Regional Mapping of Brain Glutamate Distributions Using Glutamate-Weighted Chemical Exchange Saturation Transfer Imaging
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Preparation
2.2. Data Acquisition
2.3. Data Analysis
2.4. Statistical Analysis
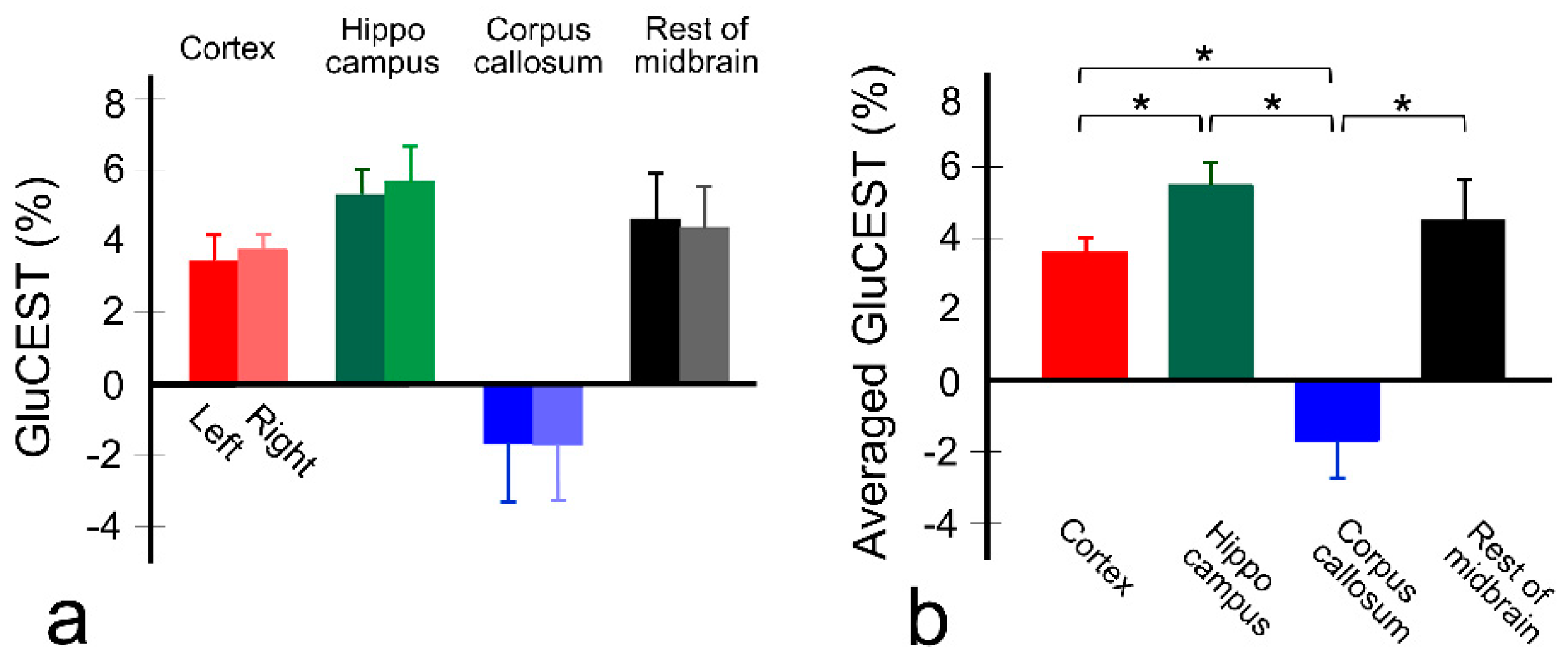
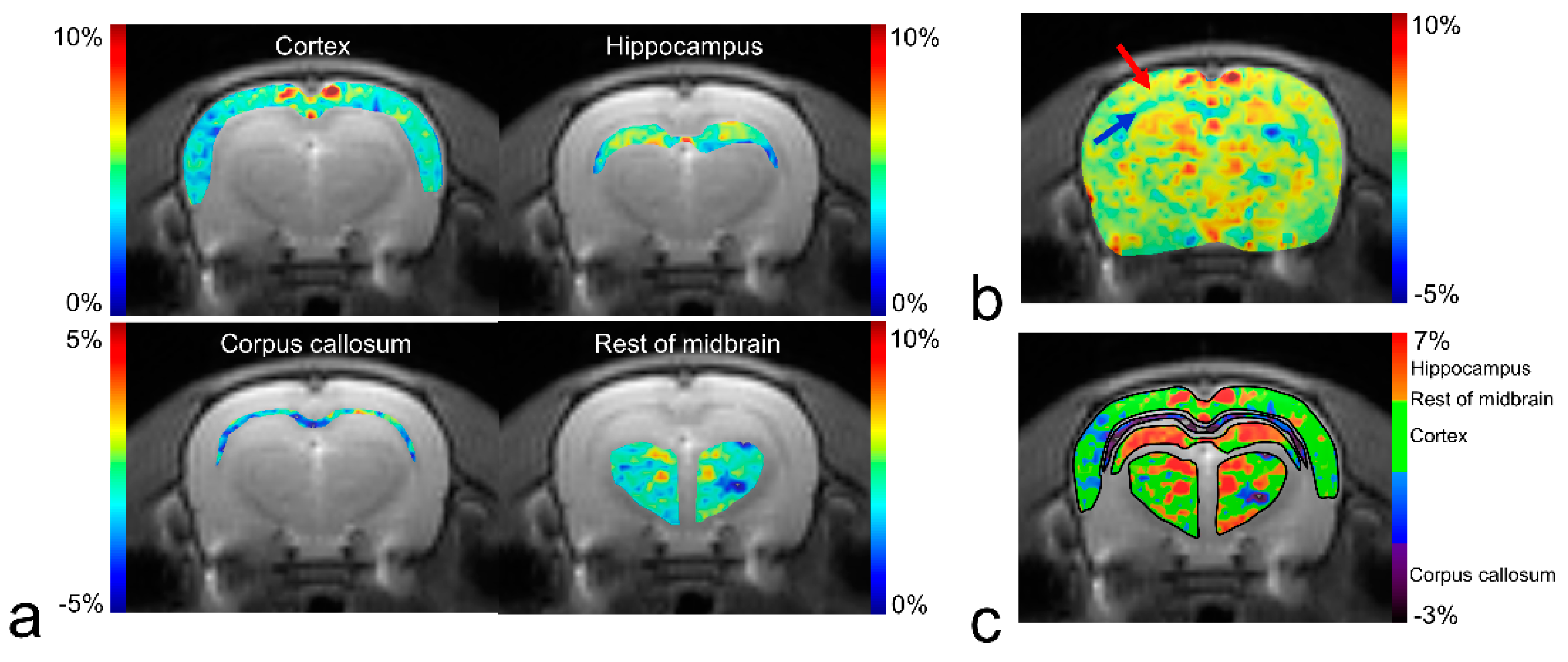
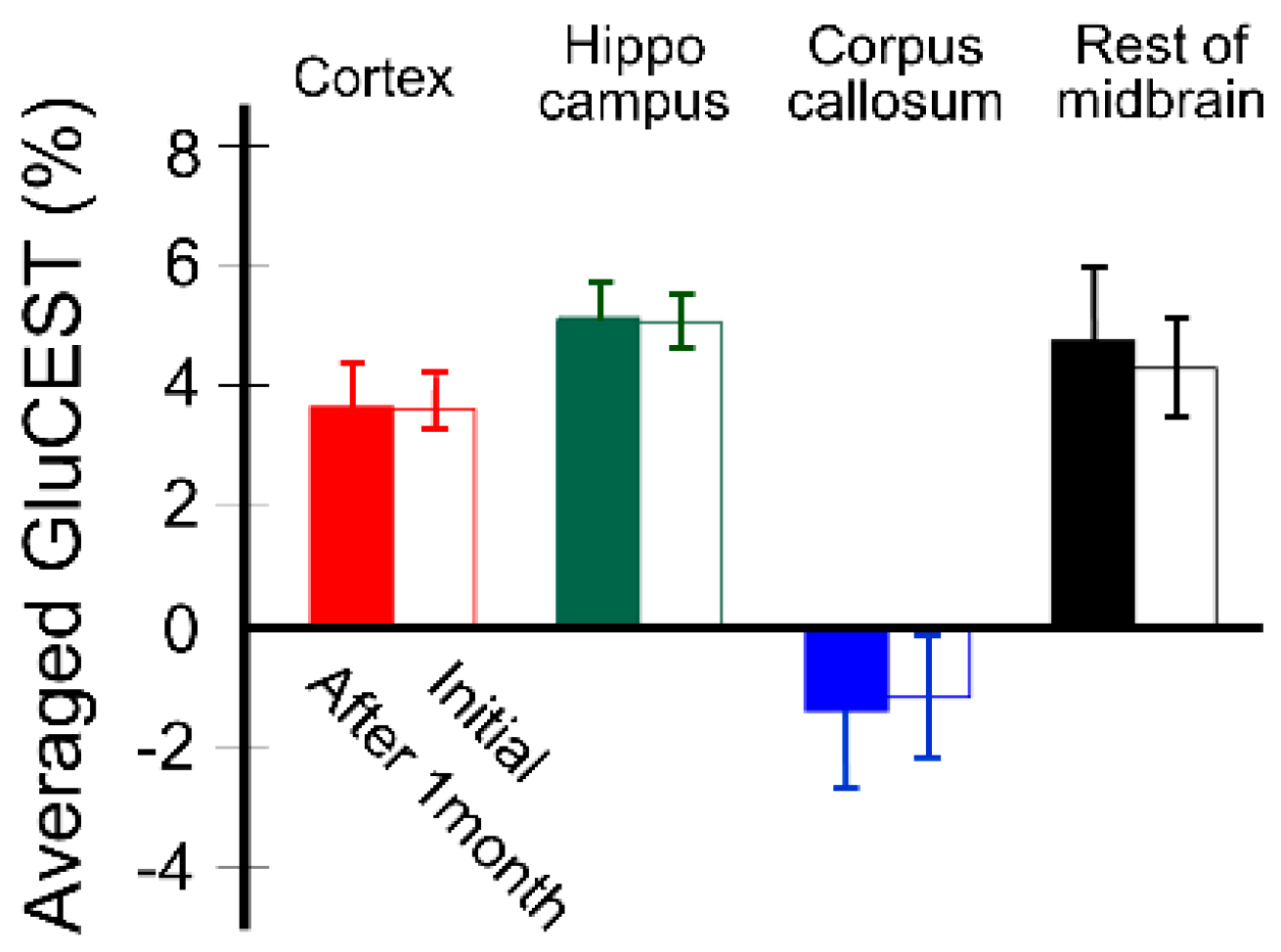
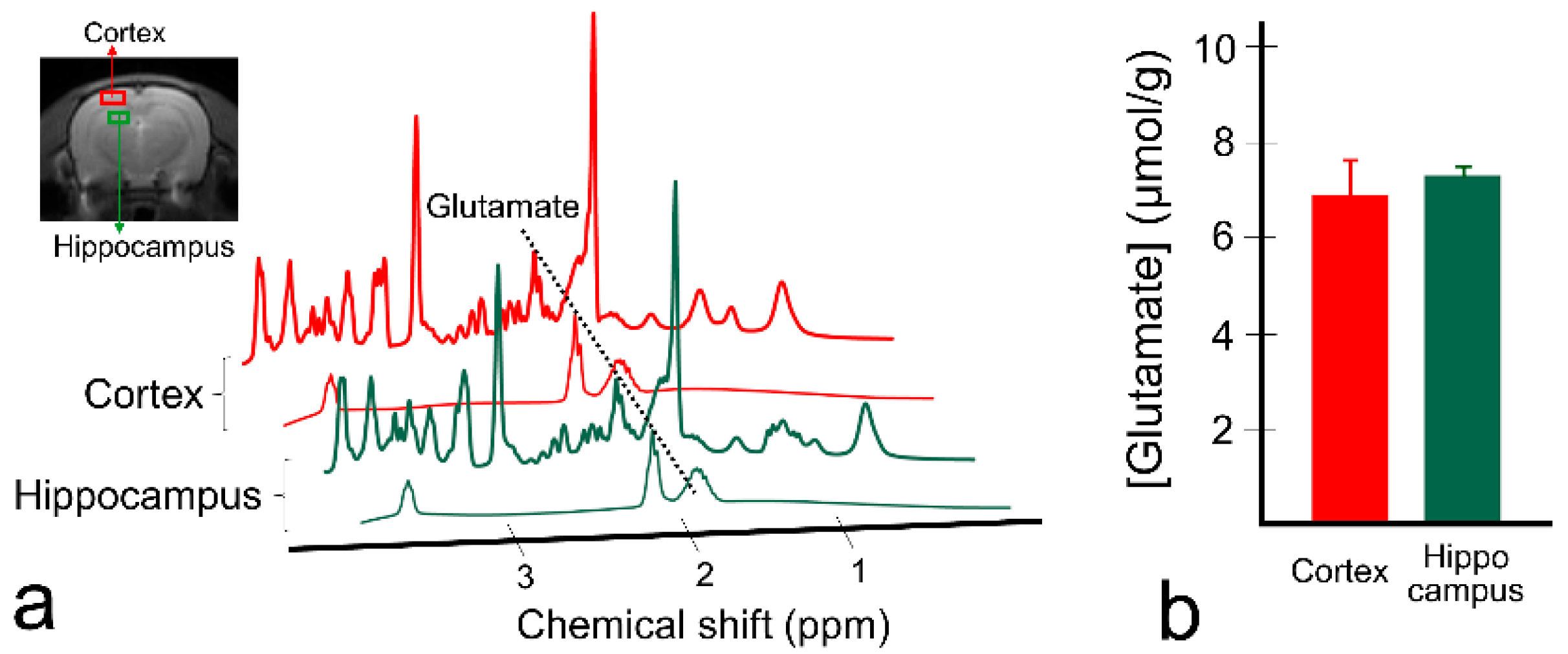
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nakanishi, S. Molecular Diversity of Glutamate Receptors and Implications for Brain-Function. Science 1992, 258, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Nedergaard, M.; Takano, T.; Hansen, A.J. Beyond the role of glutamate as a neurotransmitter. Nat. Rev. Neurosci. 2002, 3, 748–755. [Google Scholar] [CrossRef] [PubMed]
- Fonnum, F. Glutamate a Neurotransmitter in Mammalian Brain. J. Neurochem. 1984, 42, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Silvestrin, R.B.; Bambini-Junior, V.; Galland, F.; Bobermim, L.D.; Quincozes-Santos, A.; Abib, R.T.; Zanotto, C.; Batassini, C.; Brolese, G.; Goncalves, C.A.; et al. Animal model of autism induced by prenatal exposure to valproate: Altered glutamate metabolism in the hippocampus. Brain Res. 2013, 1495, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Horder, J.; Petrinovic, M.M.; Mendez, M.A.; Bruns, A.; Takumi, T.; Spooren, W.; Barker, G.J.; Kunnecke, B.; Murphy, D.G. Glutamate and GABA in autism spectrum disorder-a translational magnetic resonance spectroscopy study in man and rodent models. Transl. Psychiatry 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Modrego, P.J.; Fayed, N.; Salinas, G.R. Brain Glutamate Levels Are Decreased in Alzheimer’s Disease. A Magnetic Resonance Spectroscopy Study. Eur. J. Neurol. 2011, 18, 73. [Google Scholar]
