Multiparametric MRI and Radiomics in Prostate Cancer: A Review of the Current Literature
Abstract
:1. Introduction
2. Radiomics Analysis
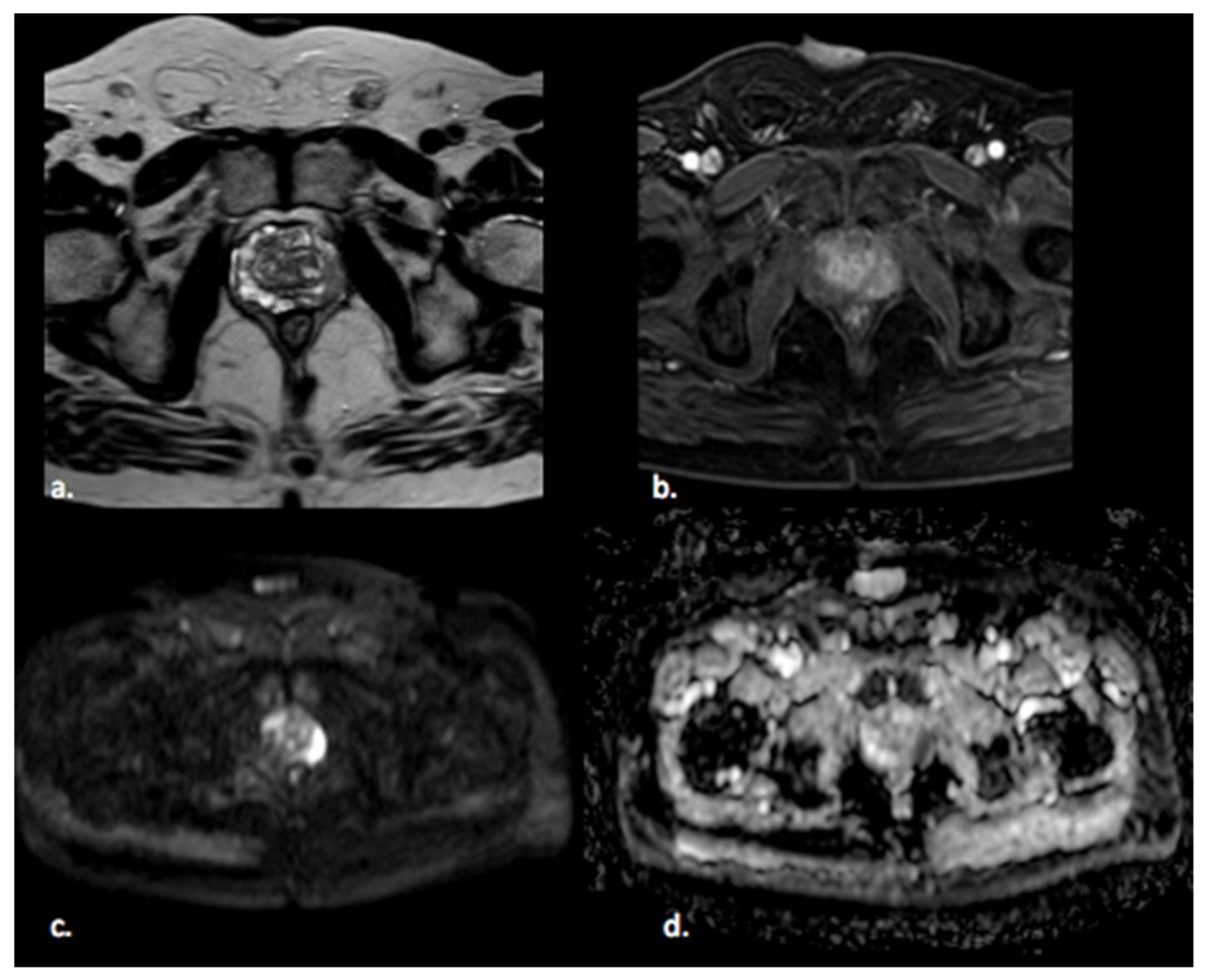
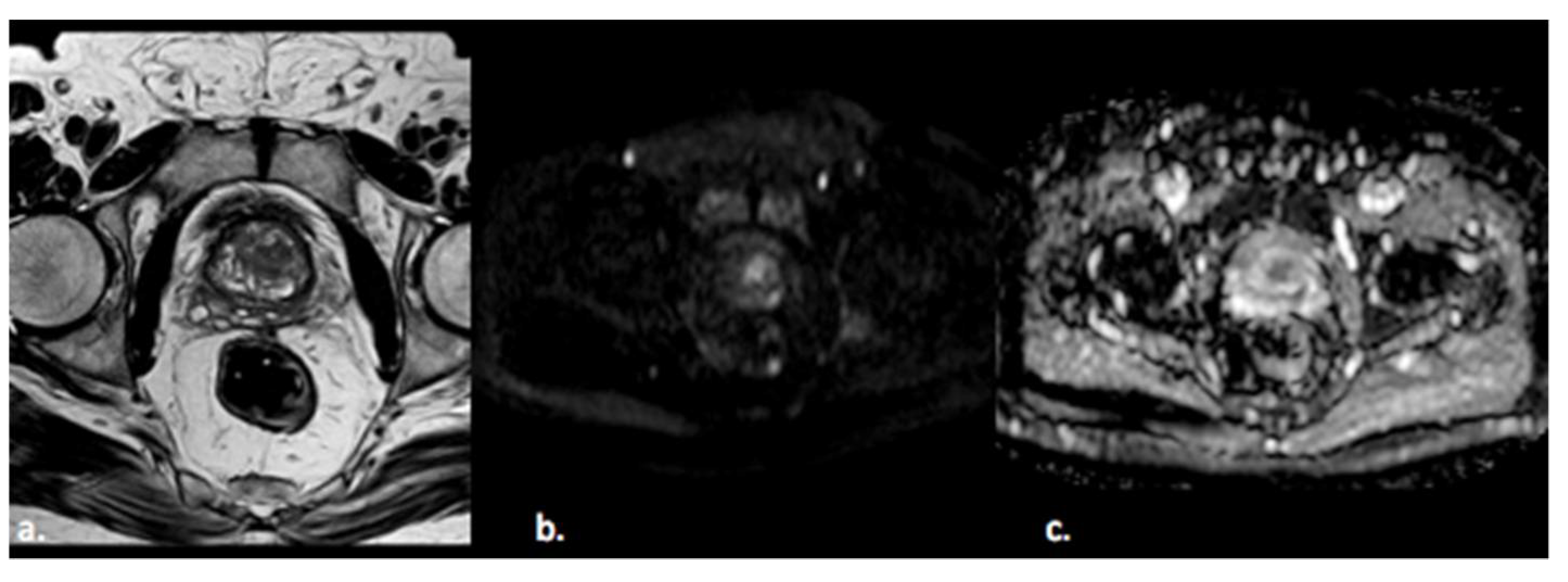
2.1. Step 1—Segmentation
2.2. Step 2—Image Processing
2.3. Step 3—Feature Extraction
2.4. Step 4—Feature Selection
2.5. Step 5—Development of Predictive Model
3. Radiomics in Prostate Cancer
3.1. Detection of Prostate Cancer
3.2. Diagnosis of Prostate Cancer
3.3. Grading and Aggressiveness
3.4. Radiomics and PI-RADS Score
3.5. Treatment Evaluation and Prediction of Biochemical Recurrence
4. Limitations and Future Applications
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ginsburg, S.B.; Ms, A.A.; Pahwa, S.; Gulani, V.; Ponsky, L.; Aronen, H.J.; Boström, P.J.; Böhm, M.; Haynes, A.; Brenner, P.; et al. Radiomic features for prostate cancer detection on MRI differ between the transition and peripheral zones: Preliminary findings from a multi-institutional study. J. Magn. Reson. Imaging 2017, 46, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.K.; Abreu, A.L.D.C.; Palmer, S. Utility of Ultrasound in the Diagnosis, Treatment, and Follow-up of Prostate Cancer: State of the Art. J. Nucl. Med. 2016, 57, 13S–18S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rooij, M.; Hamoen, E.H.J.; Fütterer, J.J.; Barentsz, J.O.; Rovers, M.M. Accuracy of multiparametric MRI for prostate cancer detection: A meta-analysis. Am. J. Roentgenol. 2014, 202, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Turkbey, B.; Rosenkrantz, A.B.; Haider, M.A.; Padhani, A.; Villeirs, G.; Macura, K.J.; Weinreb, J.C. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur. Urol. 2019, 76, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Oberlin, D.T.; Casalino, D.D.; Miller, F.H.; Meeks, J.J. Dramatic increase in the utilization of multiparametric magnetic resonance imaging for detection and management of prostate cancer. Abdom. Radiol. 2017, 42, 1255–1258. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.U.; Lind, K.E.; Garg, K.; Crawford, D.; Werahera, P.N.; Pokharel, S.S. Assessment of PI-RADS v2 categories ≥ 3 for diagnosis of clinically significant prostate cancer. Abdom. Radiol. 2019, 44, 705–712. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Vernuccio, F.; Cannella, R.; Comelli, A.; Salvaggio, G.; Lagalla, R.; Midiri, M. Radiomica e intelligenza artificiale: Nuove frontiere in medicina. Recent Prog. Med. 2020, 111, 130–135. [Google Scholar] [CrossRef]
