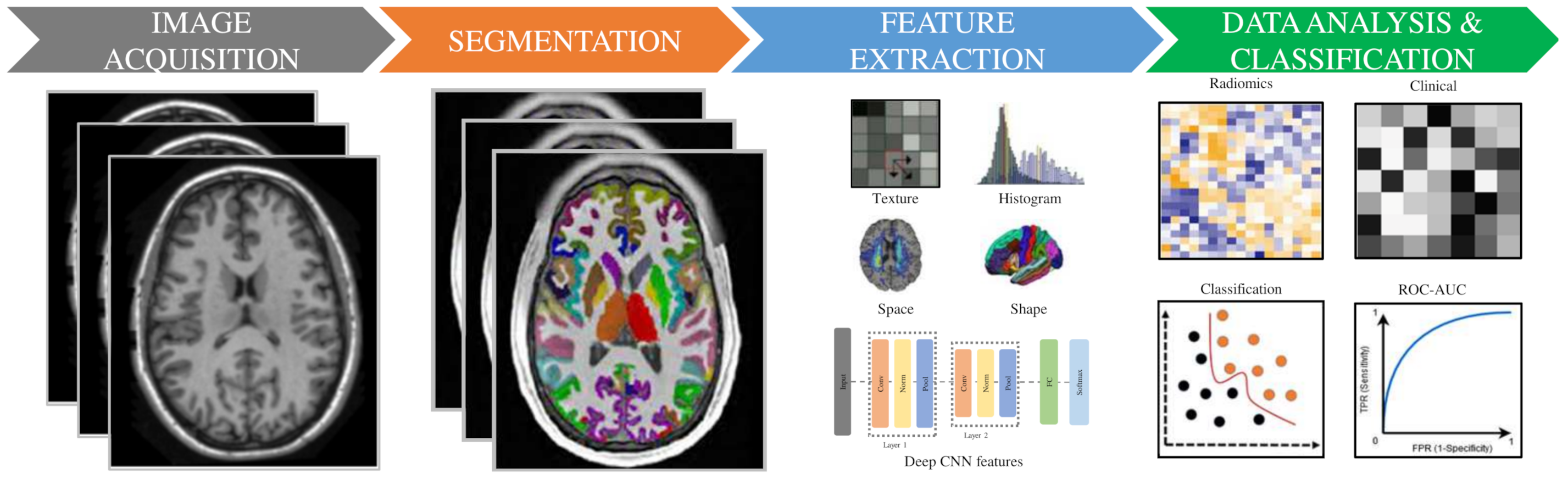
3. Radiomic Methodology
3.1. Image Acquisition and Preprocessing
3.2. Normalization/Standardization
3.3. Segmentation/Labeling
3.4. Features Extraction
3.5. Feature Selection
3.6. Statistical Analysis and Classification Models
4. Explainable Artificial Intelligence
5. Discussion
- MRI-based models for the diagnosis of ASD are more suitable for clinical trials than eye tracking and CT image analysis. MRI can provide more detail of the brain.
- The brain of ASD patients can be heterogeneous in many locations (e.g., hippocampus, amygdala, etc.). The variation could be captured by shape features (e.g., volume, thickness, etc.).
- Deep learning is still challenging to diagnose ASD patients due to the lack of benchmark datasets [156].
- XAI could be the solution as a diagnostic model for ASD. However, it needs more investigation in real-world scenarios.
- The public dataset needs to be continually expanded to avoid inappropriate studies due to insufficient data. In addition, it needs to be ensured that there is no error in results due to age, gender, etc. [157].
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Hyman, S.L.; Levy, S.E.; Myers, S.M. Identification, evaluation, and management of children with autism spectrum disorder. Pediatrics 2020, 145, e20193447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van’t Hof, M.; Tisseur, C.; van Berckelear-Onnes, I.; van Nieuwenhuyzen, A.; Daniels, A.M.; Deen, M.; Hoek, H.W.; Ester, W.A. Age at autism spectrum disorder diagnosis: A systematic review and meta-analysis from 2012 to 2019. Autism 2021, 25, 862–873. [Google Scholar] [CrossRef] [PubMed]
- Tao, L. Research Progress on Early Recognition of Childhood Autism Spectrum Disorder. China Matern. Child Health Care 2020, 35, 1554–1558. [Google Scholar]
- Toma, C. Genetic variation across phenotypic severity of autism. Trends Genet. 2020, 36, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Al-Ameen, S.A.; Tawfeeq, F.K.; Shihab, K.A. Estimation of Some Biochemical and Immunological Parameters of Autism Spectrum Disorder. Biochem. Cell. Arch. 2020, 20, 1601–1604. [Google Scholar]
- Haigh, S.M.; Keller, T.A.; Minshew, N.J.; Eack, S.M. Reduced white matter integrity and deficits in neuropsychological functioning in adults with autism spectrum disorder. Autism Res. 2020, 13, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Center, W.C.B.M. China Autism Education and Rehabilitation Industry Development Report; Beijing Normal University Press: Beijing, China, 2015. [Google Scholar]
- Lord, C. The future of autism: Global & local achievements & challenges. Indian J. Med. Res. 2020, 151, 263. [Google Scholar]
- Kim, S.H.; Bal, V.H.; Benrey, N.; Choi, Y.B.; Guthrie, W.; Colombi, C.; Lord, C. Variability in autism symptom trajectories using repeated observations from 14 to 36 months of age. J. Am. Acad. Child Adolesc. Psychiatry 2018, 57, 837–848. [Google Scholar] [CrossRef]
- Crowell, J.A.; Keluskar, J.; Gorecki, A. Parenting behavior and the development of children with autism spectrum disorder. Compr. Psychiatry 2019, 90, 21–29. [Google Scholar] [CrossRef]
- Arbanas, G. Diagnostic and Statistical Manual of Mental Disorders (DSM-5). Codas 2015, 51, 61–64. [Google Scholar]
- Lord, C.; Risi, S.; Lambrecht, L.; Cook, E.H.; Leventhal, B.L.; DiLavore, P.C.; Pickles, A.; Rutter, M. The Autism Diagnostic Observation Schedule—Generic: A standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 2000, 30, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Farooq, A.; Ahmed, S. Sociocultural Barriers to Early Diagnosis of Autism Spectrum Disorder. Life Sci. 2020, 1, 6. [Google Scholar] [CrossRef]
- Miller, L.E.; Dai, Y.G.; Fein, D.A.; Robins, D.L. Characteristics of toddlers with early versus later diagnosis of autism spectrum disorder. Autism 2021, 25, 416–428. [Google Scholar] [CrossRef] [PubMed]
- Jayawardana, Y.; Jaime, M.; Jayarathna, S. Analysis of temporal relationships between ASD and brain activity through EEG and machine learning. In Proceedings of the 2019 IEEE 20th International Conference on Information Reuse and Integration for Data Science (IRI), Los Angeles, CA, USA, 30 June–1 August 2019; pp. 151–158. [Google Scholar]
- Heunis, T.; Aldrich, C.; Peters, J.; Jeste, S.; Sahin, M.; Scheffer, C.; De Vries, P. Recurrence quantification analysis of resting state EEG signals in autism spectrum disorder—A systematic methodological exploration of technical and demographic confounders in the search for biomarkers. BMC Med. 2018, 16, 101. [Google Scholar] [CrossRef]
- Chaddad, A.; Desrosiers, C.; Toews, M. Multi-scale radiomic analysis of sub-cortical regions in MRI related to autism, gender and age. Sci. Rep. 2017, 7, 45639. [Google Scholar] [CrossRef]
- Vacas, J.; Antolí, A.; Sánchez-Raya, A.; Pérez-Dueñas, C. Eye Tracking Methodology for Studying Emotional Competence in Children with Autism Spectrum Disorder (ASD) and Specific Language Impairment (SLI): A Comparative Research Review. Rev. J. Autism Dev. Disord. 2021, 5, 1–15. [Google Scholar]
- Cederquist, G.Y.; Tchieu, J.; Callahan, S.J.; Ramnarine, K.; Ryan, S.; Zhang, C.; Rittenhouse, C.; Zeltner, N.; Chung, S.Y.; Zhou, T. A multiplex human pluripotent stem cell platform defines molecular and functional subclasses of autism-related genes. Cell Stem Cell 2020, 27, 35–49. [Google Scholar] [CrossRef]
- Soorya, L.; Kolevzon, A.; Zweifach, J.; Lim, T.; Dobry, Y.; Schwartz, L.; Frank, Y.; Wang, A.T.; Cai, G.; Parkhomenko, E. Prospective investigation of autism and genotype-phenotype correlations in 22q13 deletion syndrome and SHANK3 deficiency. Mol. Autism 2013, 4, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Noor, A.; Whibley, A.; Marshall, C.R.; Gianakopoulos, P.J.; Piton, A.; Carson, A.R.; Orlic-Milacic, M.; Lionel, A.C.; Sato, D.; Pinto, D. Disruption at the PTCHD1 Locus on Xp22. 11 in Autism spectrum disorder and intellectual disability. Sci. Transl. Med. 2010, 2, 49ra68. [Google Scholar] [CrossRef] [Green Version]