- Anggono, V.; Tsai, L.H.; Gotz, J. Glutamate Receptors in Alzheimer’s Disease: Mechanisms and Therapies. Neural Plasticity 2016. [Google Scholar] [CrossRef]
- Sun, D.A.; Sombati, S.; DeLorenzo, R.J. Glutamate injury-induced epileptogenesis in hippocampal neurons: An in vitro model of stroke-induced “epilepsy”. Stroke 2001, 32, 2344–2350. [Google Scholar] [CrossRef]
- Bradford, H.F. Glutamate, GABA and epilepsy. Prog. Neurobiol. 1995, 47, 477–511. [Google Scholar] [CrossRef]
- Petroff, O.A.; Mattson, R.H.; Rothman, D.L. Proton MRS: GABA and glutamate. Adv. Neurol. 2000, 83, 261–271. [Google Scholar]
- Ramadan, S.; Lin, A.; Stanwell, P. Glutamate and glutamine: A review of in vivo MRS in the human brain. NMR Biomed. 2013, 26, 1630–1646. [Google Scholar] [CrossRef] [PubMed]
- Lally, N.; An, L.; Banerjee, D.; Niciu, M.J.; Luckenbaugh, D.A.; Richards, E.M.; Roiser, J.P.; Shen, J.; Zarate, C.A., Jr.; Nugent, A.C. Reliability of 7T (1) H-MRS measured human prefrontal cortex glutamate, glutamine, and glutathione signals using an adapted echo time optimized PRESS sequence: A between- and within-sessions investigation. J. Magn. Reson. Imaging 2016, 43, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Lohith, T.G.; Tsujikawa, T.; Simeon, F.G.; Veronese, M.; Zoghbi, S.S.; Lyoo, C.H.; Kimura, Y.; Morse, C.L.; Pike, V.W.; Fujita, M.; et al. Comparison of two PET radioligands, [(11)C]FPEB and [(11)C]SP203, for quantification of metabotropic glutamate receptor 5 in human brain. J. Cereb. Blood Flow Metab. 2017, 37, 2458–2470. [Google Scholar] [CrossRef] [PubMed]
