MicroRNAs as Useful Tools to Estimate Time Since Death. A Systematic Review of Current Literature
Abstract
1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
2.2. Search Criteria and Critical Appraisal
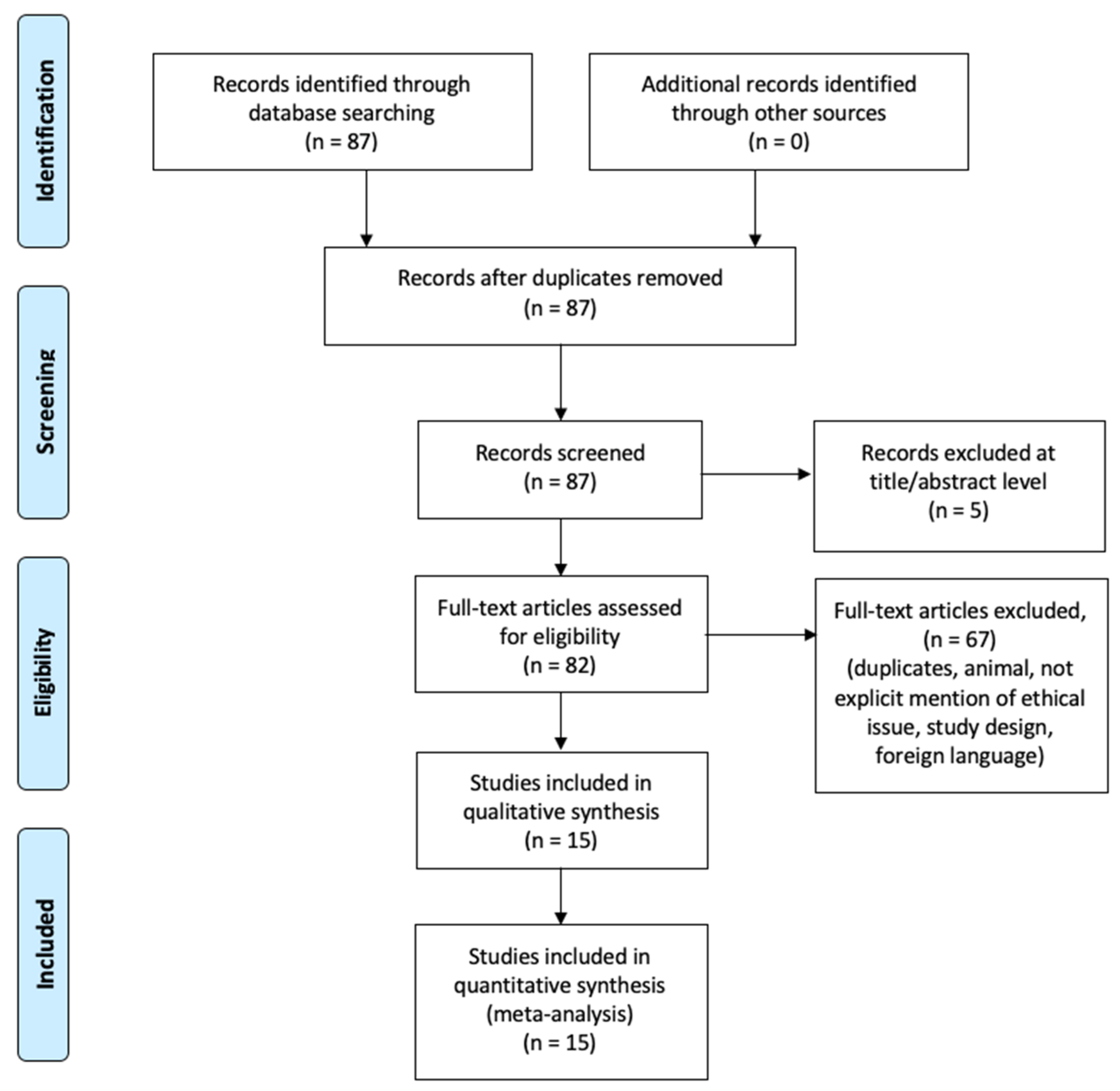
2.3. Search Results and Included Studies
- use of miRNAs in PMI determination;
- postmortem findings;
- study design.