- Napolioni, V.; Lombardi, F.; Sacco, R.; Curatolo, P.; Manzi, B.; Alessandrelli, R.; Militerni, R.; Bravaccio, C.; Lenti, C.; Saccani, M. Family-based association study of ITGB3 in autism spectrum disorder and its endophenotypes. Eur. J. Hum. Genet. 2011, 19, 353–359. [Google Scholar] [CrossRef] [Green Version]
- Nordahl, C.W.; Mello, M.; Shen, A.M.; Shen, M.D.; Vismara, L.A.; Li, D.; Harrington, K.; Tanase, C.; Goodlin-Jones, B.; Rogers, S. Methods for acquiring MRI data in children with autism spectrum disorder and intellectual impairment without the use of sedation. J. Neurodev. Disord. 2016, 8, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, B.; Borle, N.C.; Greiner, R.; Brown, M.R. A general prediction model for the detection of ADHD and Autism using structural and functional MRI. PLoS ONE 2018, 13, e0194856. [Google Scholar] [CrossRef]
- Chaddad, A.; Kucharczyk, M.J.; Daniel, P.; Sabri, S.; Jean-Claude, B.J.; Niazi, T.; Abdulkarim, B. Radiomics in glioblastoma: Current status and challenges facing clinical implementation. Front. Oncol. 2019, 9, 374. [Google Scholar] [CrossRef] [Green Version]
- Chaddad, A.; Daniel, P.; Desrosiers, C.; Toews, M.; Abdulkarim, B. Novel radiomic features based on joint intensity matrices for predicting glioblastoma patient survival time. IEEE J. BioMed Health Inform. 2018, 23, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Daniel, P.; Niazi, T. Radiomics evaluation of histological heterogeneity using multiscale textures derived from 3D wavelet transformation of multispectral images. Front. Oncol. 2018, 8, 96. [Google Scholar] [CrossRef]
- Chaddad, A.; Desrosiers, C.; Toews, M. Radiomic analysis of multi-contrast brain MRI for the prediction of survival in patients with glioblastoma multiforme. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 4035–4038. [Google Scholar] [CrossRef]
- Chaddad, A.; Zinn, P.O.; Colen, R.R. Brain tumor identification using Gaussian Mixture Model features and Decision Trees classifier. In Proceedings of the 2014 48th Annual Conference on Information Sciences and Systems (CISS), Princeton, NJ, USA, 19–21 March 2014; pp. 1–4. [Google Scholar]
- Chaddad, A.; Daniel, P.; Sabri, S.; Desrosiers, C.; Abdulkarim, B. Integration of radiomic and multi-omic analyses predicts survival of newly diagnosed IDH1 wild-type glioblastoma. Cancers 2019, 11, 1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaddad, A.; Desrosiers, C.; Abdulkarim, B.; Niazi, T. Predicting the Gene Status and Survival Outcome of Lower Grade Glioma Patients With Multimodal MRI Features. IEEE Access 2019, 7, 75976–75984. [Google Scholar] [CrossRef]
- Chaddad, A.; Desrosiers, C.; Toews, M.; Abdulkarim, B. Predicting survival time of lung cancer patients using radiomic analysis. Oncotarget 2017, 8, 104393. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, F.; Zhang, J.; Chaddad, A.; Guo, F.; Zhang, W.; Zhang, J.; Evans, A. AutoEncoder for Neuroimage. In Proceedings of the International Conference on Database and Expert Systems Applications, Linz, Austria, 27–30 September 2021; pp. 84–90. [Google Scholar]
- Chaddad, A.; Desrosiers, C.; Niazi, T. Deep radiomic analysis of MRI related to Alzheimer’s disease. IEEE Access 2018, 6, 58213–58221. [Google Scholar] [CrossRef]
- Chaddad, A.; Tanougast, C. Quantitative evaluation of robust skull stripping and tumor detection applied to axial MR images. Brain Inform. 2016, 3, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Zhu, Z.; Cao, M.; Liu, B.; Guo, J.; Wan, G. SMRI study of early brain overdevelopment in children with autism. Magn. Reson. Imaging 2020, 11, 264–269. [Google Scholar]
- Xiuyan, W. Prediction Research on Autism Based on Structural Magnetic Resonance Imaging. Ph.D. Thesis, Beijing Jiaotong University, Beijing, China, 2018. [Google Scholar]
- Smith, E.; Thurm, A.; Greenstein, D.; Farmer, C.; Swedo, S.; Giedd, J.; Raznahan, A. Cortical thickness change in autism during early childhood. Hum. Brain Mapp. 2016, 37, 2616–2629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolff, J.J.; Jacob, S.; Elison, J.T. The journey to autism: Insights from neuroimaging studies of infants and toddlers. Dev. Psychopathol. 2018, 30, 479–495. [Google Scholar] [CrossRef]
- Lainhart, J.E.; Piven, J.; Wzorek, M.; Landa, R.; Santangelo, S.L.; Coon, H.; Folstein, S.E. Macrocephaly in children and adults with autism. J. Am. Acad. Child Adolesc. Psychiatry 1997, 36, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Sacco, R.; Gabriele, S.; Persico, A.M. Head circumference and brain size in autism spectrum disorder: A systematic review and meta-analysis. Psychiatry Res. Neuroimaging 2015, 234, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Elison, J.T.; Paterson, S.J.; Wolff, J.J.; Reznick, J.S.; Sasson, N.J.; Gu, H.; Botteron, K.N.; Dager, S.R.; Estes, A.M.; Evans, A.C. White matter microstructure and atypical visual orienting in 7-month-olds at risk for autism. Am. J. Psychiatry 2013, 170, 899–908. [Google Scholar] [CrossRef] [Green Version]
- Hazlett, H.C.; Gu, H.; Munsell, B.C.; Kim, S.H.; Styner, M.; Wolff, J.J.; Elison, J.T.; Swanson, M.R.; Zhu, H.; Botteron, K.N. Early brain development in infants at high risk for autism spectrum disorder. Nature 2017, 542, 348–351. [Google Scholar] [CrossRef]
- Nassar, N.; Dixon, G.; Bourke, J.; Bower, C.; Glasson, E.; De Klerk, N.; Leonard, H. Autism spectrum disorders in young children: Effect of changes in diagnostic practices. Int. J. Epidemiol. 2009, 38, 1245–1254. [Google Scholar] [CrossRef] [PubMed]
- Wang, R. Morphological Brain Network Research on Childhood Autism. Ph.D. Thesis, University of Electronic Science and Technology of China, Chengdu, China.