- Kil, K.E.; Zhu, A.; Zhang, Z.; Choi, J.K.; Kura, S.; Gong, C.; Brownell, A.L. Development of [(123)I]IPEB and [(123)I]IMPEB as SPECT Radioligands for Metabotropic Glutamate Receptor Subtype 5. ACS Med. Chem. Lett. 2014, 5, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Majo, V.J.; Prabhakaran, J.; Mann, J.J.; Kumar, J.S. PET and SPECT tracers for glutamate receptors. Drug Discov. Today 2013, 18, 173–184. [Google Scholar] [CrossRef]
- Cai, K.; Singh, A.; Roalf, D.R.; Nanga, R.P.; Haris, M.; Hariharan, H.; Gur, R.; Reddy, R. Mapping glutamate in subcortical brain structures using high-resolution GluCEST MRI. NMR Biomed. 2013, 26, 1278–1284. [Google Scholar] [CrossRef]
- Van Zijl, P.C.M.; Yadav, N.N. Chemical Exchange Saturation Transfer (CEST): What is in a Name and What Isn’t? Magn. Reson. Med. 2011, 65, 927–948. [Google Scholar] [CrossRef]
- Ward, K.M.; Aletras, A.H.; Balaban, R.S. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST). J. Magn. Reson. 2000, 143, 79–87. [Google Scholar] [CrossRef]
- Zhou, J.Y.; Payen, J.F.; Wilson, D.A.; Traystman, R.J.; van Zijl, P.C.M. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat. Med. 2003, 9, 1085–1090. [Google Scholar] [CrossRef]
- Haris, M.; Cai, K.J.; Singh, A.; Hariharan, H.; Reddy, R. In vivo mapping of brain myo-inositol. Neuroimage 2011, 54, 2079–2085. [Google Scholar] [CrossRef]
- Van Zijl, P.C.; Jones, C.K.; Ren, J.; Malloy, C.R.; Sherry, A.D. MRI detection of glycogen in vivo by using chemical exchange saturation transfer imaging (glycoCEST). Proc. Natl. Acad. Sci. USA 2007, 104, 4359–4364. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.J.; Haris, M.; Singh, A.; Kogan, F.; Greenberg, J.H.; Hariharan, H.; Detre, J.A.; Reddy, R. Magnetic resonance imaging of glutamate. Nat. Med. 2012, 18, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Kogan, F.; Haris, M.; Singh, A.; Cai, K.J.; Debrosse, C.; Nanga, R.P.R.; Hariharan, H.; Reddy, R. Method