Domain Structures and Inter-Domain Interactions Defining the Holoenzyme Architecture of Archaeal D-Family DNA Polymerase
Abstract
:1. Introduction
2. Background
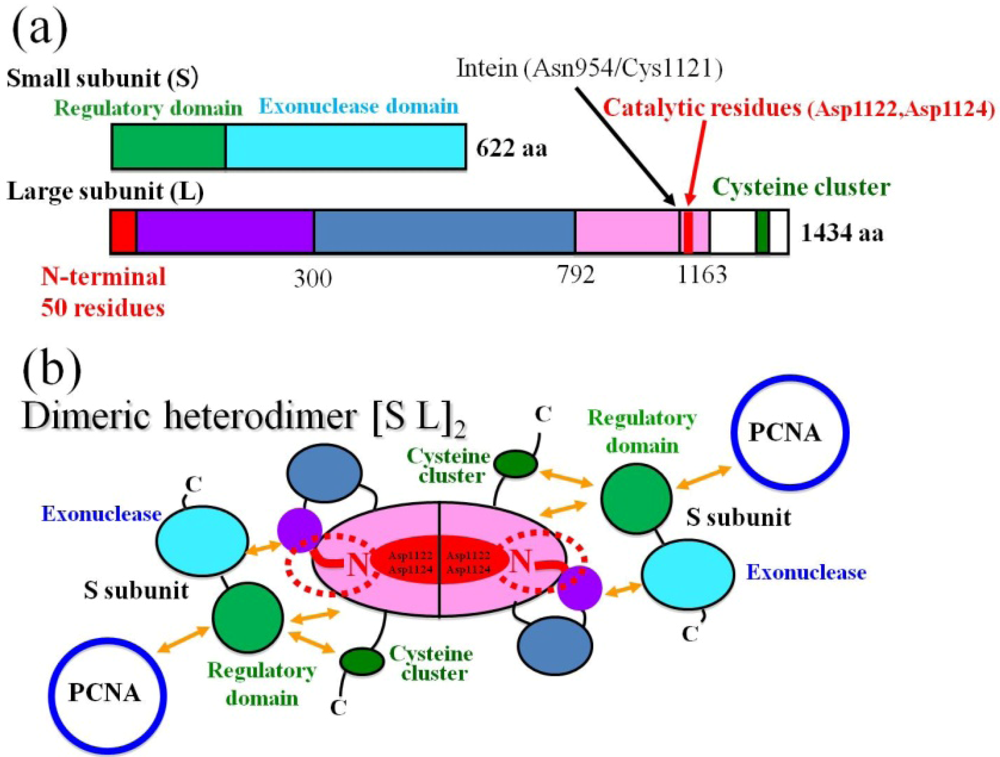
2.1. Functional Information for PolD
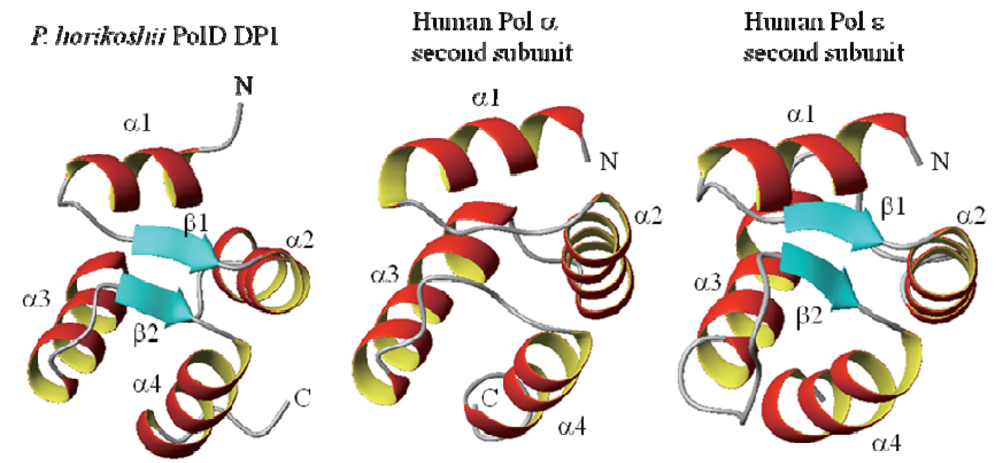
2.2. Structural Information for Isolated Domains and Fragments
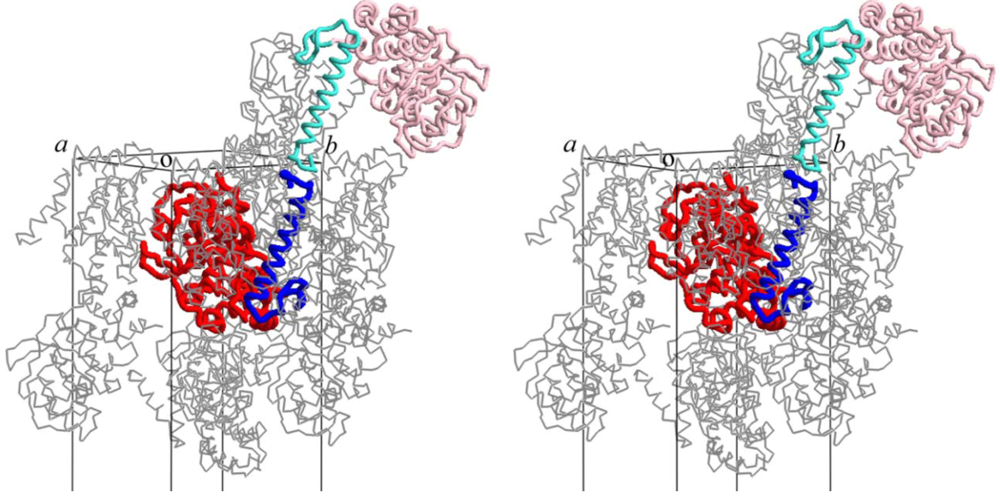
3. A Model for the PolD Holoenzyme
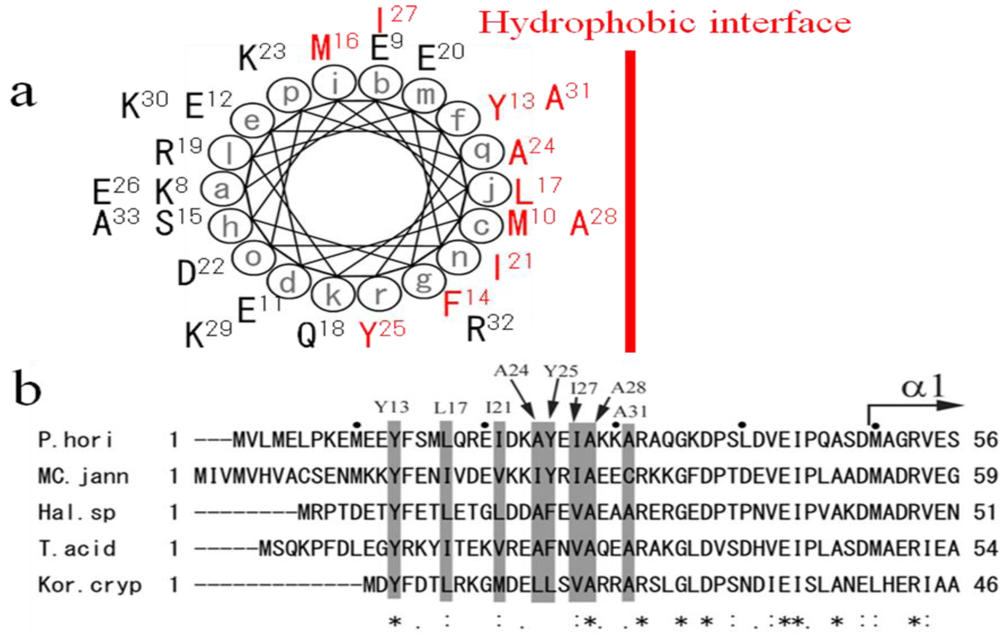
4. The Amphipathic Nature of the ~50 N-Terminal Residues Conserved Completely in PolD of Archaea
5. Conclusions
Acknowledgments
Conflict of Interest
References
- Leipe, D.D.; Aravind, L.; Koonin, E.V. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999, 27, 3389–3401. [Google Scholar] [CrossRef]
- Bailey, S.; Wing, R.A.; Steitz, T.A. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell 2006, 893–904. [Google Scholar] [CrossRef]
- Burgers, P.M.; Koonin, E.V.; Bruford, E.; Blanco, L.; Burtis, K.C.; Christman, M.F.; Copeland, W.C.; Friedberg, E.C.; Hanaoka, F.; Hinkle, D.C.; et al. Eukaryotic DNA polymerases: Proposal for a revised nomenclature. J. Biol. Chem. 2001, 276, 43487–43490. [Google Scholar] [CrossRef]
- Grabowski, B.; Kelman, Z. Archaeal DNA replication: Eukaryal proteins in a bacterial context. Annu. Rev. Microbiol. 2003, 57, 487–516. [Google Scholar] [CrossRef]
- Pavlov, Y.I.; Shcherbakova, P.V.; Rogozin, I.B. Roles of DNA polymerase in replication, repair, and recombination in eukaryotes. Int. Rev. Cytol. 2006, 255, 41–132. [Google Scholar] [CrossRef]
- Hubscher, U.; Maga, G.; Spadari, S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 2002, 71, 133–163. [Google Scholar] [CrossRef]
- Ishino, Y.; Komori, K.; Cann, I.K.; Koga, Y. A novel DNA polymerase family found in Archaea. J. Bacteriol. 1998, 180, 2232–2236. [Google Scholar]
