Structure of the Inhibited Smooth Muscle Myosin and Its Implications on the Regulation of Insect Striated Muscle Myosin
Abstract
1. Introduction
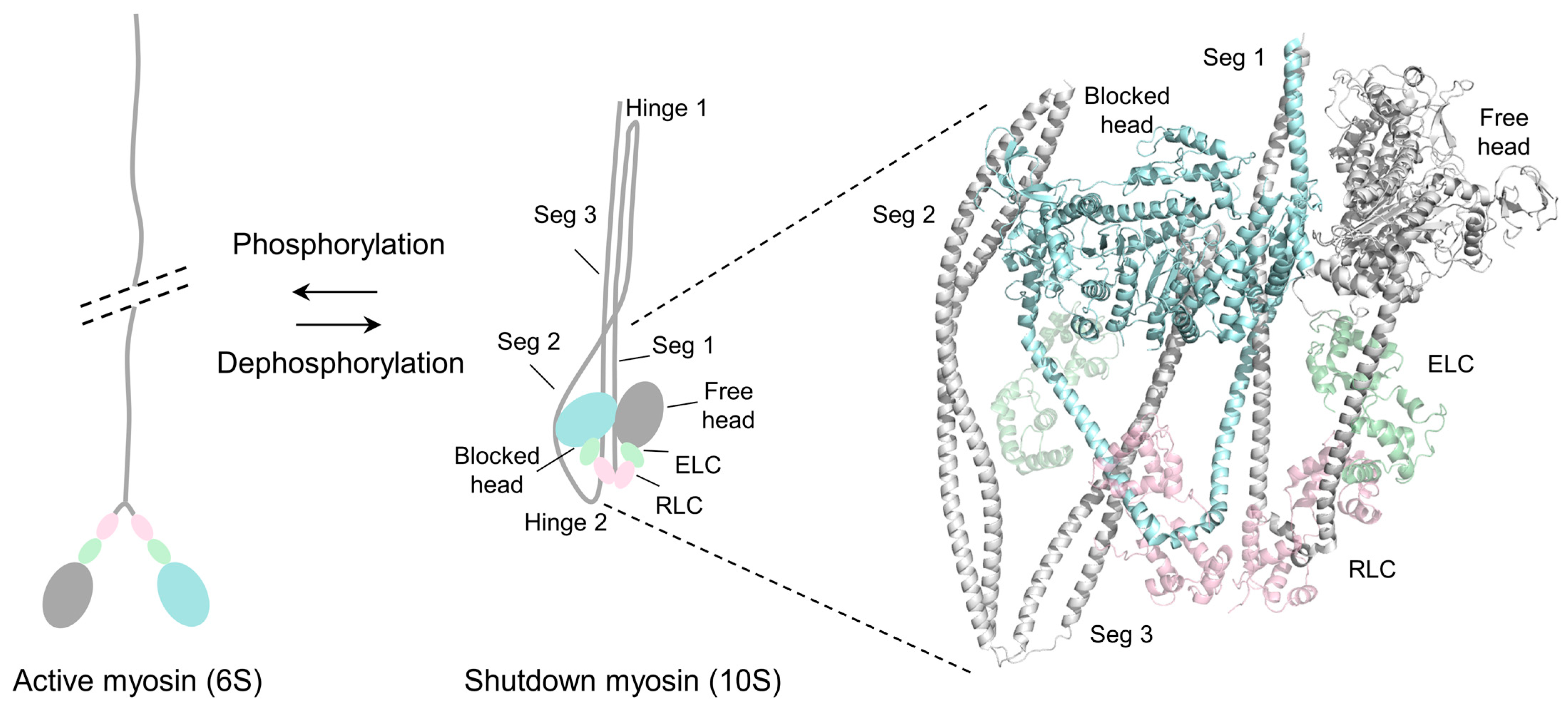
2. Structure of the Inhibited Smooth Muscle Myosin
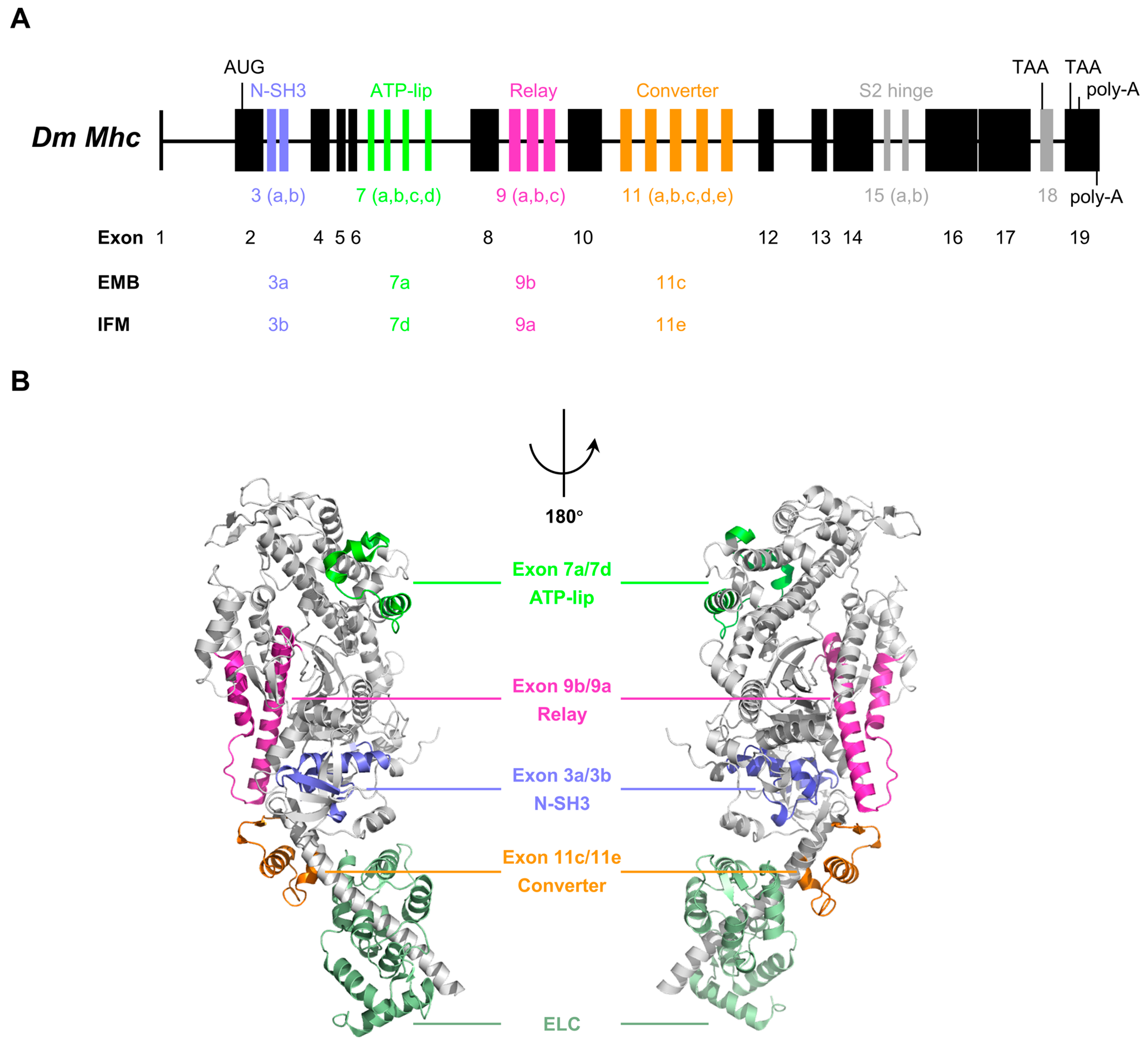