2.4. Risk of Bias
3. Results
4. Discussion
4.1. Current Methods of PMI Estimation
4.2. Postmortem Changes in Biomolecules
4.3. RNA
4.4. Current Literature about miRNAs in PMI
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| PICO | Participants: Intervention, Control, and Outcomes |
| EMBASE | Excerpta Medica Database |
| PCR | Polymerase chain reaction |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| mRNA | Messenger RNA |
| rRNA | Ribosomal RNA |
| miRNA | MicroRNA |
References
- Buchan, M.J.; Anderson, G.S. Time since death: A review of the current status of methods used in the later postmortem interval. J. Can. Soc. Forensic Sci. 2001, 34, 1–22. [Google Scholar] [CrossRef]
- Sachs, J.S. Corpse: Nature, Forensics, and the Struggle to Pinpoint Time of Death, 2nd ed.; Perseus Publishing: New York, NY, USA, 2002; pp. 1–123. [Google Scholar]
- Madea, B. Importance of supravitality in forensic medicine. Forensic Sci. Int. 1994, 69, 221–241. [Google Scholar] [CrossRef]
- Payne-James, J.; Smock, W.; Busuttil, A. Forensic Medicine: Clinical and Pathological Aspects; Greenwich Medical Media: London, UK, 2002; pp. 1–832. [Google Scholar]
- Vass, A.A.; Barshick, S.A.; Sega, G.; Caton, J.; Skeen, J.T.; Love, J.C.; Sinstelien, J.A. Decomposition chemistry of human remains: A new methodology for determining the postmortem interval. J. Forensic Sci. 2002, 47, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Kassam, N.; Gauthier, M.L.; O’Day, D.H. Postmortem changes in calmodulin binding proteins in muscle and lung. Forensic Sci. Int. 2003, 131, 140–147. [Google Scholar] [CrossRef]
- Inoue, H.; Kimura, A.; Tuji, T. Degradation profile of mRNA in a dead rat body: Basic semi-quantification study. Forensic Sci. Int. 2002, 130, 127–132. [Google Scholar] [CrossRef]
- Larkin, B.; Iaschi, S.; Dadour, I.; Tay, G.K. Using accumulated degree-days to estimate postmortem interval from the DNA yield of porcine skeletal muscle. Forensic Sci. Med. Pathol. 2010, 6, 83–92. [Google Scholar] [CrossRef]
- Birdsill, A.C.; Walker, D.G.; Lue, L.F.; Sue, L.I.; Beach, T.G. Postmortem interval effect on RNA and gene expression in human brain tissue. Cell Tissue Bank. 2011, 12, 311–318. [Google Scholar] [CrossRef]
- Partemi, S.; Berne, P.M.; Batlle, M.; Berruezo, A.; Mont, L.; Riuró, H.; Ortiz, J.T.; Roig, E.; Pascali, V.L.; Brugada, R.; et al. Analysis of mRNA from human heart tissue and putative applications in forensic molecular pathology. Forensic Sci. Int. 2010, 203, 99–105. [Google Scholar] [CrossRef]
- Bauer, M.; Gramlich, I.; Polzin, S.; Patzelt, D. Quantification of mRNA degradation as possible indicator of postmortem interval--a pilot study. Leg. Med. 2003, 5, 220–227. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in C. elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Wegman, D.W.; Cherney, L.T.; Yousef, G.M.; Krylov, S.N. Universal drag tag for direct quantitative analysis of multiple microRNAs. Anal. Chem. 2013, 85, 6518–6523. [Google Scholar] [CrossRef]
- Lees, T.; Nassif, N.; Simpson, A.; Shad-Kaneez, F.; Martiniello-Wilks, R.; Lin, Y.; Jones, A.; Qu, X.; Lal, S. Recent advances in molecular biomarkers for diabetes mellitus: A systematic review. Biomarkers 2017, 22, 604–613. [Google Scholar] [CrossRef]
- Croce, C.M.; Calin, G.A. miRNAs, cancer, and stem cell division. Cell 2005, 122, 6–7. [Google Scholar] [CrossRef]