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine Learning for Medical Imaging. RadioGraphics 2017, 37, 505–515. [Google Scholar] [CrossRef]
- Chartrand, G.; Cheng, P.M.; Vorontsov, E.; Drozdzal, M.; Turcotte, S.; Pal, C.J.; Kadoury, S.; Tang, A. Deep Learning: A Primer for Radiologists. RadioGraphics 2017, 37, 2113–2131. [Google Scholar] [CrossRef] [Green Version]
- Van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging-”how-to” guide and critical reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Haarburger, C.; Müller-Franzes, G.; Weninger, L.; Kuhl, C.; Truhn, D.; Merhof, D. Radiomics feature reproducibility under inter-rater variability in segmentations of CT images. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Galloway, M.M. Texture analysis using gray level run lengths. Comput. Graph. Image Process. 1975, 4, 172–179. [Google Scholar] [CrossRef]
- Cheng, R.; Roth, H.R.; Lay, N.; Lu, L.; Turkbey, B.; Gandler, W.; McCreedy, E.S.; Pohida, T.; Pinto, P.A.; Choyke, P.; et al. Automatic magnetic resonance prostate segmentation by deep learning with holistically nested networks. J. Med. Imaging 2017, 4, 041302. [Google Scholar] [CrossRef]
- Chan, I.; Wells, W.; Mulkern, R.V.; Haker, S.; Zhang, J.; Zou, K.H.; Maier, S.E.; Tempany, C.M.C. Detection of prostate cancer by integration of line-scan diffusion, T2-mapping and T2-weighted magnetic resonance imaging; a multichannel statistical classifier. Med. Phys. 2003, 30, 2390–2398. [Google Scholar] [CrossRef] [PubMed]
- Giannini, V.; Mazzetti, S.; Vignati, A.; Russo, F.; Bollito, E.; Porpiglia, F.; Stasi, M.; Regge, D. A fully automatic computer aided diagnosis system for peripheral zone prostate cancer detection using multi-parametric magnetic resonance imaging. Comput. Med Imaging Graph. 2015, 46, 219–226. [Google Scholar] [CrossRef]
- Wibmer, A.; Hricak, H.; Gondo, T.; Matsumoto, K.; Veeraraghavan, H.; Fehr, D.; Zheng, J.; Goldman, D.; Moskowitz, C.; Fine, S.W.; et al. Haralick texture analysis of prostate MRI: Utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur. Radiol. 2015, 25, 2840–2850. [Google Scholar] [CrossRef]
