Feasibility of Rapid Diagnostic Technology for SARS-CoV-2 Virus Using a Trace Amount of Saliva
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample
2.2. Primer and Probe
2.3. Reverse Transcription-Polymerase Chain Reaction
2.4. User-Friendliness of PCR1100
2.5. Possible Usage of a Trace Amount of Saliva without Pretreatment as a Sample
2.6. Possible Usage of Mouthwash as a Sample Instead of Saliva
2.7. Influence of Sample Collection Conditions on qRT-PCR
2.8. Detection of SARS-CoV-2 Synthetic RNA in Mouthwash Sample
2.9. Detection of SARS-CoV-2 Viral RNA in Mouthwash Obtained from COVID-19 Patients
3. Results
3.1. Analytical Limits of Detection (LoD)
3.2. PCR1100 User-Friendliness
3.3. qPCR Using a Trace Amount of Saliva as a Sample
3.4. Detection of SARS-CoV-2 Synthetic RNA
3.5. Detection of SARS-CoV-2 Viral RNA from COVID-19 Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7144809/ (accessed on 10 December 2020). [CrossRef] [Green Version]
- Evans, R.W. Diagnostic Testing for SARS-CoV-2; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Ravi, N.; Cortade, D.L.; Ng, E.; Wang, S.X. Diagnostics for SARS-CoV-2 Detection: A Comprehensive Review of the FDA-EUA COVID-19 Testing Landscape. Biosens. Bioelectron. 2020, 165, 112454. [Google Scholar] [CrossRef] [PubMed]
- Dinnes, J.; Deeks, J.J.; Adriano, A.; Berhane, S.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; Beese, S.; et al. Rapid, Point-of-Care Antigen and Molecular-Based Tests for Diagnosis of SARS-CoV-2 Infection. Cochrane Database Syst. Rev. 2020, 8, CD013705. Available online: https://www.cochranelibrary.com/cdsr/doi/10.1002/14651858.CD013705/full (accessed on 10 December 2020). [PubMed]
- Jung, Y.J.; Park, G.S.; Moon, J.H.; Ku, K.; Beak, S.H.; Kim, S.; Park, E.C.; Park, D.; Lee, J.H.; Byeon, C.W.; et al. Comparative Analysis of Primer-Probe Sets for the Laboratory Confirmation of SARS-CoV-2. bioRxiv 2020, 11, 2513–2523. [Google Scholar] [CrossRef] [Green Version]
- Shirato, K.; Nao, N.; Kawase, M.; Kageyama, T. An Ultra-Rapid Real-Time RT-PCR Method Using PCR1100 for Detecting Human Orthopneumovirus. Jpn. J. Infect. Dis. 2020, 73, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Porte, L.; Legarraga, P.; Vollrath, V.; Aguilera, X.; Munita, J.M.; Araos, R.; Pizarro, G.; Vial, P.; Iruretagoyena, M.; Dittrich, S.; et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int. J. Infect. Dis. 2020, 99, 328–333. [Google Scholar] [CrossRef]
- Joung, C.J.; Ladha, A.; Saito, M.; Segel, M.; Bruneau, R.; Huang, M.W.; Kim, N.-G.; Yu, X.; Li, J.; Walker, B.D.; et al. Point-of-Care Testing for COVID-19 Using Sherlock Diagnostics. medRxiv 2020. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7273289/ (accessed on 10 December 2020).