- Pinchi, E.; Frati, P.; Aromatario, M.; Cipolloni, L.; Fabbri, M.; La Russa, R.; Maiese, A.; Neri, M.; Santurro, A.; Scopetti, M.; et al. miR-1, miR-499 and miR-208 are sensitive markers to diagnose sudden death due to early acute myocardial infarction. J. Cell. Mol. Med. 2019, 23, 6005–6016. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, I.; Eran, A.; Nishino, I.; Moggio, M.; Lamperti, C.; Amato, A.A.; Lidov, H.G.; Kang, P.B.; North, K.N.; Mitrani-Rosenbaum, S.; et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc. Natl. Acad. Sci. USA 2007, 104, 17016–17021. [Google Scholar] [CrossRef] [PubMed]
- Pinchi, E.; Frati, A.; Cantatore, S.; D’Errico, S.; Russa, R.; Maiese, A.; Palmieri, M.; Pesce, A.; Viola, R.V.; Frati, P.; et al. Acute spinal cord injury: A systematic review investigating miRNA families involved. Int. J. Mol. Sci. 2019, 20, 1841. [Google Scholar] [CrossRef] [PubMed]
- Hanson, E.K.; Lubenow, H.; Ballantyne, J. Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal. Biochem. 2009, 387, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Neri, M.; Fabbri, M.; D’Errico, S.; Di Paolo, M.; Frati, P.; Gaudio, R.M.; La Russa, R.; Maiese, A.; Marti, M.; Pinchi, E.; et al. Regulation of miRNAs as new tool for cutaneous vitality lesions demonstration in ligature marks in deaths by hanging. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Odriozola, A.; Riancho, J.A.; de la Vega, R.; Agudo, G.; García-Blanco, A.; de Cos, E.; Fernández, F.; Sañudo, C.; Zarrabeitia, M.T. MiRNA analysis in vitreous humor to determine the time of death: A proof-of-concept pilot study. Int. J. Leg. Med. 2013, 127, 573–578. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: Explanation and elaboration. BMJ 2009, 339, b2700. [Google Scholar] [CrossRef]
- Wang, H.; Mao, J.; Lib, Y.B.; Luo, H.; Wu, J.; Liao, M.; Liang, W.; Zhang, L. 5 miRNA expression analysis in postmortem interval (PMI) within 48h. Forensic Sci. Int. Genet. Suppl. Ser. 2013, 4, e190–e191. [Google Scholar] [CrossRef]
- Tu, C.; Du, T.; Ye, X.; Shao, C.; Xie, J.; Shen, Y. Using miRNAs and circRNAs to estimate PMI in advanced stage. Leg. Med. 2019, 38, 51–57. [Google Scholar] [CrossRef]
- Li, W.C.; Ma, K.J.; Zhang, P.; Wang, H.J.; Shen, Y.W.; Zhou, Y.Q.; Zhao, Z.Q.; Ma, D.; Chen, L. Estimation of postmortem interval using microRNA and 18S rRNA degradation in rat cardiac muscle. Fa Yi Xue Za Zhi 2010, 26, 413–417. [Google Scholar]
- Pan, H.; Zhang, H.; Lü, Y.H.; Ma, J.L.; Ma, K.J.; Chen, L. Correlation between five RNA markers of rat’s skin and PMI at different temperatures. Fa Yi Xue Za Zhi 2014, 30, 245–249. [Google Scholar] [PubMed]
- Lv, Y.H.; Ma, K.J.; Zhang, H.; He, M.; Zhang, P.; Shen, Y.W.; Jiang, N.; Ma, D.; Chen, L. A time course study demonstrating mRNA, microRNA, 18S rRNA, and U6 snRNA changes to estimate PMI in deceased rat’s spleen. J. Forensic Sci. 2014, 59, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Pan, H.; Zeng, Y.; Lv, Y.; Zhang, H.; Xue, A.; Jiang, J.; Ma, K.; Chen, L. Exploration of the R code-based mathematical model for PMI estimation using profiling of RNA degradation in rat brain tissue at different temperatures. Forensic Sci. Med. Pathol. 2015, 11, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Lü, Y.H.; Li, Z.H.; Tuo, Y.; Liu, L.; Li, K.; Bian, J.; Ma, J.L.; Chen, L. Correlation between RNA Degradation Patterns of Rat’s Brain and Early PMI at Different Temperatures. Fa Yi Xue Za Zhi 2016, 32, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.H.; Ma, J.L.; Pan, H.; Zhang, H.; Li, W.C.; Xue, A.M.; Wang, H.J.; Ma, K.J.; Chen, L. RNA degradation as described by a mathematical model for postmortem interval determination. J. Forensic Leg. Med. 2016, 44, 43–52. [Google Scholar] [CrossRef]