- Cameron, A.; Khalvati, F.; Haider, M.A.; Wong, A. MAPS: A Quantitative Radiomics Approach for Prostate Cancer Detection. IEEE Trans. Biomed. Eng. 2016, 63, 1145–1156. [Google Scholar] [CrossRef]
- Bleker, J.; Kwee, T.C.; Dierckx, R.A.J.O.; de Jong, I.J.; Huisman, H.; Yakar, D. Multiparametric MRI and auto-fixed volume of inter-est-based radiomics signature for clinically significant peripheral zone prostate cancer. Eur. Radiol. 2020, 30, 1313–1324. [Google Scholar] [CrossRef] [Green Version]
- Khalvati, F.; Wong, A.; Haider, M.A. Automated prostate cancer detection via comprehensive multi-parametric magnetic resonance imaging texture feature models. BMC Med. Imaging 2015, 15, 27. [Google Scholar] [CrossRef] [Green Version]
- Fehr, D.; Veeraraghavan, H.; Wibmer, A.; Gondo, T.; Matsumoto, K.; Vargas, H.A.; Sala, E.; Hricak, H.; Deasy, J.O. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc. Natl. Acad. Sci. USA 2015, 112, E6265–E6273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nketiah, G.; Elschot, M.; Kim, E.; Teruel, J.R.; Scheenen, T.W.; Bathen, T.F.; Selnæs, K.M. T2-weighted MRI-derived textural features reflect prostate cancer aggressiveness: Preliminary results. Eur. Radiol. 2017, 27, 3050–3059. [Google Scholar] [CrossRef] [PubMed]
- Nketiah, G.A.; Elschot, M.; Scheenen, T.W.; Maas, M.C.; Bathen, T.F.; Selnæs, K.M. Utility of T2-weighted MRI texture analysis in assessment of peripheral zone prostate cancer aggressiveness: A single-arm, multicenter study. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Cuocolo, R.; Stanzione, A.; Ponsiglione, A.; Romeo, V.; Verde, F.; Creta, M.; La Rocca, R.; Longo, N.; Pace, L.; Imbriaco, M. Clinically significant prostate cancer detection on MRI: A radiomic shape features study. Eur. J. Radiol. 2019, 116, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Niazi, T.; Probst, S.; Bladou, F.; Anidjar, M.; Bahoric, B. Predicting Gleason Score of Prostate Cancer Patients Using Radiomic Analysis. Front. Oncol. 2018, 8, 630. [Google Scholar] [CrossRef] [Green Version]