for High-Resolution Imaging of Creatine In Vivo Using Chemical Exchange Saturation Transfer. Magn. Reson. Med. 2014, 71, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.A.; Nanga, R.P.; Das, S.; Chen, S.H.; Hadar, P.N.; Pollard, J.R.; Lucas, T.H.; Shinohara, R.T.; Litt, B.; Hariharan, H.; et al. Glutamate imaging (GluCEST) lateralizes epileptic foci in nonlesional temporal lobe epilepsy. Sci. Transl. Med. 2015, 7, 309ra161. [Google Scholar] [CrossRef]
- Bagga, P.; Crescenzi, R.; Krishnamoorthy, G.; Verma, G.; Nanga, R.P.; Reddy, D.; Greenberg, J.; Detre, J.A.; Hariharan, H.; Reddy, R. Mapping the alterations in glutamate with GluCEST MRI in a mouse model of dopamine deficiency. J. Neuro. Chem. 2016, 139, 432–439. [Google Scholar] [CrossRef]
- Roalf, D.R.; Nanga, R.P.R.; Rupert, P.E.; Hariharan, H.; Quarmley, M.; Calkins, M.E.; Dress, E.; Prabhakaran, K.; Elliott, M.A.; Moberg, P.J.; et al. Glutamate imaging (GluCEST) reveals lower brain GluCEST contrast in patients on the psychosis spectrum. Mol. Psychiatry 2017, 22, 1298–1305. [Google Scholar] [CrossRef]
- Bagga, P.; Pickup, S.; Crescenzi, R.; Martinez, D.; Borthakur, A.; D’Aquilla, K.; Singh, A.; Verma, G.; Detre, J.A.; Greenberg, J.; et al. In vivo GluCEST MRI: Reproducibility, background contribution and source of glutamate changes in the MPTP model of Parkinson’s disease. Sci. Rep. 2018, 8, 2883. [Google Scholar] [CrossRef]
- Nanga, R.P.R.; DeBrosse, C.; Kumar, D.; Roalf, D.; McGeehan, B.; D’Aquilla, K.; Borthakur, A.; Hariharan, H.; Reddy, D.; Elliott, M.; et al. Reproducibility of 2D GluCEST in healthy human volunteers at 7 T. Magn. Reson. Med. 2018, 80, 2033–2039. [Google Scholar] [CrossRef]