- Lv, Y.H.; Ma, J.L.; Pan, H.; Zeng, Y.; Tao, L.; Zhang, H.; Li, W.C.; Ma, K.J.; Chen, L. Estimation of the human postmortem interval using an established rat mathematical model and multi-RNA markers. Forensic Sci. Med. Pathol. 2017, 13, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.; Du, T.; Shao, C.; Liu, Z.; Li, L.; Shen, Y. Evaluating the potential of housekeeping genes, rRNAs, snRNAs, microRNAs and circRNAs as reference genes for the estimation of PMI. Forensic Sci. Med. Pathol. 2018, 14, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Kuai, J.-X.; Liu, Y.; Zhang, Y.W. A study on the relationship between the degradation of tubulin in cardiac muscle and lung of rat and the postmortem interval. Chin. J. Forensic Med. 2008, 23, 96–98. [Google Scholar]
- Sharma, S.; Singh, D.; Kaul, D. AATF RNome has the potential to define post mortem interval. Forensic Sci. Int. 2015, 247, e21–e24. [Google Scholar] [CrossRef]
- Risoluti, R.; Canepari, S.; Frati, P.; Fineschi, V.; Materazzi, S. “2 (n) Analytical Platform” to Update Procedures in Thanatochemistry: Estimation of Post Mortem Interval in Vitreous Humor. Anal. Chem. 2019, 91, 7025–7031. [Google Scholar] [CrossRef]
- Henssge, C.; Madea, B. Estimation of the time since death. Forensic Sci. Int. 2007, 165, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Giles, S.B.; Harrison, K.; Errickson, D.; Márquez-Grant, N. The effect of seasonality on the application of accumulated degree-days to estimate the early post-mortem interval. Forensic Sci. Int. 2020, 315, 110419. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.A. A Summer Carrion Study of the Baby Pig Sus Scrofa Linnaeus. Ecology 1965, 46, 592–602. [Google Scholar] [CrossRef]
- Gill-King, H. Chemical and ultrastructural aspects of decomposition. In Forensic Taphonomy: The Postmortem Fate of Human Remains; Haglund, W.D., Sorg, M.H., Eds.; CRC Press: Boca Raton, FL, USA, 1997; pp. 93–108. [Google Scholar]
- Mann, R.W.; Bass, W.M.; Meadows, L. Time since death and decomposition of the human body: Variables and observations in case and experimental field studies. J. Forensic Sci. 1990, 35, 12806. [Google Scholar] [CrossRef]
- Mathur, A.; Agrawal, Y.K. An overview of methods used for estimation of time since death. Aust. J. Forensic Sci. 2011, 43, 275–285. [Google Scholar] [CrossRef]
- Wilk, L.S.; Hoveling, R.J.M.; Edelman, G.J.; Hardy, H.J.J.; van Schouwen, S.; van Venrooij, H.; Aalders, M.C.G. Reconstructing the time since death using noninvasive thermometry and numerical analysis. Sci. Adv. 2020, 22, eaba4243. [Google Scholar] [CrossRef]
- Fais, P.; Mazzotti, M.C.; Teti, G.; Boscolo-Berto, R.; Pelotti, S.; Falconi, M. HIF1α protein and mRNA expression as a new marker for post mortem interval estimation in human gingival tissue. J. Anat. 2018, 232, 1031–1037. [Google Scholar] [CrossRef]