- Gibani, M.M.; Toumazou, C.; Sohbati, M.; Sahoo, R.; Karvela, M.; Hon, T.K.; De Mateo, S.; Burdett, A.; Leung, K.Y.F.; Barnett, J.; et al. Assessing a Novel, Lab-Free, Point-of-Care Test for SARS-CoV-2 (CovidNudge): A Diagnostic Accuracy Study. Lancet Microbe 2020, 1, e300–e307. [Google Scholar] [CrossRef]
- Davidson, J.L.; Wang, J.; Maruthamuthu, M.K.; Dextre, A.; Pascual-Garrigos, A.; Mohan, S.; Putikam, S.V.S.; Osman, F.O.I.; McChesney, D.; Seville, J.; et al. A paper-based colorimetric molecular test for SARS-CoV-2 in saliva. Biosens. Bioelectron. X 2021, 9, 100076. [Google Scholar] [CrossRef]
- Yau, F.; Ferreira, R.; Kamali, R.; Bird, P.W.; Halliwell, R.; Patel, H.; Nicoara, D.C.; Woltmann, G.; Tang, J.W. Clinical utility of a rapid ‘on-demand’ laboratory-based SARS-CoV-2 diagnostic testing service in an acute hospital setting admitting COVID-19 patients. Clin. Infect. Pract. 2021, 12, 100086. [Google Scholar] [CrossRef]
- Taki, K.; Yokota, I.; Fukumoto, T.; Iwasaki, S.; Fujisawa, S.; Takahashi, M.; Negishi, S.; Hayasaka, K.; Sato, K.; Oguri, S.; et al. SARS-CoV-2 Detection by Fluorescence Loop-Mediated Isothermal Amplification with and Without RNA Extraction. J. Infect. Chemother. 2021, 27, 410–412. Available online: http://www.sciencedirect.com/science/article/pii/S1341321X20303998 (accessed on 10 December 2020). [CrossRef]
- Nawattanapaiboon, K.; Pasomsub, E.; Prombun, P.; Wongbunmak, A.; Jenjitwanich, A.; Mahasupachai, P.; Vetcho, P.; Chayrach, C.; Manatjaroenlap, N.; Samphaongern, C.; et al. Colorimetric Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) as a Visual Diagnostic Platform for the Detection of the Emerging Coronavirus SARS-CoV-2. Analyst 2021, 146, 471–477. Available online: https://pubs.rsc.org/en/content/articlelanding/2021/an/d0an01775b (accessed on 10 December 2020). [CrossRef]
- Xia, S.; Chen, X. Single-Copy Sensitive, Field-Deployable, and Simultaneous Dual-Gene Detection of SARS-CoV-2 RNA via Modified RT–RPA. Cell Discov. 2020, 6, 37. [Google Scholar] [CrossRef]
- Behrmann, O.; Bachmann, I.; Spiegel, M.; Schramm, M.; Abd El Wahed, A.; Dobler, G.; Dame, G.; Hufert, F.T. Rapid Detection of SARS-CoV-2 by Low Volume Real-Time Single Tube Reverse Transcription Recombinase Polymerase Amplification Using an exo Probe with an Internally Linked Quencher (exo-IQ). Clin. Chem. 2020, 66, 1047–1054. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7239256/ (accessed on 10 December 2020). [CrossRef]