- Shen, Y.; Musti, K.; Hiramoto, M.; Kikuchi, H.; Kawarabayasi, Y.; Matsui, I. Invariant Asp-1122 and Asp-1124 are essential residues for polymerization catalysis of family D DNA polymerase from Pyrococcus horikoshii. J. Biol. Chem. 2001, 276, 27376–27383. [Google Scholar]
- Henneke, G.; Flament, D.; Hübscher, U.; Querellou, J.; Raffin, J.P. The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J. Mol. Biol. 2005, 350, 53–64. [Google Scholar] [CrossRef]
- Kurr, M.; Huber, R.; Konig, H.; Jannasch, H.W.; Fricke, H.; Trincone, A.; Kristjansson, J.K.; Stetter, K.O. Methanopyrus kandleri, gen. and sp. nov. represents a novel group of hyperthermophilic methanogen, growing at 110 °C. Arch. Microbiol. 1991, 156, 239–247. [Google Scholar]
- Waters, E.; Hohn, M.J.; Ahel, I.; Graham, D.E.; Adams, M.D.; Barnstead, M.; Beeson, K.Y.; Bibbs, L.; Bolanos, R.; Keller, M.; et al. The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. USA 2003, 100, 12984–12988. [Google Scholar] [CrossRef]
- Brochier-Armanet, C.; Boussau, B.; Gribaldo, S.; Forterre, P. Mesophillic Crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 2008, 6, 245–252. [Google Scholar] [CrossRef]
- Elkins, J.G.; Podar, M.; Graham, D.E.; Makarova, K.S.; Wolf, Y.; Randau, L.; Hedlund, B.P.; Brochier-Armanet, C.; Kunin, V.; Anderson, I.; et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc. Natl. Acad. Sci. USA 2008, 105, 8102–8107. [Google Scholar] [CrossRef]
- Tahirov, T.H.; Makarova, K.S.; Rogozin, I.B.; Pavlov, Y.I.; Koonin, E.V. Evolution of DNA polymerases: An inactivated polymerase-exonuclease module in Pol ε and a chimeric origin of eukaryotic polymerases from two classes of Archaeal ancestors. Biol. Direct 2009, 4, 11–15. [Google Scholar] [CrossRef]
- Berquist, B.R.; DasSarma, P.; DasSarma, S. Essential and non-essential DNA replication genes in the model halophilic archaeon, Halobacterium sp. NRC-1. BMC Genet. 2007, 8, e31. [Google Scholar] [CrossRef]