- Tao, L.; Ma, J.; Han, L.; Xu, H.; Zeng, Y.; Yehui, L.; Li, W.; Ma, K.; Xiao, B.; Chen, L. Early postmortem interval estimation based on Cdc25b mRNA in rat cardiac tissue. Leg. Med. 2018, 35, 18–24. [Google Scholar] [CrossRef]
- Elghamry, H.A.; Mohamed, M.I.; Hassan, F.M.; Abdelfattah, D.S.; Abdelaal, A.G. Potential use of GAPDH m-RNA in estimating PMI in brain tissue of albino rats at different environmental conditions. Egypt. J. Forensic Sci. 2017, 7, 24. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, Y.; Cha, H.K.; Lim, H.Y.; Kim, H.; Chung, S.; Hwang, J.J.; Park, S.H.; Son, G.H. Cell death-associated ribosomal RNA cleavage in postmortem tissues and its forensic applications. Mol. Cells 2017, 40, 410–417. [Google Scholar]
- Madea, B. Is there recent progress in the estimation of the postmortem interval by means of thanatochemistry? Forensic Sci. Int. 2005, 151, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Lijiang, L.; Xiji, S.; Liang, R.; Hongyan, Z.; Yan, L.; Wei, L.; Cheng, Z.; Liang, L. Determination of the early time of death by computerized image analysis of DNA degradation: Which is the best quantitative indicator of DNA degradation? J. Huazhong Univ. Sci. Technol. 2007, 27, 362–366. [Google Scholar]
- Li, W.C.; Ma, K.J.; Lv, Y.H.; Zhang, P.; Pan, H.; Zhang, H.; Wang, H.J.; Ma, D.; Chen, L. Postmortem interval determination using 18S-rRNA and microRNA. Sci. Justice 2014, 54, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Poór, V.S.; Lukács, D.; Nagy, T.; Rácz, E.; Sipos, K. The rate of RNA degradation in human dental pulp reveals postmortem interval. Int. J. Leg. Med. 2016, 130, 615–619. [Google Scholar] [CrossRef]
- Sampaio-Silva, F.; Magalhães, T.; Carvalho, F.; Dinis-Oliveira, R.J.; Silvestre, R. Profiling of RNA Degradation for Estimation of Post Morterm Interval. PLoS ONE 2013, 8, e56507. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, P.; Ma, K.J.; Lv, Y.H.; Li, W.C.; Luo, C.L.; Li, L.L.; Shen, Y.W.; He, M.; Jiang, J.Q.; et al. The selection of endogenous genes in human postmortem tissues. Sci. Justice 2013, 53, 115–120. [Google Scholar] [CrossRef]
- Koppelkamm, A.; Vennemann, B.; Fracasso, T.; Lutz-Bonengel, S.; Schmidt, U.; Heinrich, M. Validation of adequate endogenous reference genes for the normalisation of qPCR gene expression data in human post mortem tissue. Int. J. Leg. Med. 2010, 124, 371–380. [Google Scholar] [CrossRef]
- Bauer, M.; Polzin, S.; Patzelt, D. Quantification of RNA degradation by semi-quantitative duplex and competitive RT-PCR: A possible indicator of the age of bloodstains? Forensic Sci. Int. 2003, 138, 94–103. [Google Scholar] [CrossRef]
- Li, C.; Wang, Q.; Zhang, Y.; Lin, H.; Zhang, J.; Huang, P.; Wang, Z. Research progress in the estimation of the postmortem interval by Chinese forensic scholars. Forensic Sci. Res. 2016, 1, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, M.; Matt, K.; Lutz-Bonengel, S.; Schmidt, U. Successful RNA extraction from various human postmortem tissues. Int. J. Leg. Med. 2007, 121, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Preece, P.; Cairns, N. Quantifying mRNA in postmortem human brain: Influence of gender, age at death, postmortem interval, brain pH, agonal state and inter-lobe mRNA variance. Brain Res. Mol. Brain Res. 2003, 118, 60–71. [Google Scholar] [CrossRef]