- Harrington, A.; Cox, B.; Snowdon, J.; Bakst, J.; Ley, E.; Grajales, P.; Maggiore, J.; Kahn, S. Comparison of Abbott ID Now and Abbott m2000 Methods for the Detection of SARS-CoV-2 from Nasopharyngeal and Nasal Swabs from Symptomatic Patients. J. Clin. Microbiol. 2020, 58, e00798-20. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7383519/ (accessed on 10 December 2020). [CrossRef] [PubMed] [Green Version]
- Renzoni, A.; Perez, F.; Ngo Nsoga, M.T.; Yerly, S.; Boehm, E.; Gayet-Ageron, A.; Kaiser, L.; Schibler, M. Analytical Evaluation of Visby Medical RT-PCR Portable Device for Rapid Detection of SARS-CoV-2. Diagnostics 2021, 11, 813. [Google Scholar] [CrossRef]
- Hou, H.; Wang, T.; Zhang, B.; Luo, Y.; Mao, L.; Wang, F.; Wu, S.; Sun, Z. Detection of IgM and IgG Antibodies in Patients with Coronavirus disease 2019. Clin. Transl. Immunol. 2020, 9, e01136. [Google Scholar] [CrossRef] [PubMed]
- Kontou, P.I.; Braliou, G.G.; Dimou, N.L.; Nikolopoulos, G.; Bagos, P.G. Antibody Tests in Detecting SARS-CoV-2 Infection: A Meta-Analysis. Diagnostics 2020, 10, 319. [Google Scholar] [CrossRef] [PubMed]
- Infantino, M.; Grossi, V.; Lari, B.; Bambi, R.; Perri, A.; Manneschi, M.; Terenzi, G.; Liotti, I.; Ciotta, G.; Taddei, C.; et al. Diagnostic Accuracy of an Automated Chemiluminescent Immunoassay for Anti-SARS-CoV-2 IgM and IgG Antibodies: An Italian Experience. J. Med. Virol. 2020, 92, 1671–1675. [Google Scholar] [CrossRef]
- Chuan, J.; Gong, B.; Shuai, P.; Zhou, Y.; Zhang, Y.; Jiang, Z.; Zhang, D.; Liu, X.; Ma, S. Detection of serum IgM and IgG for COVID-19 diagnosis. Sci. China 2020, 63, 777–780. [Google Scholar]
- Wen, T.; Huang, C.; Shi, F.J.; Zeng, X.Y.; Lu, T.; Ding, S.N.; Jiao, Y.J. Development of a Lateral Flow Immunoassay Strip for Rapid Detection of IgG Antibody Against SARS-CoV-2 Virus. Analyst 2020, 145, 5345–5352. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and Clinical Application of a Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J. Med. Virol. 2020, 92, 1518–1524. [Google Scholar] [CrossRef]
- Cheng, M.P.; Papenburg, J.; Desjardins, M.; Kanjilal, S.; Quach, C.; Libman, M.; Dittrich, S.; Yansouni, C.P. Diagnostic Testing for Severe Acute Respiratory Syndrome-Related Coronavirus 2: A Narrative Review. Ann. Intern. Med. 2020, 172, 726–734. [Google Scholar] [CrossRef] [Green Version]
- Sheridan, C. Fast, Portable Tests Come Online to Curb Coronavirus Pandemic. Nat. Biotechnol. 2020, 38, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Ji, T.; Liu, Z.; Wang, G.; Guo, X.; Akbar Khan, S.; Lai, C.; Chen, H.; Huang, S.; Xia, S.; Chen, B.; et al. Detection of COVID-19: A Review of the Current Literature and Future Perspectives. Biosens. Bioelectron. 2020, 166, 112455. [Google Scholar] [CrossRef]