- Giambelluca, D.; Cannella, R.; Vernuccio, F.; Comelli, A.; Pavone, A.; Salvaggio, L.; Galia, M.; Midiri, M.; Lagalla, R.; Salvaggio, G. PI-RADS 3 Lesions: Role of Prostate MRI Texture Analysis in the Identification of Prostate Cancer. Curr. Probl. Diagn. Radiol. 2021, 50, 175–185. [Google Scholar] [CrossRef]
- Brancato, V.; Aiello, M.; Basso, L.; Monti, S.; Palumbo, L.; Di Costanzo, G.; Salvatore, M.; Ragozzino, A.; Cavaliere, C. Evaluation of a multiparametric MRI radiomic-based approach for stratification of equivocal PI-RADS 3 and upgraded PI-RADS 4 prostatic lesions. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Shiradkar, R.; Podder, T.K.; Algohary, A.; Viswanath, S.; Ellis, R.J.; Madabhushi, A. Radiomics based targeted radiotherapy planning (Rad-TRaP): A computational framework for prostate cancer treatment planning with MRI. Radiat. Oncol. 2016, 11, 148. [Google Scholar] [CrossRef] [Green Version]
- Gnep, K.; Fargeas, A.; Gutiérrez-Carvajal, R.E.; Commandeur, F.; Mathieu, R.; Ospina, J.D.; Rolland, Y.; Rohou, T.; Vincendeau, S.; Hatt, M.; et al. Haralick textural features on T2-weighted MRI are associated with biochemical recurrence following radiotherapy for peripheral zone prostate cancer: Impact of MRI in Prostate Cancer. J. Magn. Reson. Imaging. 2017, 45, 103–117. [Google Scholar] [CrossRef]
- Lemaître, G.; Martí, R.; Freixenet, J.; Vilanova, J.C.; Walker, P.M.; Meriaudeau, F. Computer-Aided Detection and diagnosis for prostate cancer based on mono and multi-parametric MRI: A review. Comput. Biol. Med. 2015, 60, 8–31. [Google Scholar] [CrossRef] [Green Version]
- Comelli, A.; Dahiya, N.; Stefano, A.; Vernuccio, F.; Portoghese, M.; Cutaia, G.; Bruno, A.; Salvaggio, G.; Yezzi, A. Deep Learning-Based Methods for Prostate Segmentation in Magnetic Resonance Imaging. Appl. Sci. 2021, 11, 782. [Google Scholar] [CrossRef]
- Epstein, J.I.; Allsbrook, W.C., Jr.; Amin, M.B.; Egevad, L.L.; ISUP Grading Committee. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am. J. Surg. Pathol. 2005, 29, 1228–1242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, H.U.; Bosaily, A.E.-S.; Brown, L.C.; Gabe, R.; Kaplan, R.S.; Parmar, M.K.; PROMIS Study Group. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): A paired validating confirmatory study. Lancet 2017, 389, 815–822. [Google Scholar] [CrossRef] [Green Version]