- Young, S.T.; Wells, J.D.; Hobbs, G.R.; Bishop, C.P. Estimating postmortem interval using RNA degradation and morphological changes in tooth pulp. Forensic Sci. Int. 2013, 229, e1–e163. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, M.; Lutz-Bonengel, S.; Matt, K.; Schmidt, U. Real-time PCR detection of five different ‘endogenous control gene’ transcripts in forensic autopsy material. Forensic Sci. Int. Genet. 2007, 1, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Scrivano, S.; Sanavio, M.; Tozzo, P.; Caenazzo, L. Analysis of RNA in the estimation of postmortem interval: A review of current evidence. Int. J. Leg. Med. 2019, 133, 1629–1640. [Google Scholar] [CrossRef]
- Lü, Y.H.; Ma, K.J.; Li, Z.H.; Gu, J.; Bao, J.Y.; Yang, Z.F.; Gao, J.; Zeng, Y.; Tao, L.; Chen, L. Correlation between RNA Expression Level and Early PMI in Human Brain Tissue. Fa Yi Xue Za Zhi 2016, 32, 245–249. [Google Scholar] [CrossRef]
- Tozzo, P.; Scrivano, S.; Sanavio, M.; Caenazzo, L. The Role of DNA degradation in the estimation of post-mortem interval: A systematic review of the current literature. Int. J. Mol. Sci. 2020, 21, 3540. [Google Scholar] [CrossRef]
- Fineschi, V.; Picchi, M.P.; Tassini, M.; Valensin, G.; Vivi, A. 1H-NMR studies of postmortem biochemical changes in rat skeletal muscle. Forensic Sci. Int. 1990, 44, 225–236. [Google Scholar] [CrossRef]
- Zubakov, D.; Boersma, A.W.; Choi, Y.; van Kuijk, P.F.; Wiemer, E.A.; Kayser, M. MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int. J. Leg. Med. 2010, 124, 217–226. [Google Scholar] [CrossRef]
- Di Nunno, N.R.; Costantinides, F.; Bernasconi, P.; Bottin, C.; Melato, M. Is flow cytometric evaluation of DNA degradation a reliable method to investigate the early postmortem period? Am. J. Forensic Med. Pathol. 1998, 19, 50–53. [Google Scholar] [CrossRef]
- Johnson, L.A.; Ferris, J.A.J. Analysis of postmortem DNA degradation by single-cell gel electrophoresis. Forensic Sci. Int. 2002, 126, 43–47. [Google Scholar] [CrossRef]
- Chen, J.H.; Inamori-Kawamoto, O.; Michiue, T.; Ikeda, S.; Ishikawa, T.; Maeda, H. Cardiac biomarkers in blood, and pericardial and cerebrospinal fluids of forensic autopsy cases: A reassessment with special regard to postmortem interval. Leg. Med. 2015, 17, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Kawahara, K.I.; Biswas, K.K.; Ito, T.; Tancharoen, S.; Shiomi, N.; Koda, Y.; Matsuda, F.; Morimoto, Y.; Oyama, Y.; et al. HMGB1: A new marker for estimation of the postmortem interval. Exp. Ther. Med. 2010, 1, 109–111. [Google Scholar] [CrossRef] [PubMed]
- Poloz, Y.O.; O’Day, D.H. Determining time of death: Temperature-dependent postmortem changes in calcineurin A, MARCKS, CaMKII, and protein phosphatase 2A in mouse. Int. J. Leg. Med. 2009, 123, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Ali, W.; Singh, U.S.; Kumar, A.; Verma, A.; Bhattacharya, S. The effect of elapsed time on the cardiac Troponin-T (cTnT) proteolysis in case of death due to burn: A study to evaluate the potential forensic use of cTnT to determine the postmortem interval. Sci. Justice 2014, 55, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Kimura, A.; Ishida, Y.; Hayashi, T.; Nosaka, M.; Kondo, T. Estimating time of death based on the biological clock. Int. J. Leg. Med. 2011, 125, 385–391. [Google Scholar] [CrossRef] [PubMed]
| miRNA | Epicrisis of PMI | Tissues and Organs | Sample Source | Year | Authors [Reference] |
|---|---|---|---|---|---|