- Tang, Y.W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58, e00512-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vashist, S.K. In Vitro Diagnostic Assays for COVID-19: Recent Advances and Emerging Trends. Diagnostics 2020, 10, 202. [Google Scholar] [CrossRef] [Green Version]
- Adachi, D.; Johnson, G.; Draker, R.; Ayers, M.; Mazzulli, T.; Talbot, P.J.; Tellier, R. Comprehensive Detection and Identification of Human Coronaviruses, Including the SARS-Associated Coronavirus, with a Single RT-PCR Assay. J. Virol. Methods 2004, 122, 29–36. [Google Scholar] [CrossRef]
- Czumbel, L.M.; Kiss, S.; Farkas, N.; Mandel, I.; Hegyi, A.; Nagy, Á.; Lohinai, Z.; Szakács, Z.; Hegyi, P.; Steward, M.C.; et al. Saliva as a Candidate for COVID-19 Diagnostic Testing: A Meta-Analysis. Front. Med. 2020, 7, 465. [Google Scholar] [CrossRef]
- Sapkota, D.; Søland, T.M.; Galtung, H.K.; Sand, L.P.; Giannecchini, S.; To, K.K.W.; Mendes-Correa, M.C.; Giglio, D.; Hasséus, B.; Braz-Silva, P.H. COVID-19 Salivary Signature: Diagnostic and Research Opportunities. J. Clin. Pathol. 2020, 74, 344–349. [Google Scholar] [CrossRef]
- Marty, F.M.; Chen, K.; Verrill, K.A. How to Obtain a Nasopharyngeal Swab Specimen. N. Engl. J. Med. 2020, 382, e76. [Google Scholar] [CrossRef]
- Fernandes, L.L.; Pacheco, V.B.; Borges, L.; Athwal, H.K.; de Paula Eduardo, F.; Bezinelli, L.; Correa, L.; Jimenez, M.; Dame-Teixeira, N.; Lombaert, I.M.A.; et al. Saliva in the Diagnosis of COVID-19: A Review and New Research Directions. J. Dent. Res. 2020, 99, 1435–1443. [Google Scholar] [CrossRef]
- Barat, B.; Das, S.; De Giorgi, V.; Henderson, D.K.; Kopka, S.; Lau, A.F.; Miller, T.; Moriarty, T.; Palmore, T.N.; Sawney, S.; et al. Pooled Saliva Specimens for SARS-CoV-2 Testing. J. Clin. Microbiol. 2021, 59, e02486-20. [Google Scholar] [CrossRef] [PubMed]
- Braz-Silva, P.H.; Mamana, A.C.; Romano, C.M.; Felix, A.C.; de Paula, A.V.; Fereira, N.E.; Buss, L.F.; Tozetto-Mendoza, T.R.; Caixeta, R.A.V.; Leal, F.E.; et al. Performance of at-Home Self-Collected Saliva and Nasal-Oropharyngeal Swabs in the Surveillance of COVID-19. J. Oral Microbiol. 2020, 13, 1858002. [Google Scholar] [CrossRef] [PubMed]
- Mesoraca, A.; Margiotti, K.; Viola, A.; Cima, A.; Sparacino, D.; Giorlandino, C. Evaluation of SARS-CoV-2 Viral RNA in Fecal Samples. Virol. J. 2020, 17, 86. [Google Scholar] [CrossRef]
- Pasomsub, E.; Watcharananan, S.P.; Boonyawat, K.; Janchompoo, P.; Wongtabtim, G.; Suksuwan, W.; Sungkanuparph, S.; Phuphuakrat, A. Saliva Sample as a Non-Invasive Specimen for the Diagnosis of Coronavirus disease 2019: A Cross-Sectional Study. Clin. Microbiol. Infect. 2020, 27, 285.e1–285.e4. [Google Scholar] [CrossRef]