- Vignati, A.; Mazzetti, S.; Giannini, V.; Russo, F.; Bollito, E.; Porpiglia, F.; Stasi, M.; Regge, D. Texture features on T2-weighted magnetic resonance imaging: New potential biomarkers for prostate cancer aggressiveness. Phys. Med. Biol. 2015, 60, 2685–2701. [Google Scholar] [CrossRef] [PubMed]
- Min, X.; Li, M.; Dong, D.; Feng, Z.; Zhang, P.; Ke, Z.; You, H.; Han, F.; Ma, H.; Tian, J.; et al. Multi-parametric MRI-based radiomics signature for discriminating between clinically significant and insignificant prostate cancer: Cross-validation of a machine learning method. Eur. J. Radiol. 2019, 115, 16–21. [Google Scholar] [CrossRef] [Green Version]
- Toivonen, J.; Perez, I.M.; Movahedi, P.; Merisaari, H.; Pesola, M.; Taimen, P.; Bostrom, P.J.; Pohjankukka, J.; Kiviniemi, A.; Panikkala, T.; et al. Radiomics and machine learning of mul-tisequence multiparametric prostate MRI: Towards improved non-invasive prostate cancer characterization. PLoS ONE 2019, 14, e0217702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Li, M.; Gu, Y.; Zhang, Y.; Yang, S.; Wei, C.; Wu, J.; Li, X.; Zhao, W.; Shen, J. Prostate Cancer Differentiation and Aggressiveness: Assessment with a Radiomic-Based Model vs. PI-RADS v2. J. Magn. Reson. Imaging 2019, 49, 875–884. [Google Scholar] [CrossRef] [Green Version]
- Chaddad, A.; Kucharczyk, M.J.; Niazi, T. Multimodal Radiomic Features for the Predicting Gleason Score of Prostate Cancer. Cancers 2018, 10, 249. [Google Scholar] [CrossRef] [Green Version]
- Kan, Y.; Zhang, Q.; Hao, J.; Wang, W.; Zhuang, J.; Gao, J.; Huang, H.; Liang, J.; Marra, G.; Calleris, G.; et al. Clinico-radiological characteristic-based machine learning in reducing unnecessary prostate biopsies of PI-RADS 3 lesions with dual validation. Eur. Radiol. 2020, 30, 6274–6284. [Google Scholar] [CrossRef]
- Wang, J.; Wu, C.-J.; Bao, M.-L.; Zhang, J.; Wang, X.-N.; Zhang, Y.-D. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. Eur. Radiol. 2017, 27, 4082–4090. [Google Scholar] [CrossRef]
- Hou, Y.; Bao, M.-L.; Wu, C.-J.; Zhang, J.; Zhang, Y.-D.; Shi, H.-B. A radiomics machine learning-based redefining score robustly identifies clinically significant prostate cancer in equivocal PI-RADS score 3 lesions. Abdom. Radiol. 2020, 45, 4223–4234. [Google Scholar] [CrossRef]