| miR-206 | downregulated after 24 h | Liver | mice | 2013 | Wang, H.; et al. [30] |
| miR-150 | downregulated after 24 h | Liver | mice | 2013 | Wang, H.; et al. [30] |
| miR-133 | downregulated after 24 h | Liver | mice | 2013 | Wang, H.; et al. [30] |
| miR-122 | downregulated after 24 h | Liver | mice | 2013 | Wang, H.; et al. [30] |
| miR-195 | upregulated for first 24 h, then downregulated | Liver | mice | 2013 | Wang, H.; et al. [30] |
| miR-122 | stable in heart tissue and liver; downregulated in skeletal muscle | heart, liver and skeletal muscle | mice | 2018 | Tu, C.; et al. [31] |
| miR-133a | stable in heart tissue; downregulated in skeletal muscle and liver | heart, liver and skeletal muscle | mice | 2018 | Tu, C.; et al. [31] |
| miR-1 | stable for fist 120 h then downregulated | Heart | rat | 2010 | Li, W.C.; et al. [32] |
| miR-2 | stable for fist 120 h then downregulated | Heart | rat | 2010 | Li, W.C.; et al. [32] |
| miR-203 | miR-203 levels in this study were taken as internal reference because of his good stability. | Skin | rat | 2014 | Pan, H.; et al. [33] |
| miR-125b | the Ct values of miRs fluctuated slightly within 36 h and then increased slowly at 25 °C, with the same trend observed within 144 h at 4 °C. microRNAs are less susceptible to degradation induced by PMI and environmental conditions because of its short length of 21–25 bp. | Spleen | rat | 2014 | Lv, Y.H.; et al. [34] |
| miR-143 | the Ct values of miRs fluctuated slightly within 36 h and then increased slowly at 25 °C, with the same trend observed within 144 h at 4 °C. microRNAs are less susceptible to degradation induced by PMI and environmental conditions because of its short length of 21–25 bp. | Spleen | rat | 2014 | Lv, Y.H.; et al. [34] |
| miR-9 | stable up to 144 h post mortem | Brain | rat | 2015 | Ma, J.; et al. [35] |
| miR-125b | stable up to 144 h post mortem | Brain | rat | 2015 | Ma, J.; et al. [35] |
| miR-9 | stable up to 24 h post mortem | Brain | rat | 2016 | Lü, Y.H.; et al. [36] |
| miR-125b | stable up to 24 h post mortem | Brain | rat | 2016 | Lü, Y.H.; et al. [36] |
| miR-195 | chosen as control markers for muscle tissues because stable up to 144 h, then downregulated | lung; skeletal muscle | rat | 2016 | Lv, Y.H.; et al. [37] |
| miR-200c | chosen as control markers for muscle tissues because stable up to 144 h, then downregulated | lung; skeletal muscle | rat | 2016 | Lv, Y.H.; et al. [37] |
| miR-201 | chosen as control markers for muscle tissues because stable up to 144 h, then downregulated | lung; skeletal muscle | rat | 2016 | Lv, Y.H.; et al. [37] |
| miR-206 | chosen as control markers for muscle tissues because stable up to 144 h, then downregulated | lung; skeletal muscle | rat | 2016 | Lv, Y.H.; et al. [37] |
| miR-9 | stable enough in order to be reference markers up to 22.5 h | Brain | dead body | 2016 | Lü, Y.H.; et al. [34] |
| miR-125b | stable enough in order to be reference markers up to 22.5 h | Brain | dead body | 2016 | Lü, Y.H.; et al. [34] |
| miR-1 | myocardium-specific fairly stable over 5 or more days, even at 35 °C | myocardium, liver, brain | dead body | 2017 | Lv, Y.H.; et al. [38] |
| miR-133a | myocardium-specific fairly stable over 5 or more days, even at 35 °C | myocardium, liver, brain | dead body | 2017 | Lv, Y.H.; et al. [38] |