- Peng, L.; Liu, J.; Xu, W.; Luo, Q.; Chen, D.; Lei, Z.; Huang, Z.; Li, X.; Deng, K.; Lin, B.; et al. SARS-CoV-2 Can Be Detected in Urine, Blood, Anal Swabs, and Oropharyngeal Swabs Specimens. J. Med. Virol. 2020, 92, 1676–1680. [Google Scholar] [CrossRef]
- Perchetti, G.A.; Nalla, A.K.; Huang, M.L.; Zhu, H.; Wei, Y.; Stensland, L.; Loprieno, M.A.; Jerome, K.R.; Greninger, A.L. Validation of SARS-CoV-2 Detection Across Multiple Specimen Types. J. Clin. Virol. 2020, 128, 104438. [Google Scholar] [CrossRef]
- Rao, M.; Rashid, F.A.; Sabri, F.S.A.H.; Jamil, N.N.; Zain, R.; Hashim, R.; Amran, F.; Kok, H.T.; Samad, M.A.A.; Ahmad, N. Comparing Nasopharyngeal Swab and Early Morning Saliva for the Identification of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2021, 72, e352–e356. [Google Scholar] [CrossRef] [PubMed]
- Senok, A.; Alsuwaidi, H.; Atrah, Y.; Al Ayedi, O.; Al Zahid, J.; Han, A.; Al Marzooqi, A.; Al Heialy, S.; Altrabulsi, B.; AbdelWareth, L.; et al. Saliva as an Alternative Specimen for Molecular COVID-19 Testing in Community Settings and Population-Based Screening. Infect. Drug Resist. 2020, 13, 3393–3399. [Google Scholar] [CrossRef]
- Moreira, V.M.; Mascarenhas, P.; Botelho, J.; Mendes, J.J.; Taveira, N.; Almeida, M.G. Diagnosis of SARS-Cov-2 Infection by RT-PCR Using Specimens Other Than Naso- and Oropharyngeal Swabs: A Systematic Review and Meta-Analysis. Diagnostics 2021, 11, 363. [Google Scholar] [CrossRef] [PubMed]
- Sahajpal, N.S.; Mondal, A.K.; Ananth, S.; Njau, A.; Ahluwalia, P.; Newnam, G.; Lozoya-Colinas, A.; Hud, N.V.; Kota, V.; Ross, T.M.; et al. SalivaSTAT: Direct-PCR and Pooling of Saliva Samples Collected in Healthcare and Community Setting for SARS-CoV-2 Mass Surveillance. Diagnostics 2021, 11, 904. [Google Scholar] [CrossRef]
- Han, P.; Ivanovski, S. Saliva—Friend and Foe in the COVID-19 Outbreak Diagnostics. Diagnostics 2020, 10, 290. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.-W.; Tsang, O.T.-Y.; Yip, C.C.-Y.; Chan, K.H.; Wu, T.C.; Chan, J.M.-C.; Leung, W.S.; Chik, T.S.-H.; Choi, C.Y.-C.; Kandamby, D.H.; et al. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin. Infect. Dis. 2020, 71, 841–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azzi, L.; Carcano, G.; Gianfagna, F.; Grossi, P.; Gasperina, D.D.; Genoni, A.; Fasano, M.; Sessa, F.; Tettamanti, L.; Carinci, F.; et al. Saliva is a Reliable Tool to Detect SARS-CoV-2. J. Infect. 2020, 81, e45–e50. [Google Scholar] [CrossRef]