- Zhou, R.; Bagga, P.; Nath, K.; Hariharan, H.; Mankoff, D.A.; Reddy, R. Glutamate-Weighted Chemical Exchange Saturation Transfer Magnetic Resonance Imaging Detects Glutaminase Inhibition in a Mouse Model of Triple-Negative Breast Cancer. Cancer Res. 2018, 78, 5521–5526. [Google Scholar] [CrossRef]
- Lee, D.H.; Lee, D.W.; Kwon, J.I.; Woo, C.W.; Kim, S.T.; Kim, J.K.; Kim, K.W.; Woo, D.C. Retrospective Brain Motion Correction in Glutamate Chemical Exchange Saturation Transfer (GluCEST) MRI. Mol. Imaging Biol. 2019. [Google Scholar] [CrossRef]
- Lee, D.H.; Woo, C.W.; Kwon, J.I.; Chae, Y.J.; Ham, S.J.; Suh, J.Y.; Kim, S.T.; Kim, J.K.; Kim, K.W.; Woo, D.C. Cerebral mapping of glutamate using chemical exchange saturation transfer imaging in a rat model of stress-induced sleep disturbance at 7.0T. J. Magn. Reson. Imaging 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Shen, Z.; Chen, Y.; Dai, Z.; Zhang, X.; Mao, Y.; Zhang, B.; Zeng, H.; Chen, P.; Wu, R. Mapping the Changes of Glutamate Using Glutamate Chemical Exchange Saturation Transfer (GluCEST) Technique in a Traumatic Brain Injury Model: A Longitudinal Pilot Study. ACS Chem. Neurosci. 2019, 10, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Cai, K.J.; Haris, M.; Hariharan, H.; Reddy, R. On B1 inhomogeneity correction of in vivo human brain glutamate chemical exchange saturation transfer contrast at 7T. Magn. Reson. Med. 2013, 69, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Gillen, J.; Landman, B.A.; Zhou, J.; van Zijl, P.C. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn. Reson. Med. 2009, 61, 1441–1450. [Google Scholar] [CrossRef] [PubMed]