- Shen, M.D.; Kim, S.H.; McKinstry, R.C.; Gu, H.; Hazlett, H.C.; Nordahl, C.W.; Emerson, R.W.; Shaw, D.; Elison, J.T.; Swanson, M.R.; et al. Increased extra-axial cerebrospinal fluid in high-risk infants who later develop autism. Biol. Psychiatry 2017, 82, 186–193. [Google Scholar] [CrossRef]
- Wolff, J.J.; Gerig, G.; Lewis, J.D.; Soda, T.; Styner, M.A.; Vachet, C.; Botteron, K.N.; Elison, J.T.; Dager, S.R.; Estes, A.M. Altered corpus callosum morphology associated with autism over the first 2 years of life. Brain 2015, 138, 2046–2058. [Google Scholar] [CrossRef] [Green Version]
- Wolff, J.J.; Swanson, M.R.; Elison, J.T.; Gerig, G.; Pruett, J.R.; Styner, M.A.; Vachet, C.; Botteron, K.N.; Dager, S.R.; Estes, A.M. Neural circuitry at age 6 months associated with later repetitive behavior and sensory responsiveness in autism. Mol. Autism 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piven, J.; Saliba, K.; Bailey, J.; Arndt, S. An MRI study of autism: The cerebellum revisited. Neurology 1997, 49, 546–551. [Google Scholar] [CrossRef]
- Courchesne, E.; Karns, C.; Davis, H.; Ziccardi, R.; Carper, R.; Tigue, Z.; Chisum, H.; Moses, P.; Pierce, K.; Lord, C. Unusual brain growth patterns in early life in patients with autistic disorder: An MRI study. Neurology 2001, 57, 245–254. [Google Scholar] [CrossRef]
- Nordahl, C.W.; Iosif, A.M.; Young, G.S.; Hechtman, A.; Heath, B.; Lee, J.K.; Libero, L.; Reinhardt, V.P.; Winder-Patel, B.; Amaral, D.G.; et al. High Psychopathology Subgroup in Young Children With Autism: Associations With Biological Sex and Amygdala Volume. J. Am. Acad. Child Adolesc. Psychiatry 2020, 59, 1353–1363.e2. [Google Scholar] [CrossRef] [Green Version]
- Reinhardt, V.P.; Iosif, A.M.; Libero, L.; Heath, B.; Rogers, S.J.; Ferrer, E.; Nordahl, C.; Ghetti, S.; Amaral, D.; Solomon, M. Understanding hippocampal development in young children with autism spectrum disorder. J. Am. Acad. Child Adolesc. Psychiatry 2020, 59, 1069–1079. [Google Scholar] [CrossRef]
- Li, G.; Chen, M.H.; Li, G.; Wu, D.; Sun, Q.; Shen, D.; Wang, L. A Preliminary Volumetric Mri Study of Amygdala and Hippocampal Subfields in Autism During Infancy. IEEE Int. Symp. Biomed. Imaging 2019, 2019, 1052–1056. [Google Scholar]
- Chaddad, A.; Desrosiers, C.; Hassan, L.; Tanougast, C. Hippocampus and amygdala radiomic biomarkers for the study of autism spectrum disorder. BMC Neurosci. 2017, 18, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosl, W.J.; Tager-Flusberg, H.; Nelson, C.A. EEG analytics for early detection of autism spectrum disorder: A data-driven approach. Sci. Rep. 2018, 8, 1–20. [Google Scholar] [CrossRef]
- Vicnesh, J.; Wei, J.K.E.; Oh, S.L.; Arunkumar, N.; Abdulhay, E.; Ciaccio, E.J.; Acharya, U.R. Autism spectrum disorder diagnostic system using HOS bispectrum with EEG signals. Int. J. Environ. Res. Public Health 2020, 17, 971. [Google Scholar]
- Peya, Z.J.; Akhand, M.; Srabonee, J.F.; Siddique, N. EEG Based Autism Detection Using CNN Through Correlation Based Transformation of Channels’ Data. In Proceedings of the 2020 IEEE Region 10 Symposium (TENSYMP), Dhaka, Bangladesh, 5–7 June 2020; pp. 1278–1281. [Google Scholar]
- Moriuchi, J.M.; Klin, A.; Jones, W. Mechanisms of diminished attention to eyes in autism. Am. J. Psychiatry 2017, 174, 26–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pino, M.C.; Vagnetti, R.; Valenti, M.; Mazza, M. Comparing virtual vs. real faces expressing emotions in children with autism: An eye-tracking study. Educ. Inf. Technol. 2021, 1–16. [Google Scholar] [CrossRef]
- Albajara Sáenz, A.; Van Schuerbeek, P.; Baijot, S.; Septier, M.; Deconinck, N.; Defresne, P.; Delvenne, V.; Passeri, G.; Raeymaekers, H.; Slama, H. Disorder-specific brain volumetric abnormalities in attention-deficit/hyperactivity disorder relative to autism spectrum disorder. PLoS ONE 2020, 15, e0241856. [Google Scholar] [CrossRef]
- Mellema, C.; Treacher, A.; Nguyen, K.; Montillo, A. Multiple Deep Learning Architectures Achieve Superior Performance Diagnosing Autism Spectrum Disorder Using Features Previously Extracted From Structural And Functional Mri. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1891–1895. [Google Scholar] [CrossRef]
- Wang, W.; Wu, X.; Yuan, X.; Gao, Z. An experiment-based review of low-light image enhancement methods. IEEE Access 2020, 8, 87884–87917. [Google Scholar] [CrossRef]
- Salem, N.; Malik, H.; Shams, A. Medical image enhancement based on histogram algorithms. Procedia Comput. Sci. 2019, 163, 300–311. [Google Scholar] [CrossRef]
- Bao, S.; Bermudez, C.; Huo, Y.; Parvathaneni, P.; Rodriguez, W.; Resnick, S.M.; D’Haese, P.F.; McHugo, M.; Heckers, S.; Dawant, B.M. Registration-based image enhancement improves multi-atlas segmentation of the thalamic nuclei and hippocampal subfields. Magn. Reson. Imaging 2019, 59, 143–152. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, Z.; Li, H.; Wu, X.; Qiao, S.; Sun, J. A new approach for medical image enhancement based on luminance-level modulation and gradient modulation. Biomed. Signal Process. Control 2019, 48, 189–196. [Google Scholar] [CrossRef]
- Yugander, P.; Tejaswini, C.; Meenakshi, J.; Varma, B.S.; Jagannath, M. MR image enhancement using adaptive weighted mean filtering and homomorphic filtering. Procedia Comput. Sci. 2020, 167, 677–685. [Google Scholar] [CrossRef]
- Sagheer, S.V.M.; George, S.N. A review on medical image denoising algorithms. Biomed. Signal Process. Control 2020, 61, 102036. [Google Scholar] [CrossRef]
- Park, W.J.; Schauder, K.B.; Zhang, R.; Bennetto, L.; Tadin, D. High internal noise and poor external noise filtering characterize perception in autism spectrum disorder. Sci. Rep. 2017, 7, 17584. [Google Scholar] [CrossRef] [Green Version]