- Rouillon, C.; Henneke, G.; Flament, D.; Querellou, J.; Raffin, J.P. DNA polymerase switching on homotrimeric PCNA at the replication fork of the Euryarchaea Pyrococcus abyssi. J. Mol. Biol. 2007, 350, 53–64. [Google Scholar]
- Shen, Y.; Tang, X.-F.; Yokoyama, H.; Matsui, E.; Matsui, I. A 21-amino acid peptide from the cysteine cluster II of the family D DNA polymerase from Pyrococcus horikoshii stimulate its nuclease activity which is Mre-11-like and prefers manganese ion as the cofactor. Nucleic Acids Res. 2004, 32, 158–168. [Google Scholar] [CrossRef]
- Aravind, L.; Koonin, E.V. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 1998, 26, 3746–3752. [Google Scholar] [CrossRef]
- Gueguen, Y.; Rolland, J.; Lecompte, O.; Azam, P.; Romancer, G.L.; Flament, D.; Raffin, J.P.; Dietrich, J. Characterization of two DNA polymerases from the hyperthermophilic euryarchaeon Pyrococcus abyssi. Eur. J. Biochem. 2001, 268, 5961–5969. [Google Scholar] [CrossRef]
- Hopfner, K.; Karcher, A.; Craig, L.; Woo, T.T.; Carney, J.P.; Tainer, J.A. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 2001, 105, 473–485. [Google Scholar] [CrossRef]
- Uemori, T.; Sato, Y.; Kato, I.; Doi, H.; Ishino, Y. A novel DNA polymerase in the hyperthermophilic archaeon, Pyrococcus furiosus: Gene cloning, expression, and characterization. Genes Cells 1997, 2, 499–512. [Google Scholar] [CrossRef]
- Shen, Y.; Tang, X.-F.; Matsui, I. Subunit interaction and regulation of activity through terminal domains of the family D DNA polymerase from Pyrococcus horikoshii. J. Biol. Chem. 2003, 278, 21247–21257. [Google Scholar] [CrossRef]
- Tang, X.-F.; Shen, Y.; Matsui, E.; Matsui, I. Domain topology of the DNA polymerase D complex from hyperthermophilic archaeon Pyrococcus horikoshii. Biochemistry 2004, 43, 11818–11827. [Google Scholar] [CrossRef]