- Wels, M.G.; Lades, F.; Muehlberg, A.; Suehling, M. General purpose radiomics for multi-modal clinical research. In Proceedings of the Volume 10950, Medical Imaging 2019: Computer-Aided Diagnosis, 13 March 2019, San Diego, CA, USA; p. 1095046. [CrossRef]
- Rodrigues, G.; Warde, P.; Pickles, T.; Crook, F.J.; Brundage, M.; Souhami, M.L.; Lukka, H.; on behalf of the Genitourinary Radiation Oncologists of Canada. Pre-treatment risk stratification of prostate cancer patients: A critical review. Can. Urol. Assoc. J. 2012, 6, 121–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdollahi, H.; Tanha, K.; Mofid, B.; Razzaghdoust, A.; Saadipoor, A.; Khalafi, L.; Bakhshandeh, M.; Mahdavi, S.R. MRI Radiomic Analysis of IMRT-Induced Bladder Wall Changes in Prostate Cancer Patients: A Relationship with Radiation Dose and Toxicity. J. Med. Imaging Radiat. Sci. 2019, 50, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Dotan, Z.A.; Bianco, F.J., Jr.; Rabbani, F.; Eastham, J.A.; Fearn, P.; Scher, H.I.; Kelly, K.W.; Chen, H.-N.; Schöder, H.; Hricak, H.; et al. Pattern of Prostate-Specific Antigen (PSA) Failure Dictates the Probability of a Positive Bone Scan in Patients with an Increasing PSA After Radical Prostatectomy. J. Clin. Oncol. 2005, 23, 1962–1968. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.S.F.; Aerts, H.J.W.L. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150–R166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skvortsov, S.; Schäfer, G.; Stasyk, T.; Fuchsberger, C.; Bonn, G.K.; Bartsch, G.; Klocker, H.; Huber, L.A. Proteomics profiling of micro-dissected low- and high-grade prostate tumors identifies Lamin A as a discriminatory biomarker. J. Proteome. Res. 2011, 10, 259–268. [Google Scholar] [CrossRef]
- Alongi, P.; Stefano, A.; Comelli, A.; Laudicella, R.; Scalisi, S.; Arnone, G.; Barone, S.; Spada, M.; Purpura, P.; Bartolotta, T.V.; et al. Radiomics analysis of 18F-Choline PET/CT in the prediction of disease outcome in high-risk prostate cancer: An explorative study on machine learning feature classification in 94 patients. Eur. Radiol. 2021, 31, 4595–4605. [Google Scholar] [CrossRef] [PubMed]
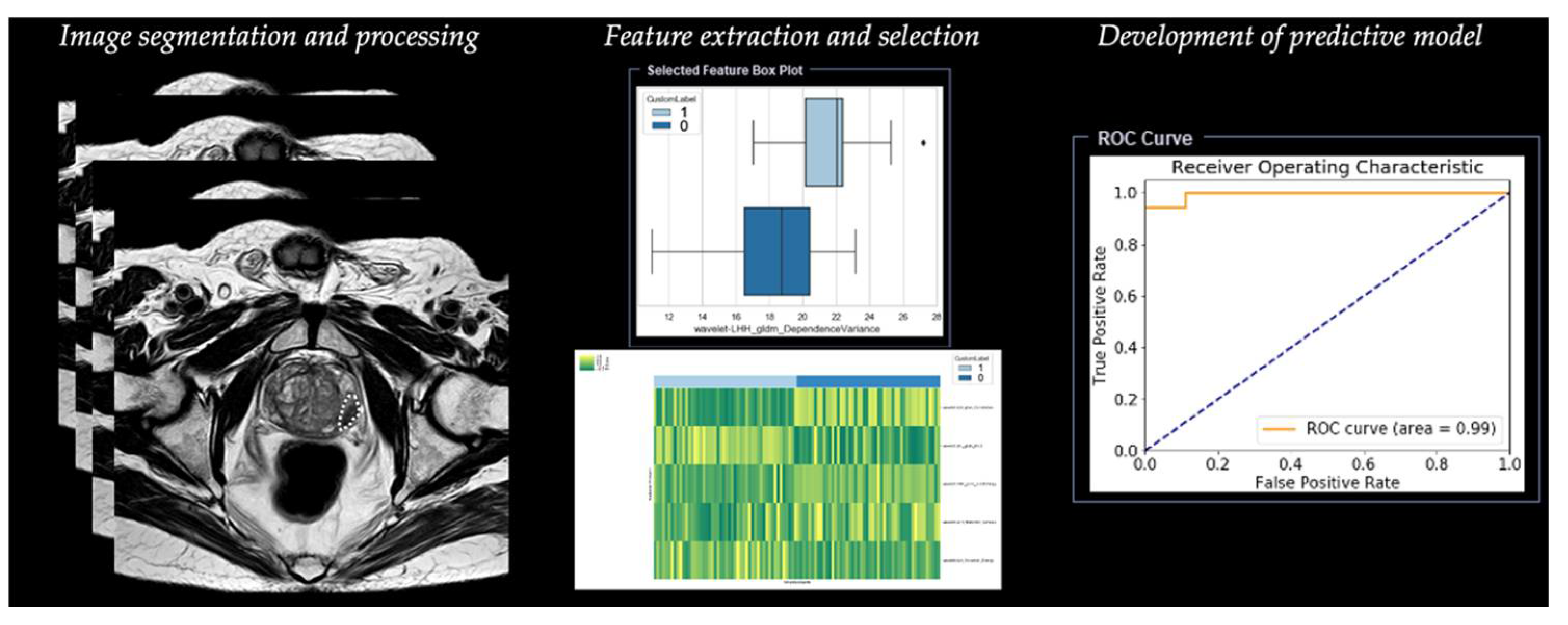
| Step Number | Type of Process | Description of the Step |
|---|---|---|