- Yankowitz, L.D.; Herrington, J.D.; Yerys, B.E.; Pereira, J.A.; Pandey, J.; Schultz, R.T. Evidence against the “normalization” prediction of the early brain overgrowth hypothesis of autism. Mol. Autism 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Hoeksma, M.R.; Kenemans, J.L.; Kemner, C.; van Engeland, H. Variability in spatial normalization of pediatric and adult brain images. Clin. Neurophysiol. 2005, 116, 1188–1194. [Google Scholar] [CrossRef]
- Delisle, P.L.; Anctil-Robitaille, B.; Desrosiers, C.; Lombaert, H. Realistic Image Normalization for Multi-Domain Segmentation. Med. Image Anal. 2021, 74, 102191. [Google Scholar] [CrossRef]
- Ahammed, M.S.; Niu, S.; Ahmed, M.R.; Dong, J.; Gao, X.; Chen, Y. DarkASDNet: Classification of ASD on Functional MRI Using Deep Neural Network. Front. Neuroinform. 2021, 20. [Google Scholar] [CrossRef]
- Dekhil, O.; Ali, M.; Haweel, R.; Elnakib, Y.; Ghazal, M.; Hajjdiab, H.; Fraiwan, L.; Shalaby, A.; Soliman, A.; Mahmoud, A. A comprehensive framework for differentiating autism spectrum disorder from neurotypicals by fusing structural MRI and resting state functional MRI. In Seminars in Pediatric Neurology; Elsevier: Amsterdam, The Netherlands, 2020; Volume 34, p. 100805. [Google Scholar]
- Mendes, S.L.; Pinaya, W.H.L.; Pan, P.; Sato, J.R. Estimating Gender and Age from Brain Structural MRI of Children and Adolescents: A 3D Convolutional Neural Network Multitask Learning Model. Comput. Intell. Neurosci. 2021, 2021, 5550914. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, J.; Friston, K.J. Voxel-based morphometry—The methods. Neuroimage 2000, 11, 805–821. [Google Scholar] [CrossRef] [Green Version]
- Haweel, R.; Shalaby, A.; Mahmoud, A.; Seada, N.; Ghoniemy, S.; Ghazal, M.; Casanova, M.F.; Barnes, G.N.; El-Baz, A. A robust DWT–CNN-based CAD system for early diagnosis of autism using task-based fMRI. Med. Phys. 2021, 48, 2315–2326. [Google Scholar] [CrossRef]
- K, D.; Murthy Oruganti, V.R. A Machine Learning Approach for Diagnosing Neurological Disorders using Longitudinal Resting-State fMRI. In Proceedings of the 2021 11th International Conference on Cloud Computing, Data Science Engineering (Confluence), Noida, India, 28–29 January 2021; pp. 494–499. [Google Scholar] [CrossRef]
- Dekhil, O.; Ali, M.; El-Nakieb, Y.; Shalaby, A.; Soliman, A.; Switala, A.; Mahmoud, A.; Ghazal, M.; Hajjdiab, H.; Casanova, M.F. A personalized autism diagnosis CAD system using a fusion of structural MRI and resting-state functional MRI data. Front. Psychiatry 2021, 10, 392. [Google Scholar] [CrossRef]
- Yang, X.; Islam, M.S.; Khaled, A.M.A. Functional connectivity magnetic resonance imaging classification of autism spectrum disorder using the multisite ABIDE dataset. In Proceedings of the 2019 IEEE EMBS International Conference on Biomedical Health Informatics (BHI), Chicago, IL, USA, 19–22 May 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Gao, J.; Chen, M.; Li, Y.; Gao, Y.; Li, Y.; Cai, S.; Wang, J. Multisite Autism Spectrum Disorder Classification Using Convolutional Neural Network Classifier and Individual Morphological Brain Networks. Front. Neurosci. 2021, 14, 1473. [Google Scholar] [CrossRef]
- Squarcina, L.; Nosari, G.; Marin, R.; Castellani, U.; Bellani, M.; Bonivento, C.; Fabbro, F.; Molteni, M.; Brambilla, P. Automatic classification of autism spectrum disorder in children using cortical thickness and support vector machine. Brain Behav. 2021, 11, e2238. [Google Scholar] [CrossRef] [PubMed]
- Heinsfeld, A.S.; Franco, A.R.; Craddock, R.C.; Buchweitz, A.; Meneguzzi, F. Identification of autism spectrum disorder using deep learning and the ABIDE dataset. Neuroimage Clin. 2018, 17, 16–23. [Google Scholar] [CrossRef]
- Alvarez-Jimenez, C.; Munera Garzon, N.; Zuluaga, M.A.; Velasco, N.F.; Romero, E. Autism spectrum disorder characterization in children by capturing local regional brain changes in MRI. Med. Phys. 2020, 47, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Madine, M.; Rekik, I.; Werghi, N. Diagnosing Autism Using T1-W MRI With Multi-Kernel Learning and Hypergraph Neural Network. In Proceedings of the 2020 IEEE International Conference on Image Processing (ICIP), Virtual, 25–28 September 2020; pp. 438–442. [Google Scholar] [CrossRef]
- Wang, C.; Xiao, Z.; Wu, J. Functional connectivity-based classification of autism and control using SVM-RFECV on rs-fMRI data. Phys. Medica 2019, 65, 99–105. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, J.; Li, Y.; Shi, J.; Zou, Y.; Guo, H.; Li, Y.; Yao, Z.; Wang, Y.; Hu, B. A novel pipeline leveraging surface-based features of small subcortical structures to classify individuals with autism spectrum disorder. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 104, 109989. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Zhang, J.; Wu, X. An fMRI feature selection method based on a minimum spanning tree for identifying patients with autism. Symmetry 2020, 12, 1995. [Google Scholar] [CrossRef]
- You, Y.; Liu, H.; Zhang, S.; Shao, L. Classification of Autism Based on fMRI Data with Feature-Fused Convolutional Neural Network. In Cyberspace Data and Intelligence, and Cyber-Living, Syndrome, and Health; Springer: Berlin/Heidelberg, Germany, 2020; pp. 77–88. [Google Scholar]
- Byeon, K.; Kwon, J.; Hong, J.; Park, H. Artificial Neural Network Inspired by Neuroimaging Connectivity: Application in Autism Spectrum Disorder. In Proceedings of the 2020 IEEE International Conference on Big Data and Smart Computing (BigComp), Busan, Korea, 19–22 February 2020; pp. 575–578. [Google Scholar] [CrossRef]
- Soeiro, J.; Dias, L.; Silva, A.; Tomé, A. Radiomic Analysis of Brain MRI: A Case Study in Autism Spectrum Disorder. Available online: https://recpad2021.uevora.pt/wp-content/uploads/2020/10/RECPAD_2020_paper_9.pdf (accessed on 23 August 2021).
- Ma, X.; Wang, X.H.; Li, L. Identifying individuals with autism spectrum disorder based on the principal components of whole-brain phase synchrony. Neurosci. Lett. 2021, 742, 135519. [Google Scholar] [CrossRef]