- Matsui, I.; Urushibata, Y.; Shen, Y.; Matsui, E.; Yokoyama, H. Novel structure of an N-terminal domain that is crucial for the dimeric assembly and DNA-binding of an archaeal DNA polymerase D large subunit from Pyrococcus horikoshi. FEBS Lett. 2011, 585, 452–458. [Google Scholar] [CrossRef]
- Yamasaki, K.; Urushibata, Y.; Yamasaki, T.; Arisaka, F.; Matsui, I. Solution structure of the N-terminal domain of the archaeal D-family DNA polymerase small subunit reveals evolutionary relationship to eukaryotic B-family polymerase. FEBS Lett. 2010, 584, 3370–3375. [Google Scholar] [CrossRef]
- Holm, L.; Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993, 233, 123–138. [Google Scholar] [CrossRef]
- Nuutinen, T.; Tossavainen, H.; Fredriksson, K.; Pirilä, P.; Permi, P.; Pospiech, H.; Syvaoja, J.E. The solution structure of the amino-terminal domain of human DNA polymerase ε subunit B is homologous to C-domains of AAA+ proteins. Nucleic Acids Res. 2008, 36, 5102–5110. [Google Scholar] [CrossRef]
- Jeruzalmi, D.; Yurieva, O.; Zhao, Y.; Young, M.; Stewart, J.; Hingorani, M.; O’Donnel, M.; Kuriyan, J. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E.coli DNA polymerase III. Cell 2001, 106, 417–428. [Google Scholar] [CrossRef]
- Putnam, C.D.; Clancy, S.B.; Tsuruta, H.; Gonzalez, S.; Wetmur, J.G.; Tainer, J.A. Structure and mechanism of the RuvB Holliday junction branch migration motor. J. Mol. Biol. 2001, 311, 297–310. [Google Scholar] [CrossRef]
- Neuwald, A.F.; Aravind, L.; Spouge, J.L.; Koonin, E.V. AAA+: A class of chaperonine-like ATPases associated with the assemply, operation, and disassembly of protein complexes. Genome Res. 1999, 9, 27–43. [Google Scholar]
- Baranovskiy, A.G.; Babayeva, N.D.; Liston, V.G.; Rogozin, I.B.; Koonin, E.V.; Pavlov, Y.I.; Vassylyev, D.G.; Tahirov, T.H. X-ray structure of the complex of regulatory subunits of human DNA polymerase delta. Cell Cycle 2008, 7, 3026–3036. [Google Scholar]