- Kogan, F.; Singh, A.; Debrosse, C.; Haris, M.; Cai, K.; Nanga, R.P.; Elliott, M.; Hariharan, H.; Reddy, R. Imaging of glutamate in the spinal cord using GluCEST. Neuroimage 2013, 77, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Inglese, M.; Rusinek, H.; George, I.C.; Babb, J.S.; Grossman, R.I.; Gonen, O. Global average gray and white matter N-acetylaspartate concentration in the human brain. Neuroimage 2008, 41, 270–276. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, J. Simultaneous Quantification of Glutamate and Glutamine by J-modulated Spectroscopy at 3 Tesla. Magn. Reson. Med. 2016, 76, 725–732. [Google Scholar] [CrossRef]
- Ametamey, S.M.; Treyer, V.; Streffer, J.; Wyss, M.T.; Schmidt, M.; Blagoev, M.; Hintermann, S.; Auberson, Y.; Gasparini, F.; Fischer, U.C.; et al. Human PET studies of metabotropic glutamate receptor subtype 5 with 11C-ABP688. J. Nucl. Med. 2007, 48, 247–252. [Google Scholar]
- Zhao, F.; Zhang, L.; Zhu, A.; Shi, G.; Tian, Y. In vivo monitoring of local pH values in a live rat brain based on the design of a specific electroactive molecule for H(+). Chem. Commun. 2016, 52, 3717–3720. [Google Scholar] [CrossRef]
- Hassel, B.; Boldingh, K.A.; Narvesen, C.; Iversen, E.G.; Skrede, K.K. Glutamate transport, glutamine synthetase and phosphate-activated glutaminase in rat CNS white matter. A quantitative study. J. Neurochem. 2003, 87, 230–237. [Google Scholar] [CrossRef]
- De Graaf, R.A.; Mason, G.F.; Patel, A.B.; Rothman, D.L.; Behar, K.L. Regional glucose metabolism and glutamatergic neurotransmission in rat brain in vivo. Proc. Natl. Acad. Sci. USA 2004, 101, 12700–12705. [Google Scholar] [CrossRef] [PubMed]
- Heo, H.Y.; Zhang, Y.; Lee, D.H.; Jiang, S.; Zhao, X.; Zhou, J. Accelerating chemical exchange saturation transfer (CEST) MRI by combining compressed sensing and sensitivity encoding techniques. Magn. Reson. Med. 2017, 77, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Heo, H.Y.; Lee, D.H.; Jiang, S.; Zhao, X.; Bottomley, P.A.; Zhou, J. Chemical exchange saturation transfer (CEST) imaging with fast variably-accelerated sensitivity encoding (vSENSE). Magn. Reson. Med. 2017, 77, 2225–2238. [Google Scholar] [CrossRef] [PubMed]