- Husna, R.N.S.; Syafeeza, A.; Hamid, N.A.; Wong, Y.; Raihan, R.A. Functional Magnetic Resonance Imaging for Autism Spectrum Disorder Detection Using Deep Learning. J. Teknol. 2021, 83, 45–52. [Google Scholar] [CrossRef]
- Sherkatghanad, Z.; Akhondzadeh, M.; Salari, S.; Zomorodi-Moghadam, M.; Abdar, M.; Acharya, U.R.; Khosrowabadi, R.; Salari, V. Automated detection of autism spectrum disorder using a convolutional neural network. Front. Neurosci. 2020, 13, 1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raki, M.; Cabezas, M.; Kushibar, K.; Oliver, A.; Llad, X. Improving the detection of autism spectrum disorder by combining structural and functional MRI information. Neuroimage Clin. 2020, 25, 102181. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.M.; Gallo, S.; Cerliani, L.; Zhutovsky, P.; El-Gazzar, A.; van Wingen, G. Classifying autism spectrum disorder using the temporal statistics of resting-state functional MRI data with 3D convolutional neural networks. Front. Psychiatry 2020, 11, 440. [Google Scholar] [CrossRef]
- Shrivastava, S.; Mishra, U.; Singh, N.; Chandra, A.; Verma, S. Control or Autism—Classification using Convolutional Neural Networks on Functional MRI. In Proceedings of the 2020 11th International Conference on Computing, Communication and Networking Technologies (ICCCNT), Kharagapur, India, 1–3 July 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Liu, J.; Sheng, Y.; Lan, W.; Guo, R.; Wang, Y.; Wang, J. Improved ASD classification using dynamic functional connectivity and multi-task feature selection. Pattern Recognit. Lett. 2020, 138, 82–87. [Google Scholar] [CrossRef]
- Zhang, Z.; Zheng, W. The Discriminative Power of White Matter Microstructures for Autism Diagnosis. IFAC-PapersOnLine 2020, 53, 446–451. [Google Scholar] [CrossRef]
- Bayram, M.A.; İlyas, Ö.; Temurtaş, F. Deep Learning Methods for Autism Spectrum Disorder Diagnosis Based on fMRI Images. Sak. Univ. J. Comput. Inf. Sci. 2021, 4, 142–155. [Google Scholar]
- Almuqhim, F.; Saeed, F. ASD-SAENet: A Sparse Autoencoder, and Deep-Neural Network Model for Detecting Autism Spectrum Disorder (ASD) Using fMRI Data. Front. Comput. Neurosci. 2021, 15, 27. [Google Scholar] [CrossRef]
- Ali, M.T.; Elnakieb, Y.A.; Shalaby, A.; Mahmoud, A.; Switala, A.; Ghazal, M.; Khelifi, A.; Fraiwan, L.; Barnes, G.; El-Baz, A. Autism Classification Using SMRI: A Recursive Features Selection Based on Sampling from Multi-Level High Dimensional Spaces. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Virtual, 13–16 April 2021; pp. 267–270. [Google Scholar] [CrossRef]
- Lu, P.; Li, X.; Hu, L.; Lu, L. Integrating genomic and resting State fMRI for efficient autism spectrum disorder classification. InMultimedia Tools and Applications; Springer: Berlin/Heidelberg, Germany, 2021; pp. 1–12. [Google Scholar]
- Chaitra, N.; Vijaya, P.; Deshpande, G. Diagnostic prediction of autism spectrum disorder using complex network measures in a machine learning framework. Biomed. Signal Process. Control 2020, 62, 102099. [Google Scholar] [CrossRef]
- Haweel, R.; Shalaby, A.; Mahmoud, A.; Ghazal, M.; Seada, N.; Ghoniemy, S.; Barnes, G.; El-Baz, A. A Novel Dwt-Based Discriminant Features Extraction From Task-Based Fmri: An Asd Diagnosis Study Using Cnn. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 28–31 March 2021; pp. 196–199. [Google Scholar] [CrossRef]
- Al-Hiyali, M.I.; Yahya, N.; Faye, I.; Khan, Z.; Alsaih, K. Classification of BOLD FMRI Signals using Wavelet Transform and Transfer Learning for Detection of Autism Spectrum Disorder. In Proceedings of the 2020 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES), Langkawi, Malaysia, 1–3 March 2021; pp. 94–98. [Google Scholar] [CrossRef]
- Li, Z. Segmentation and Recognition of MRI Images of the Brain. Ph.D. Thesis, Jilin University, Changchun, China, 2020. [Google Scholar]
- Yang, X.; Zhao, X.; Tjio, G.; Chen, C.; Wang, L.; Wen, B.; Su, Y. Opencc—An open Benchmark data set for Corpus Callosum Segmentation and Evaluation. In Proceedings of the 2020 IEEE International Conference on Image Processing (ICIP), Virtual, 25–28 October 2020; pp. 3020–3024. [Google Scholar]
- Pieper, S.; Halle, M.; Kikinis, R. 3D Slicer. In Proceedings of the 2004 2nd IEEE International Symposium on Biomedical imaging: Nano to Macro (IEEE Cat No. 04EX821), Arlington, VA, USA, 18 April 2004; pp. 632–635. [Google Scholar]
- Dolz, J.; Desrosiers, C.; Ayed, I.B. 3D fully convolutional networks for subcortical segmentation in MRI: A large-scale study. NeuroImage 2018, 170, 456–470. [Google Scholar] [CrossRef] [Green Version]
- Fischl, B. FreeSurfer. Neuroimage 2012, 62, 774–781. [Google Scholar] [CrossRef] [Green Version]
- Hegarty, J.P.; II, L.C.L.; Raman, M.M.; Hallmayer, J.F.; Cleveland, S.C.; Wolke, O.N.; Phillips, J.M.; Reiss, A.L.; Hardan, A.Y. Genetic and environmental influences on corticostriatal circuits in twins with autism. J. Psychiatry Neurosci. JPN 2020, 45, 188. [Google Scholar] [CrossRef]
- Wu, C.; Zheng, H.; Wu, H.; Tang, Y.; Li, F.; Wang, D. Age-related Brain Morphological Alteration of Medication-naive Boys With High Functioning Autism. Acad. Radiol. 2020. [Google Scholar] [CrossRef]
- Kumar, G.; Bhatia, P.K. A Detailed Review of Feature Extraction in Image Processing Systems. In Proceedings of the 2014 Fourth International Conference on Advanced Computing Communication Technologies, Rohtak, India, 8–9 February 2014; pp. 5–12. [Google Scholar] [CrossRef]
- Subbaraju, V.; Suresh, M.B.; Sundaram, S.; Narasimhan, S. Identifying differences in brain activities and an accurate detection of autism spectrum disorder using resting state functional-magnetic resonance imaging: A spatial filtering approach. Med. Image Anal. 2017, 35, 375–389. [Google Scholar] [CrossRef]