| 1 | Segmentation | Manual, automatic, or semiautomatic segmentation of the images to define the region or volume of interest |
| 2 | Image processing | Processing of images to increase reproducibility |
| 3 | Feature extraction | Feature descriptors are used to quantify characteristics of the gray levels within the region or volume of interest |
| 4 | Feature selection | Selection of the most useful features and exclusion of nonreproducible features to create a statistical model |
| 5 | Development of predictive model | Development of a classifier with different machine learning algorithms |
| Type of Study | Authors, Year | Description |
|---|---|---|
| Detection of PCa | Chan et al., 2003 [19] | Development of one of the first predictive models using support vector machine (AUC 0.71–0.80) |
| Giannini et al., 2015 [20] | Parametric color-coded map of the prostate based on the probability of each voxel to be tumoral (AUC 0.83–0.98) | |
| Diagnosis of PCa | Ginsburg et al., 2017 [2] | Evaluation of different radiomic features in PZ and TZ tumors (AUC 0.61–0.71) |
| Wibmer et al., 2015 [21] | Haralick texture analysis to differentiate clinically significant and not clinically significant PCa | |
| Cameron et al., 2016 [22] | Development of a comprehensive feature model consisting of an initial tumor candidate identification schema (AUC 0.81–0.93) | |
| Bleker et al., 2020 [23] | Development of a model that quantifies the phenotype of clinically significant PCa in PZ based on T2W and DWI images (AUC 0.75–0.98) | |
| Khalvati et al., 2015 [24] | New automatic texture feature models incorporating computed high-b diffused weighted imaging (CHB-DWI; AUC 0.73–0.85) and correlated diffusion imaging (CDI; AUC 0.81–0.90) to improve differentiation of tumoral and healthy tissue | |
| Grading and aggresiveness | Fehr et al., 2015 [25] | Development of an automatic classification with a high accuracy combining ADC and T2W to evaluate the aggressiveness of PCa (AUC 0.93) |
| Nketiah et al., 2017; 2021 [26,27] | Texture features, such as homogeneity and entropy, could reveal the aggressiveness of peripheral PCa distinguishing GS 7 (3 + 4) and GS 7 (4 + 3) (AUC 0.83 vs. 0.72 of MRI parameters) | |