- Cann, I.K.; Komori, K.; Toh, H.; Kanai, S.; Ishino, Y. A heterodimeric DNA polymerase: Evidence that members of Euryarchaeota possess a distinct DNA polymerase. Proc. Natl. Acad. Sci. USA 1998, 95, 14250–14255. [Google Scholar] [CrossRef]
- Mäkiniemi, M.; Pospiech, H.; Kilpeläinen, S.; Jokela, M.; Vihinen, M.; Syväoja, J.E. A novel family of DNA-polymerase-associated B subunits. Trends Biochem. Sci. 1999, 24, 14–16. [Google Scholar] [CrossRef]
- Bryson, K.; McGuffin, L.J.; Marsden, R.L.; Ward, J.J.; Sodhi, J.S.; Jones, D.T. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005, 33, W36–W38. [Google Scholar] [CrossRef]
- Castrec, B.; Laurent, S.; Henneke, G.; Flament, D.; Raffin, J.P. The glycine-rich motif of Pyrococcus abyssi DNA polymerase is critical for protein stability. J. Mol. Biol. 2010, 396, 840–848. [Google Scholar] [CrossRef] [Green Version]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Matsui, I.; Matsui, E.; Yamasaki, K.; Yokoyama, H. Domain Structures and Inter-Domain Interactions Defining the Holoenzyme Architecture of Archaeal D-Family DNA Polymerase. Life 2013, 3, 375-385. https://doi.org/10.3390/life3030375
Matsui I, Matsui E, Yamasaki K, Yokoyama H. Domain Structures and Inter-Domain Interactions Defining the Holoenzyme Architecture of Archaeal D-Family DNA Polymerase. Life. 2013; 3(3):375-385. https://doi.org/10.3390/life3030375
Chicago/Turabian StyleMatsui, Ikuo, Eriko Matsui, Kazuhiko Yamasaki, and Hideshi Yokoyama. 2013. "Domain Structures and Inter-Domain Interactions Defining the Holoenzyme Architecture of Archaeal D-Family DNA Polymerase" Life 3, no. 3: 375-385. https://doi.org/10.3390/life3030375
APA StyleMatsui, I., Matsui, E., Yamasaki, K., & Yokoyama, H. (2013). Domain Structures and Inter-Domain Interactions Defining the Holoenzyme Architecture of Archaeal D-Family DNA Polymerase. Life, 3(3), 375-385. https://doi.org/10.3390/life3030375