- El-Gazzar, A.; Quaak, M.; Cerliani, L.; Bloem, P.; van Wingen, G.; Thomas, R.M. A hybrid 3dcnn and 3dc-lstm based model for 4d spatio-temporal fMRI data: An abide autism classification study. In OR 2.0 Context-Aware Operating Theaters and Machine Learning in Clinical Neuroimaging; Springer: Berlin/Heidelberg, Germany, 2019; pp. 95–102. [Google Scholar]
- Kou, Q. Research on Image Texture Feature Extraction Method Based on Principal Curvature. Ph.D. Thesis, China University of Mining and Technology (Jiangsu), Xuzhou, China, 2019. [Google Scholar]
- Zebari, R.; Abdulazeez, A.; Zeebaree, D.; Zebari, D.; Saeed, J. A comprehensive review of dimensionality reduction techniques for feature selection and feature extraction. J. Appl. Sci. Technol. Trends 2020, 1, 56–70. [Google Scholar] [CrossRef]
- Urbanowicz, R.J.; Meeker, M.; La Cava, W.; Olson, R.S.; Moore, J.H. Relief-based feature selection: Introduction and review. J. Biomed. Inform. 2018, 85, 189–203. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.W.; Tsai, Y.H.; Chang, F.R.; Lin, W.C. Ensemble feature selection in medical datasets: Combining filter, wrapper, and embedded feature selection results. Expert Syst. 2020, 37, e12553. [Google Scholar] [CrossRef]
- Bommert, A.; Sun, X.; Bischl, B.; Rahnenführer, J.; Lang, M. Benchmark for filter methods for feature selection in high-dimensional classification data. Comput. Stat. Data Anal. 2020, 143, 106839. [Google Scholar] [CrossRef]
- Solorio-Fernández, S.; Carrasco-Ochoa, J.A.; Martínez-Trinidad, J.F. A review of unsupervised feature selection methods. Artif. Intell. Rev. 2020, 53, 907–948. [Google Scholar] [CrossRef]
- Rahman, M.; Usman, O.L.; Muniyandi, R.C.; Sahran, S.; Mohamed, S.; Razak, R.A. A Review of machine learning methods of feature selection and classification for autism spectrum disorder. Brain Sci. 2020, 10, 949. [Google Scholar] [CrossRef] [PubMed]
- Pavithra, V.; Jayalakshmi, V. Review of Feature Selection Techniques for Predicting Diseases. In Proceedings of the 2020 5th International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 10–12 June 2020; pp. 1213–1217. [Google Scholar]
- Nielsen, A.N.; Barch, D.M.; Petersen, S.E.; Schlaggar, B.L.; Greene, D.J. Machine learning with neuroimaging: Evaluating its applications in psychiatry. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2020, 5, 791–798. [Google Scholar] [CrossRef]
- Ronicko, J.F.A.; Thomas, J.; Thangavel, P.; Koneru, V.; Langs, G.; Dauwels, J. Diagnostic classification of autism using resting-state fMRI data improves with full correlation functional brain connectivity compared to partial correlation. J. Neurosci. Methods 2020, 345, 108884. [Google Scholar] [CrossRef]
- Haweel, R.; Dekhil, O.; Shalaby, A.; Mahmoud, A.; Ghazal, M.; Khalil, A.; Keynton, R.; Barnes, G.; El-Baz, A. A novel framework for grading autism severity using task-based fmri. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1404–1407. [Google Scholar]
- Mostafa, S.; Tang, L.; Wu, F.X. Diagnosis of autism spectrum disorder based on eigenvalues of brain networks. IEEE Access 2019, 7, 128474–128486. [Google Scholar] [CrossRef]
- Huang, Z.A.; Zhu, Z.; Yau, C.H.; Tan, K.C. Identifying autism spectrum disorder from resting-state fMRI using deep belief network. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 2847–2861. [Google Scholar] [CrossRef]
- Abdullah, A.A.; Rijal, S.; Dash, S.R. Evaluation on Machine Learning Algorithms for Classification of Autism Spectrum Disorder (ASD). J. Phys. Conf. Ser. 2019, 1372, 012052. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, J.; Dvornek, N.C.; Zhao, Q.; Li, X.; Ventola, P.; Duncan, J.S. Prediction of treatment outcome for autism from structure of the brain based on sure independence screening. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 404–408. [Google Scholar]
- Chen, H.; Duan, X.; Liu, F.; Lu, F.; Ma, X.; Zhang, Y.; Uddin, L.Q.; Chen, H. Multivariate classification of autism spectrum disorder using frequency-specific resting-state functional connectivity—A multi-center study. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2016, 64, 1–9. [Google Scholar] [CrossRef]
- Jahedi, A. Novel Random Forest Methods and Algorithms for Autism Spectrum Disorders Research. Ph.D. Thesis, The Claremont Graduate University, Claremont, CA, USA, 2020. [Google Scholar]
- Grossard, C.; Dapogny, A.; Cohen, D.; Bernheim, S.; Juillet, E.; Hamel, F.; Hun, S.; Bourgeois, J.; Pellerin, H.; Serret, S. Children with autism spectrum disorder produce more ambiguous and less socially meaningful facial expressions: An experimental study using random forest classifiers. Mol. Autism 2020, 11, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Amit, Y.; Geman, D. Shape quantization and recognition with randomized trees. Neural Comput. 1997, 9, 1545–1588. [Google Scholar] [CrossRef] [Green Version]
- Berrar, D. Cross-Validation. In Encyclopedia of Bioinformatics and Computational Biology; Academic Press: Waltham, MA, USA, 2019; pp. 542–545. [Google Scholar]
- Choi, J.Y. Radiomics and deep learning in clinical imaging: What should we do? Nucl. Med. Mol. Imaging 2018, 52, 89–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aickin, M.; Gensler, H. Adjusting for multiple testing when reporting research results: The Bonferroni vs. Holm methods. Am. J. Public Health 1996, 86, 726–728. [Google Scholar] [CrossRef] [Green Version]
- Raj, S.; Masood, S. Analysis and detection of autism spectrum disorder using machine learning techniques. Procedia Comput. Sci. 2020, 167, 994–1004. [Google Scholar] [CrossRef]
- Dominic, N.; Cenggoro, T.W.; Budiarto, A.; Pardamean, B. Transfer learning using inception-ResNet-v2 model to the augmented neuroimages data for autism spectrum disorder classification. Commun. Math. Biol. Neurosci. 2021. Available online: http://www.scik.org/index.php/cmbn/article/view/5565 (accessed on 23 August 2021).