- Meng, L.; Hua, F.; Bian, Z. Coronavirus Disease 2019 (COVID-19): Emerging and Future Challenges for Dental and Oral Medicine. J. Dent. Res. 2020, 99, 481–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, X.; Xu, X.; Li, Y.; Cheng, L.; Zhou, X.; Ren, B. Transmission Routes of 2019-nCoV and Controls in Dental Practice. Int. J. Oral Sci. 2020, 12, 9. [Google Scholar] [CrossRef]
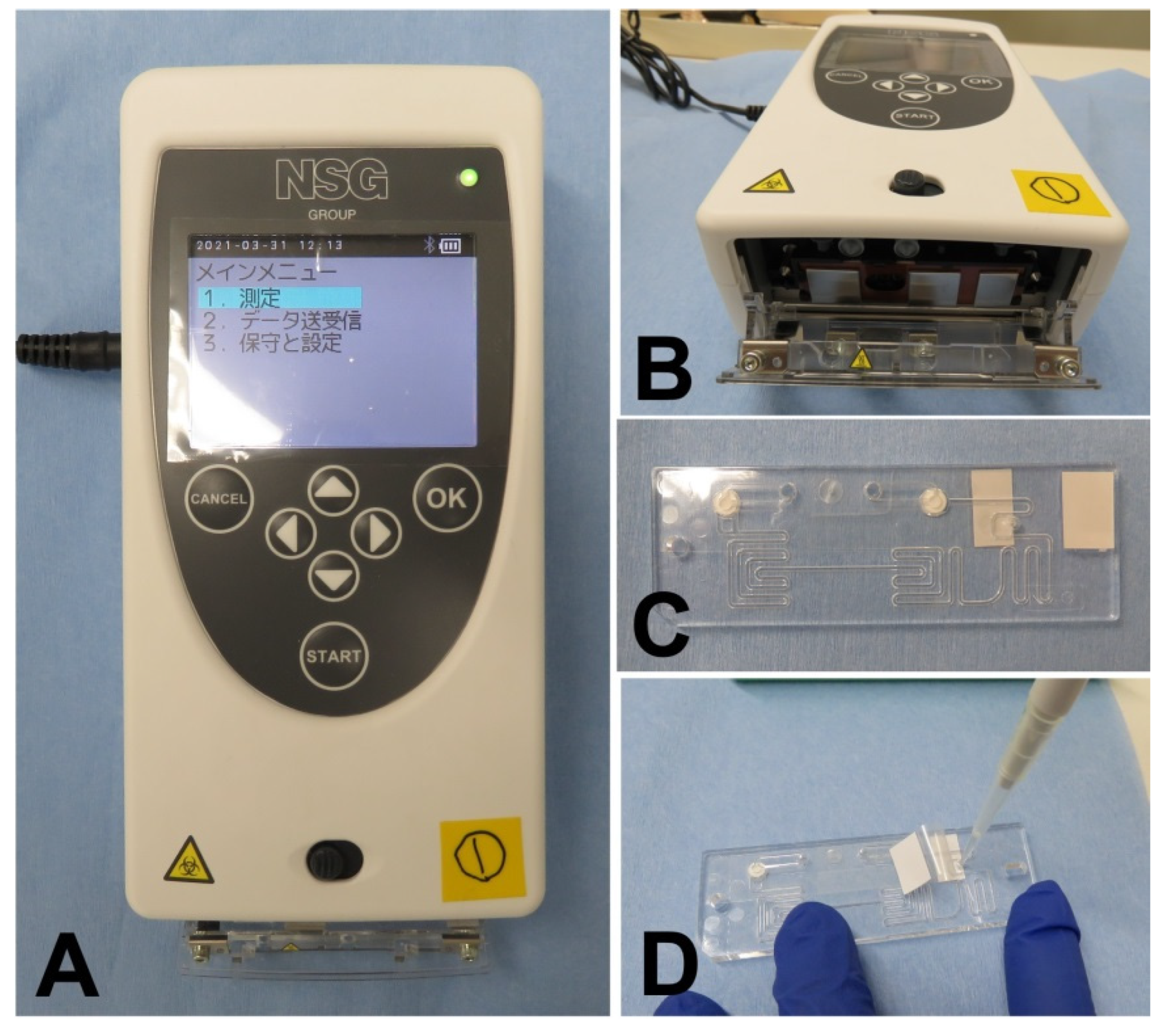
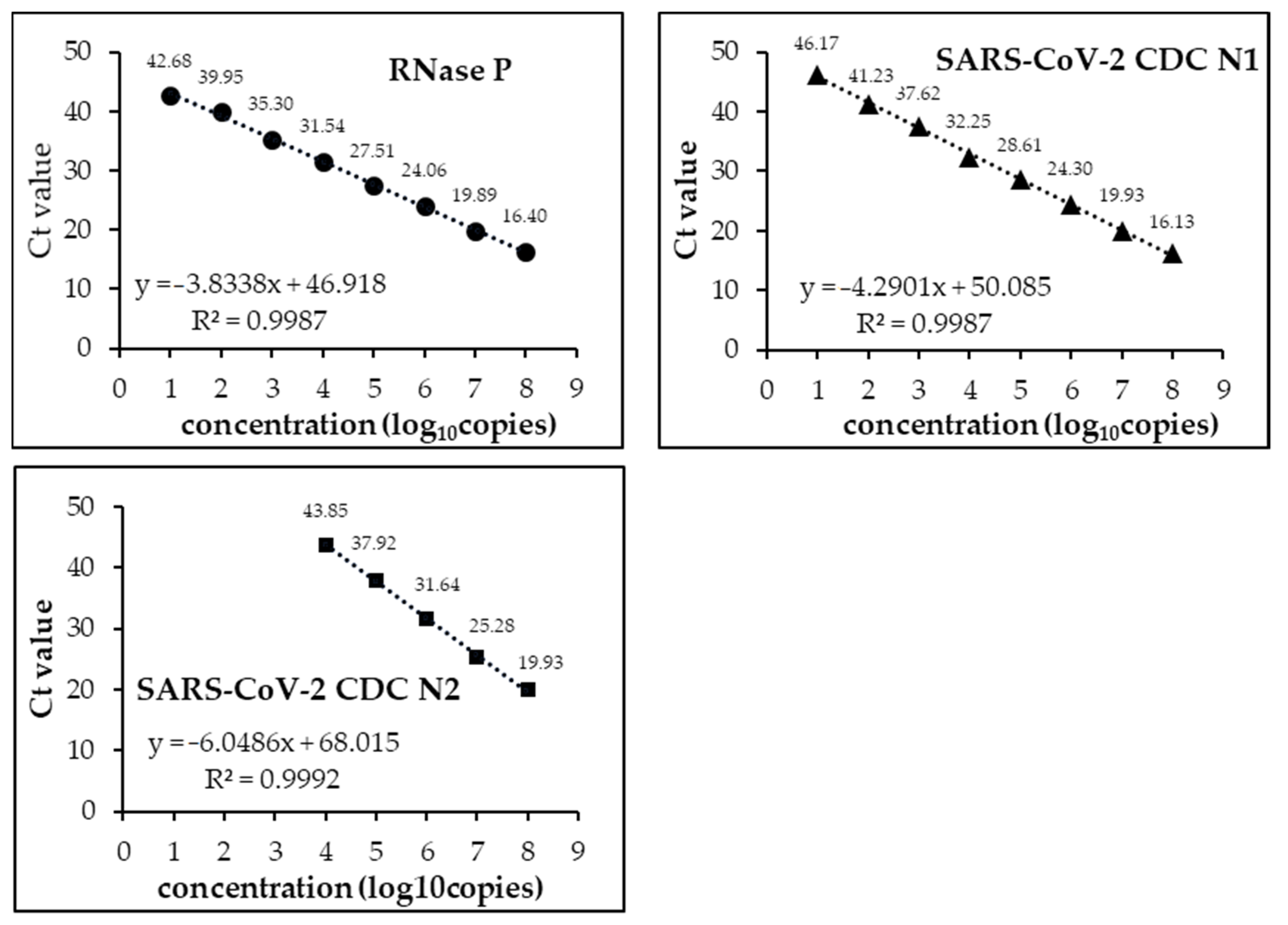
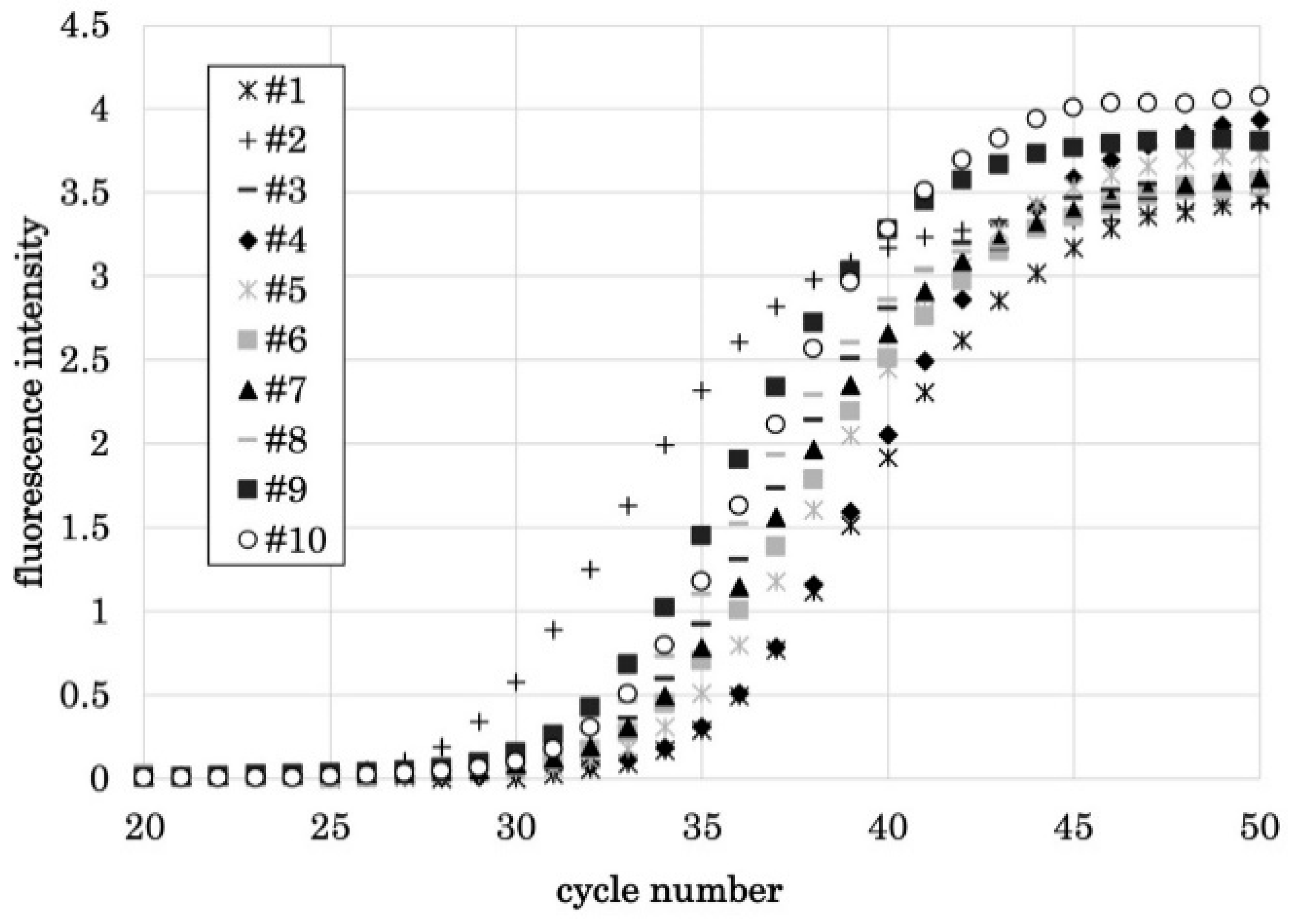

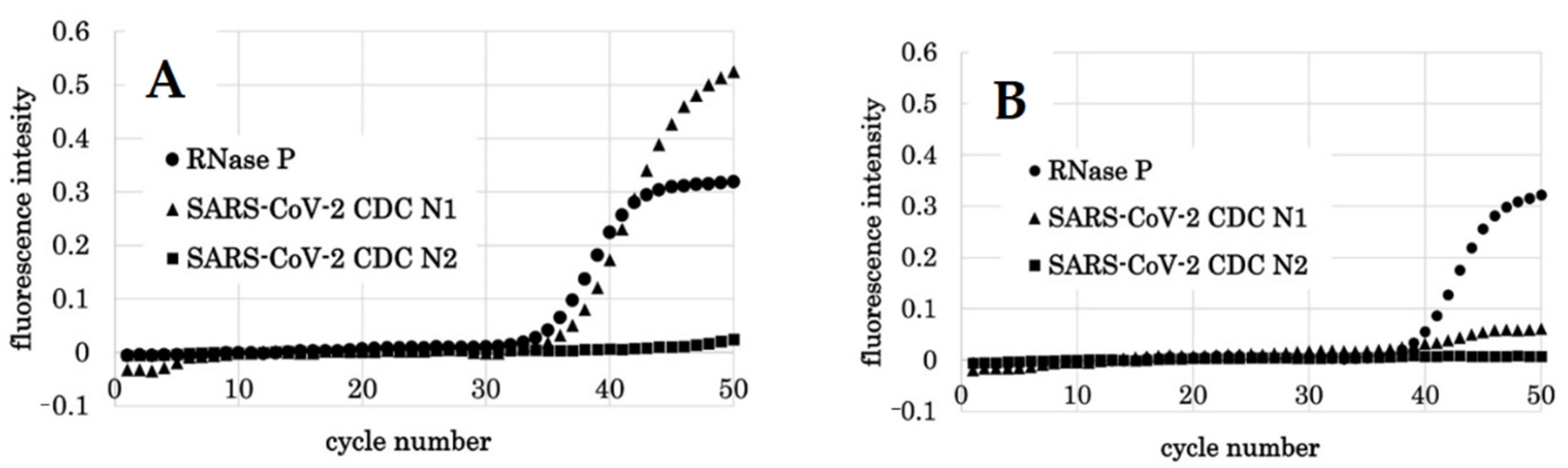

| Target Name | Sequence | Final Concentration | Amplicon | |
|---|---|---|---|---|
| hRNase P | forward reverse probe | 5′-AGA TTT GGA CCT GCG AGC G-3′ 5′-GAG CGG CTG TCT CCA CAA GT-3′ 5′-FAM-TTC TGA CCT GAA GGC TCT GCG CG-BHQ1-3′ | 0.40 μM | 65 bp |
| 0.40 μM | ||||
| 0.20 μM | ||||
| 2019-nCoV_Nl (CDC N1) | forward reverse probe | 5′-GAC CCC AAA ATC AGC GAA AT-3′ 5′-TCT GGT TAC TGC CAG TTG AAT CTG-3′ 5′-HEX-ACC CCG CAT TAC GTT TGG TGG ACC-BHQ1-3′ | 0.20 μM | 72 bp |
| 0.20 μM | ||||
| 0.20 μM | ||||
| 2019-nCoV_N2 (CDC N2) | forward reverse probe | 5′-TTA CAA ACA TTG GCC GCA AA-3′ 5′-GCG CGA CAT TCC GAA GAA-3′ 5′-Cy5-ACA ATT TGC CCC CAG CGC TTC AG-BHQ3-3′ | 0.40 μM | 67 bp |
| 0.40 μM | ||||
| 0.20 μM | ||||
| Control RNA | Sequnce | |||
| hRNase P (ll0 bp) | GGACUUCAGCAUGGCGGUGUUUGCAGAUUUGGACCUGCGAGCGGGUUCUGACCUGAAGGCUCUGCGCGGACUUGUGGAGACAGCCGCUCAUUGUGAGUUGCCCCGGCUUC | |||
| Nl (120 bp) | ACAAACUAAAAUGUCUGAUAAUGGACCCCAAAAUCAGCGAAAUGCACCCCGCAUUACGUUUGGUGGACCCUCAGAUUCAACUGGCAGUAACCAGAAUGGAGAACGCAGUGGGGCGCGAUC | |||
| N2 (200 bp) | UGGUCCAGAACAAACCCAAGGGAAAUUUUGGGGACCAGGAACUAAUCAGACAAGGAACUGAUUACAAACAUUGGCCGCAAAUUGCACAAUUUGCCCCCAGCGCUUCAGCGUUCUUCGGAAUGUCGCGCAUUGGCAUGGAAGUCACACCUUCGGGAACGUGGUUGACCUACACAGGUGCCAUCAAAUUGGAUGACAAAGAUC | |||
| Number | Occupation | Age |
|---|---|---|
| 1 | Dental hygienist | 43 |
| 2 | Dental hygienist | 26 |
| 3 | Dental hygienist | 29 |
| 4 | Nurse | 47 |
| 5 | Dental hygienist | 36 |
| 6 | Nurse | 53 |
| 7 | Dental hygienist | 37 |
| 8 | Nurse | 45 |
| 9 | Nurse | 49 |
| 10 | Nurse | 44 |
| Sample | Detection Status | Ct Value |
|---|---|---|
| Saliva (undiluted solution) | Not detected | - |
| Saliva (diluted 2-fold with pure water) | Detected | 23.34 |
| Representative saliva (for 5 people) (diluted 2-fold with pure water) | Detected | 28.50 |
| Sample | Detection Status | Ct Value |
|---|---|---|
| Saliva (diluted 2-fold with PBS) | Detected | 36.08 |
| Saliva (diluted 4-fold with PBS) | Detected | 42.34 |
| Sample (Collect 3 µL from Mouthwash and Perform qPCR) | Detection Status | Ct Value | ||||
|---|---|---|---|---|---|---|
| Wash | mouth | with | 1 mL | of 10-fold diluted PBS for 10 s | Detected | 37.30 |