- Debnath, A.; Hariharan, H.; Nanga, R.P.R.; Reddy, R.; Singh, A. Glutamate-Weighted CEST Contrast After Removal of Magnetization Transfer Effect in Human Brain and Rat Brain with Tumor. Mol. Imaging Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zu, Z. Towards the molecular origin of glutamate CEST (GluCEST) imaging in rat brain. Magn. Reson. Med. 2020, 83, 1405–1417. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, D.-W.; Woo, C.-W.; Woo, D.-C.; Kim, J.K.; Kim, K.W.; Lee, D.-H. Regional Mapping of Brain Glutamate Distributions Using Glutamate-Weighted Chemical Exchange Saturation Transfer Imaging. Diagnostics 2020, 10, 571. https://doi.org/10.3390/diagnostics10080571
Lee D-W, Woo C-W, Woo D-C, Kim JK, Kim KW, Lee D-H. Regional Mapping of Brain Glutamate Distributions Using Glutamate-Weighted Chemical Exchange Saturation Transfer Imaging. Diagnostics. 2020; 10(8):571. https://doi.org/10.3390/diagnostics10080571
Chicago/Turabian StyleLee, Do-Wan, Chul-Woong Woo, Dong-Cheol Woo, Jeong Kon Kim, Kyung Won Kim, and Dong-Hoon Lee. 2020. "Regional Mapping of Brain Glutamate Distributions Using Glutamate-Weighted Chemical Exchange Saturation Transfer Imaging" Diagnostics 10, no. 8: 571. https://doi.org/10.3390/diagnostics10080571
APA StyleLee, D.-W., Woo, C.-W., Woo, D.-C., Kim, J. K., Kim, K. W., & Lee, D.-H. (2020). Regional Mapping of Brain Glutamate Distributions Using Glutamate-Weighted Chemical Exchange Saturation Transfer Imaging. Diagnostics, 10(8), 571. https://doi.org/10.3390/diagnostics10080571