- Goldfeld, Z.; Polyanskiy, Y. The information bottleneck problem and its applications in machine learning. IEEE J. Sel. Areas Inf. Theory 2020, 1, 19–38. [Google Scholar] [CrossRef]
- Mateen, B.A.; Liley, J.; Denniston, A.K.; Holmes, C.C.; Vollmer, S.J. Improving the quality of machine learning in health applications and clinical research. Nat. Mach. Intell. 2020, 2, 554–556. [Google Scholar] [CrossRef]
- Chen, X.; Wang, X.; Zhang, K.; Zhang, R.; Fung, K.M.; Thai, T.C.; Moore, K.; Mannel, R.S.; Liu, H.; Zheng, B. Recent advances and clinical applications of deep learning in medical image analysis. arXiv 2021, arXiv:2105.13381. [Google Scholar]
- Castelvecchi, D. Can we open the black box of AI? Nat. News 2016, 538, 20. [Google Scholar] [CrossRef] [Green Version]
- Fellous, J.M.; Sapiro, G.; Rossi, A.; Mayberg, H.; Ferrante, M. Explainable Artificial Intelligence for Neuroscience: Behavioral Neurostimulation. Front. Neurosci. 2019, 13, 1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, M.; Webster, P.J.; Li, X.; Wang, S. Deep Neural Network Reveals the World of Autism From a First-Person Perspective. Autism Res. 2021, 14, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Hassan, L.; Desrosiers, C. Deep Radiomic Analysis for Predicting Coronavirus Disease 2019 in Computerized Tomography and X-ray Images. IEEE Trans. Neural Netw. Learn. Syst. 2021, 1–9. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Taylor, G.W.; Fergus, R. Adaptive deconvolutional networks for mid and high level feature learning. In Proceedings of the 2011 International Conference on Computer Vision, Barcelona, Spain, 6–13 November 2011; pp. 2018–2025. [Google Scholar]
- Zhang, Q.; Wu, Y.N.; Zhu, S.C. Interpretable convolutional neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8827–8836. [Google Scholar]
- Chaddad, A.; Toews, M.; Desrosiers, C.; Niazi, T. Deep Radiomic Analysis Based on Modeling Information Flow in Convolutional Neural Networks. IEEE Access 2019, 7, 97242–97252. [Google Scholar] [CrossRef]
- Mayor-Torres, J.M.; Ravanelli, M.; Medina-DeVilliers, S.E.; Lerner, M.D.; Riccardi, G. Interpretable SincNet-based Deep Learning for Emotion Recognition from EEG brain activity. arXiv 2021, arXiv:2107.10790. [Google Scholar]
- Biswas, M.; Kaiser, M.S.; Mahmud, M.; Al Mamun, S.; Hossain, M.; Rahman, M.A. An XAI Based Autism Detection: The Context Behind the Detection. In Proceedings of the International Conference on Brain Informatics, Virtual, 17–19 September 2021; pp. 448–459. [Google Scholar]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4768–4777. [Google Scholar]
- Tjoa, E.; Guan, C. A Survey on Explainable Artificial Intelligence (XAI): Toward Medical XAI. IEEE Trans. Neural Netw. Learn. Syst. 2020, 1–21. [Google Scholar] [CrossRef]
- Parekh, V.S.; Jacobs, M.A. Radiomic Synthesis Using Deep Convolutional Neural Networks. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1114–1117. [Google Scholar] [CrossRef] [Green Version]
- Cui, L.B.; Xu, X.; Cao, F. Building the precision medicine for mental disorders via radiomics/machine learning and neuroimaging. Front. Neurosci. 2021, 15, 650. [Google Scholar] [CrossRef] [PubMed]
- Thabtah, F. Machine learning in autistic spectrum disorder behavioral research: A review and ways forward. Inform. Health Soc. Care 2019, 44, 278–297. [Google Scholar] [CrossRef]
- de Belen, R.A.J.; Bednarz, T.; Sowmya, A.; Del Favero, D. Computer vision in autism spectrum disorder research: A systematic review of published studies from 2009 to 2019. Transl. Psychiatry 2020, 10, 1–20. [Google Scholar] [CrossRef]
| Work | Data Source | Cases Number | Data Type | FEM | Classifer Model | Acc | Sen | Spec | AUC |
|---|---|---|---|---|---|---|---|---|---|
| [76] | FSL | 50 ASD and 50 HC | TfMRI | SPF | DWT-CNN | 80% | 84% | 76% | - |
| [77] | ABIDE-I+II | 23 ASD and 15 HC | Rs-fMRI | SPF | SVM | 80.76% | - | - | - |
| [78] | NDAR | 185 subjects | sMRI-fMRI | SF | RF | 80.8% | 84.9% | 79.2% | 81.92% |
| [79] | ABIDE | 505 ASD and 530 HC | Rs-fMRI | SPF | Ridge Return | 71.98% | - | - | - |
| [80] | ABIDE | 518 ASD and 567 HC | rs-fMRI | SF | CNN | 71.8% | 81.25% | 68.75% | 67% |
| [81] | Private | 40 ASD and 36 HC | MRI | SF | SVM | 84.2% | 80% | 88.9% | - |
| [82] | ABIDE-I | 505 ASD and 530 HC | rs-fMRI | OF | DNN | 70% | 74% | 63% | - |
| [24] | ADHD-200 | 279 ASD and 279 HC | fMRI | TF | SVM | 64.91% | 44.16% | 81.91% | - |
| [83] | ABIDE-I+II | 76 ASD and 75 HC | MRI | TF and SF | SVM | 64.3% | 77% | 82% | 69% |