| Wash | mouth | with | 2 mL | of 10-fold diluted PBS for 10 s | Detected | 38.30 |
| Wash | mouth | with | 1 mL | of 10-fold diluted PBS for 15 s | Detected | 34.43 |
| Wash | mouth | with | 2 mL | of 10-fold diluted PBS for 15 s | Detected | 33.63 |
| Wash | mouth | with | 2 mL | of saline for 15 s (subject 1) | Detected | 37.74 |
| Wash | mouth | with | 2 mL | of saline for 15 s (subject 2) | Detected | 39.03 |
| Wash | mouth | with | 2 mL | of saline for 15 s (subject 3) | Detected | 35.76 |
| Wash | mouth | with | 2 mL | of saline for 15 s (subject 1) | Detected | 35.45 |
| (After freezing for 2 weeks) | ||||||
| Wash | mouth | with | 2 mL | of saline for 15 s (subject 2) | Detected | 37.98 |
| (After freezing for 2 weeks) | ||||||
| Wash | mouth | with | 2 mL | of saline for 15 s (subject 3) | Detected | 35.14 |
| (After freezing for 2 weeks) | ||||||
| Conditions for Sample Collection | Diluted Saliva | Mouthwash | |
|---|---|---|---|
| 2-Fold with PBS | 4-Fold with PBS | ||
| Normal time (3 h after eating and brushing) | 36.08 | 42.34 | 37.12 |
| After meals (30 min) | 39.44 | 38.63 | 37.54 |
| After brushing (15 min) | 42.15 | 40.60 | 41.15 |
| Sample | Conditions | Ct Value | ||
|---|---|---|---|---|
| RNase P | CDC Nl | CDC N2 | ||
| Positive control | Synthetic RNA of 107 copies each in pure water | 21.09 | 18.95 | 24.74 |
| Subject 1 | Premix containing synthetic RNA+ Mouthwash 3 µL | 38.40 | 18.96 | 37.54 |
| Subject 1 | Premix + Mouthwash 3 µL containing synthetic RNA | 33.97 | 31.44 | >50 |
| Subject 2 | Premix containing synthetic RNA+ Mouthwash 3 µL | 36.99 | 19.61 | 26.37 |
| Subject 2 | Premix + Mouthwash 3 µL containing synthetic RNA | 33.19 | 26.27 | 43.83 |
| Subject 3 | Premix containing synthetic RNA+ Mouthwash 3 µL | 35.62 | 19.92 | 26.87 |
| Subject 3 | Premix + Mouthwash 3 µL containing synthetic RNA | 32.39 | 27.50 | 43.59 |
| Subject 4 | Premix containing synthetic RNA+ Mouthwash 3 µL | 39.84 | 18.68 | 25.92 |
| Subject 4 | Premix + Mouthwash 3 µL containing synthetic RNA | 36.36 | 26.60 | 42.41 |
| Subjects | Period from Onset of Symptom (Days)/Detect (Ct Value) or Not (-) | Symptom (Period, Days) | Past History | ||
|---|---|---|---|---|---|
| Age Sex | 1st | 2nd | 3rd | ||
| 1. 38 M | 13/44.6 | 20/(-) | Fever (6), malaise, dysgeusia | HT, DM, HL, CCF | |
| 2. 90 M | 7/29.3 | 14/34.5 | 21/38.8 | Fever (8), breathing difficulty | HT, DM, CKD |
| 3. 56 F | 18/(-) | 25/(-) | Fever, respiratory management (7) | n.p. | |
| 4. 34 M | ll/(-) | Fever (10), dysgeusia | n.p. | ||
| 5. 34 F | 7/38.1 | 14/(-) | Fever (5), breathing difficulty | DM, HT, obesity | |