| miR-122 | degrading after 4 days of PMI particularly at high temperatures | myocardium, liver, brain | dead body | 2017 | Lv, Y.H.; et al. [38] |
| miR-9 | are suitable as internal reference markers of human brain tissue | myocardium, liver, brain | dead body | 2017 | Lv, Y.H.; et al. [38] |
| miR-125b | are suitable as internal reference markers of human brain tissue | myocardium, liver, brain | dead body | 2017 | Lv, Y.H.; et al. [38] |
| miR-122 | stable up to 8 days choosen as reference control up to 7.5 d of PMI | heart, liver, skeletal muscle | rat | 2019 | Tu, C.; et al. [39] |
| miR-133a | stable up to 8 days choosen as reference control up to 7.5 d of PMI | heart, liver, skeletal muscle | rat | 2019 | Tu, C.; et al. [39] |
| miR-34c | CT downregulated with increase of PMI | vitreous humor | human | 2013 | Odriozola, A.; et al. [28] |
| miR-222 | best reference gene, stable at least for 24 h | vitreous humor | human | 2013 | Odriozola, A.; et al. [28] |
| miR-888 | almost stable up to 24 h post mortem, stable at least for 24 h | vitreous humor | human | 2013 | Odriozola, A.; et al. [28] |
| miR-484 | CT downregulated with increase of PMI, stable at least for 24 h | vitreous humor | human | 2013 | Odriozola, A.; et al. [28] |
| miR-142-5p | upregulated in night time deaths, stable at least for 24 h | vitreous humor | human | 2013 | Odriozola, A.; et al. [28] |
| miR-541 | upregulated in night time deaths, stable at least for 24 h | vitreous humor | human | 2013 | Odriozola, A.; et al. [28] |
| miR-miR-1 | upregulated until 96 h | heart | rats | 2014 | Kuai, J.-X.; et al. [40] |
| miR-1 | - | muscle | rat | 2016 | Lv, Y.H.; et al. [34] |
| miR-206 | - | muscle | rat | 2016 | Lv, Y.H.; et al. [34] |
| miR-195 | - | lung | rat | 2016 | Lv, Y.H.; et al. [34] |
| miR-2909 | stable up to 48 h post death if the mouse was sacrificed at 8 PM and 12 h post death if the mouse was sacrificed at noon. They also concluded that miR-2909 is much more stable to enzymatic degradation then AATF mRNA | brain, heart, lungs, kidneys, pancreas, spleen and liver | mice | 2014 | Sharma, S.; et al. [41] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maiese, A.; Scatena, A.; Costantino, A.; Di Paolo, M.; La Russa, R.; Turillazzi, E.; Frati, P.; Fineschi, V. MicroRNAs as Useful Tools to Estimate Time Since Death. A Systematic Review of Current Literature. Diagnostics 2021, 11, 64. https://doi.org/10.3390/diagnostics11010064
Maiese A, Scatena A, Costantino A, Di Paolo M, La Russa R, Turillazzi E, Frati P, Fineschi V. MicroRNAs as Useful Tools to Estimate Time Since Death. A Systematic Review of Current Literature. Diagnostics. 2021; 11(1):64. https://doi.org/10.3390/diagnostics11010064
Chicago/Turabian StyleMaiese, Aniello, Andrea Scatena, Andrea Costantino, Marco Di Paolo, Raffaele La Russa, Emanuela Turillazzi, Paola Frati, and Vittorio Fineschi. 2021. "MicroRNAs as Useful Tools to Estimate Time Since Death. A Systematic Review of Current Literature" Diagnostics 11, no. 1: 64. https://doi.org/10.3390/diagnostics11010064
APA StyleMaiese, A., Scatena, A., Costantino, A., Di Paolo, M., La Russa, R., Turillazzi, E., Frati, P., & Fineschi, V. (2021). MicroRNAs as Useful Tools to Estimate Time Since Death. A Systematic Review of Current Literature. Diagnostics, 11(1), 64. https://doi.org/10.3390/diagnostics11010064