| [84] | ABIDE-I | 155 ASD and 186 HC | T1-MRI | SF | HGNN | 76.7% | - | - | - |
| [85] | ABIDE-I+II | 255 ASD and 276 HC | rs-fMRI | SF | SVM | 75.00–5.23% | 90.62% | 90.58% | - |
| [86] | ABIDE | 539 ASD and 573 HC | T1-MRI | SF | 6 classifiers | >80% | - | - | - |
| [87] | ABIDE | 539 ASD and 573 HC | rs-fMRI | OF | SVM | 86.7% | 87.5% | 85.7% | - |
| [88] | ABIDE | 99 ASD and 85 HC | fMRI | SPF | CNN | 68.54% | 69.49% | 67.58% | - |
| [89] | ABIDE-I | 270 ASD and 305 HC | rs-fMRI | SPF | ANN | 74.54% | 63.46% | 84.33% | - |
| [90] | ABIDE-I | 48 ASD and 24HC | MRI | TF and SF | RF | 98% | - | - | 52.5–53% |
| [91] | ABIDE | 49 ASD and 41 HC | rs-fMRI | SF | SVM | 78.89% | 85.71% | 70.73% | - |
| [92] | ABIDE | 539 ASD and 573 HC | fMRI | SF | CNN | 87% | - | - | - |
| [93] | ABIDE-I | 505 ASD and 530 HC | fMRI | SF | CNN | 70.22% | 77.46% | 61.82% | 74.86% |
| [72] | ABIDE-I | 79 ASD and 105 HC | 3D-fMRI | OF | CNN | 94.7% | - | - | 94.703% |
| [85] | ABIDE-I+II | 255 ASD and 276 HC | rs-fMRI | SF | SVM-RFECV | 75.0–95.23% | 90.62% | 90.58% | - |
| [94] | ABIDE-I | 368 ASD and 449 HC | sMRI | SF | AE, MLP | 85.06% | - | - | - |
| [95] | ABIDE-I+II | 620 ASD and 542 HC | rs-fMRI | SF | 3D-CNN, SVM | 72.3% | - | - | - |
| [96] | ABIDE-I | 505 ASD and 530 HC | rs-fMRI | OF | CNN | 82.69% | 88.23% | 88.67% | - |
| [97] | ABIDE-I | 403 ASD and 468 HC | fMRI | OF | SVM | 76.8% | 72.5% | 79.9% | 81% |
| [98] | ABIDE-II | 26 ASD and 26 HC | MRI | SF | SVM-RFE | 73% | 71% | 75% | 81% |
| [99] | ABIDE-I | 403 ASD and 468 HC | rs-fMRI | SF | RNN-LSTM | 74.74% | 72.95% | - | - |
| [100] | ABIDE-I | 505 ASD and 530 HC | fMRI | SF | SAE | 70.8% | 62.2% | 79.1% | - |
| [101] | ABIDE-I | 505 ASD and 530 HC | sMRI | SF | RFE+RF | 72% | - | - | - |
| [94] | ABIDE-I | 368 ASD and 449 HC | sMRI | SF | AE | 85.06 ± 3.52% | - | - | - |
| [102] | NDAR | 47 ASD and 24 HC | rs-fMRI | OF | SVM-RFE | 86% | 81% | 88% | - |
| [103] | ABIDE | 539 ASD and 573 HC | fMRI | SF | RCE-SVM | 70.01% | - | - | - |
| [87] | ABIDE-I | 539 ASD and573 HC | rs-fMRI | SF | SVM | 86.7% | 87.5% | 85.7% | - |
| [104] | NDAR | 33 ASD and 33 HC | fMRI | SF | 1D-CNN | 77.2% | 78.1% | 76.5% | - |
| [105] | ABIDE | 41 ASD and 41 HC | rs-fMRI | OF | KNN | 85.9% | 79.3% | 92.6% | - |
| Work | Feature Group | Feature Selection Type | Technique |
|---|---|---|---|
| [81] | SF | WM | Identify the feature group that achieves the best performance through greedy forward feature selection. |
| [87] | OF | WM | A feature selection algorithm based on a minimum spanning tree is proposed to find the optimal feature set. |
| [101] | SF | WM | Use recursion to perform feature selection. |
| [125] | OF | FM | Use Pearson correlation coefficient to filter redundant features. |
| [126] | OF | WM | Use recursive feature elimination (RFE) to rank the importance of features and then remove irrelevant features recursively. |
| [127] | OF | WM | Use the reverse order feature selection algorithm. |
| [128] | OF | WM | Adopt a restricted path depth-first search algorithm (RP-DFS). |
| [129] | OF | FM | Chi-square is used to remove non-significant features. |
| [91] | SF | EM | Use principal component analysis (PCA) to select the principal components. |
| [130] | SF | EM | Use the sure independence screening (SIS) method. Multiple features are removed in each iteration. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaddad, A.; Li, J.; Lu, Q.; Li, Y.; Okuwobi, I.P.; Tanougast, C.; Desrosiers, C.; Niazi, T. Can Autism Be Diagnosed with Artificial Intelligence? A Narrative Review. Diagnostics 2021, 11, 2032. https://doi.org/10.3390/diagnostics11112032
Chaddad A, Li J, Lu Q, Li Y, Okuwobi IP, Tanougast C, Desrosiers C, Niazi T. Can Autism Be Diagnosed with Artificial Intelligence? A Narrative Review. Diagnostics. 2021; 11(11):2032. https://doi.org/10.3390/diagnostics11112032
Chicago/Turabian StyleChaddad, Ahmad, Jiali Li, Qizong Lu, Yujie Li, Idowu Paul Okuwobi, Camel Tanougast, Christian Desrosiers, and Tamim Niazi. 2021. "Can Autism Be Diagnosed with Artificial Intelligence? A Narrative Review" Diagnostics 11, no. 11: 2032. https://doi.org/10.3390/diagnostics11112032
APA StyleChaddad, A., Li, J., Lu, Q., Li, Y., Okuwobi, I. P., Tanougast, C., Desrosiers, C., & Niazi, T. (2021). Can Autism Be Diagnosed with Artificial Intelligence? A Narrative Review. Diagnostics, 11(11), 2032. https://doi.org/10.3390/diagnostics11112032