| 6. 47 F | ll/(-) | 18/(-) | Fever (11), dysgeusia | CKD, thyroid and parathyroid cancer | |
| 7. 67 M | Exclusion | ||||
| 8. 47 M | 3/38.4 | 10/(-) | Fever (5), cough | CKD, HD, DM | |
| 9. 76 M | 9/45.2 | Fever (7), breathing difficulty | n.p. | ||
| 10. 44 M | 7/36.9 | 14/(-) | Fever (10), breathing difficulty | DM, HT, HL | |
| 11. 30 M | 10/(-) | Fever (10), breathing difficulty | n.p. | ||
| 12. 22 M | 8/35.7 | 15/(-) | Fever (l0), diarrhea | n.p. | |
| 13. 57 M | 10/(-) | cough, breathing difficulty | DM | ||
| 14. 61 M | 8/34.2 | Fever (13), dysgeusia | integration disorder syndrome | ||
| 15. 61 M | 25/(-) | Fever (15), breathing difficulty | DM, HT | ||
| 16. 34 M | 11/34.1 | Fever (11), breathing difficulty | DM | ||
| 17. 61 M | Exclusion | ||||
| 18. 54 M | Exclusion | ||||
| 19. 81 M | 1/40.2 | 8/37.7 | Fever (1), pneumonia | HT *1 | |
| 20. 60 F | 5/40.9 | 12/(-) | Fever (4), breathing difficulty | CKD, HD | |
| PCR1100 | Conventional qPCR Device | |
|---|---|---|
| Turnaround time | approximately 18 min | a few hours |
| Sample | mouthwash | nasopharyngeal swab, saliva, etc. |
| Person who carries out | skilled specialist, clinical laboratory technician | anyone |
| Ease of handling | mobile (POCT) | stationary type (cannot move) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tokuyama-Toda, R.; Muraoka, M.; Terada-Ito, C.; Ide, S.; Horiuchi, T.; Amemiya, T.; Fukuoka, A.; Hamada, Y.; Sejima, S.; Satomura, K. Feasibility of Rapid Diagnostic Technology for SARS-CoV-2 Virus Using a Trace Amount of Saliva. Diagnostics 2021, 11, 2024. https://doi.org/10.3390/diagnostics11112024
Tokuyama-Toda R, Muraoka M, Terada-Ito C, Ide S, Horiuchi T, Amemiya T, Fukuoka A, Hamada Y, Sejima S, Satomura K. Feasibility of Rapid Diagnostic Technology for SARS-CoV-2 Virus Using a Trace Amount of Saliva. Diagnostics. 2021; 11(11):2024. https://doi.org/10.3390/diagnostics11112024
Chicago/Turabian StyleTokuyama-Toda, Reiko, Masaaki Muraoka, Chika Terada-Ito, Shinji Ide, Toshikatsu Horiuchi, Tsuyoshi Amemiya, Airi Fukuoka, Yoshiki Hamada, Shunsuke Sejima, and Kazuhito Satomura. 2021. "Feasibility of Rapid Diagnostic Technology for SARS-CoV-2 Virus Using a Trace Amount of Saliva" Diagnostics 11, no. 11: 2024. https://doi.org/10.3390/diagnostics11112024
APA StyleTokuyama-Toda, R., Muraoka, M., Terada-Ito, C., Ide, S., Horiuchi, T., Amemiya, T., Fukuoka, A., Hamada, Y., Sejima, S., & Satomura, K. (2021). Feasibility of Rapid Diagnostic Technology for SARS-CoV-2 Virus Using a Trace Amount of Saliva. Diagnostics, 11(11), 2024. https://doi.org/10.3390/diagnostics11112024