| Cuocolo et al., 2019 [28] | Geometric parameters, such as surface area-to-volume ratio (SAVR), could predict clinically and non-clinically significant PCa (AUC 0.78) | |
| Chaddad et al., 2018 [29] | Model based on the joint intensity matrix (JIM) to predict the GS through different derived-features and comparing between GS groups: GS 6 (AUC 0.83), GS 3 + 4 (AUC 0.72), and GS ≥ 4 + 3 (AUC 0.77) | |
| PI-RADS score | Giambelluca et al., 2021 [30] | Development of a texture analysis model to diagnose clinically significant PCa withing PI-RADS 3 lesions larger than 5 mm on T2W (AUC = 0.77) and ADC map (AUC = 0.81) images. |
| Brancato et al., 2021 [31] | Relevant texture features for stratifying PI-RADS 3 (AUC 0.80) and PI-RADS 4 lesions (AUC 0.89) from T2W and ADC images | |
| Treatment evaluation and prediction of biochemical recurrence | Shiradkar et al., 2016 [32] | Targeted treatment radiotherapy planning based on a radiomic model, consisting of cancer detection on feature analysis, transference of delineation to CT, and generation of targeted focal radiotherapy plans |
| Gnep et al., 2017 [33] | Geometrical characteristics and Haralick texture correlate with Gleason score, and they are associated with biochemical recurrence |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Midiri, F.; Vernuccio, F.; Purpura, P.; Alongi, P.; Bartolotta, T.V. Multiparametric MRI and Radiomics in Prostate Cancer: A Review of the Current Literature. Diagnostics 2021, 11, 1829. https://doi.org/10.3390/diagnostics11101829
Midiri F, Vernuccio F, Purpura P, Alongi P, Bartolotta TV. Multiparametric MRI and Radiomics in Prostate Cancer: A Review of the Current Literature. Diagnostics. 2021; 11(10):1829. https://doi.org/10.3390/diagnostics11101829
Chicago/Turabian StyleMidiri, Federico, Federica Vernuccio, Pierpaolo Purpura, Pierpaolo Alongi, and Tommaso Vincenzo Bartolotta. 2021. "Multiparametric MRI and Radiomics in Prostate Cancer: A Review of the Current Literature" Diagnostics 11, no. 10: 1829. https://doi.org/10.3390/diagnostics11101829
APA StyleMidiri, F., Vernuccio, F., Purpura, P., Alongi, P., & Bartolotta, T. V. (2021). Multiparametric MRI and Radiomics in Prostate Cancer: A Review of the Current Literature. Diagnostics, 11(10), 1829. https://doi.org/10.3390/diagnostics11101829