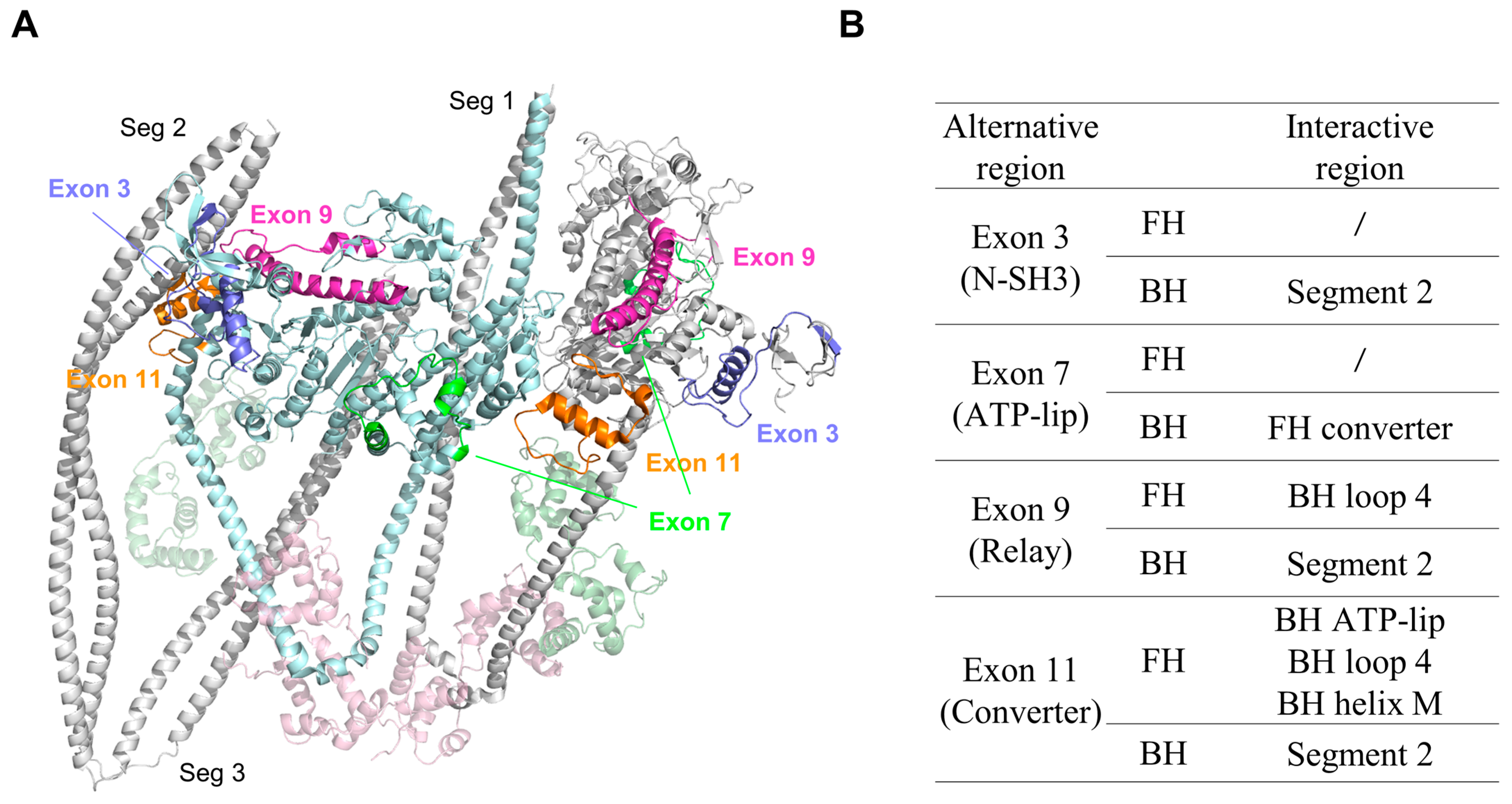
3. Regulatory Role of the Alternative Regions in Insect Striated Muscle Myosin
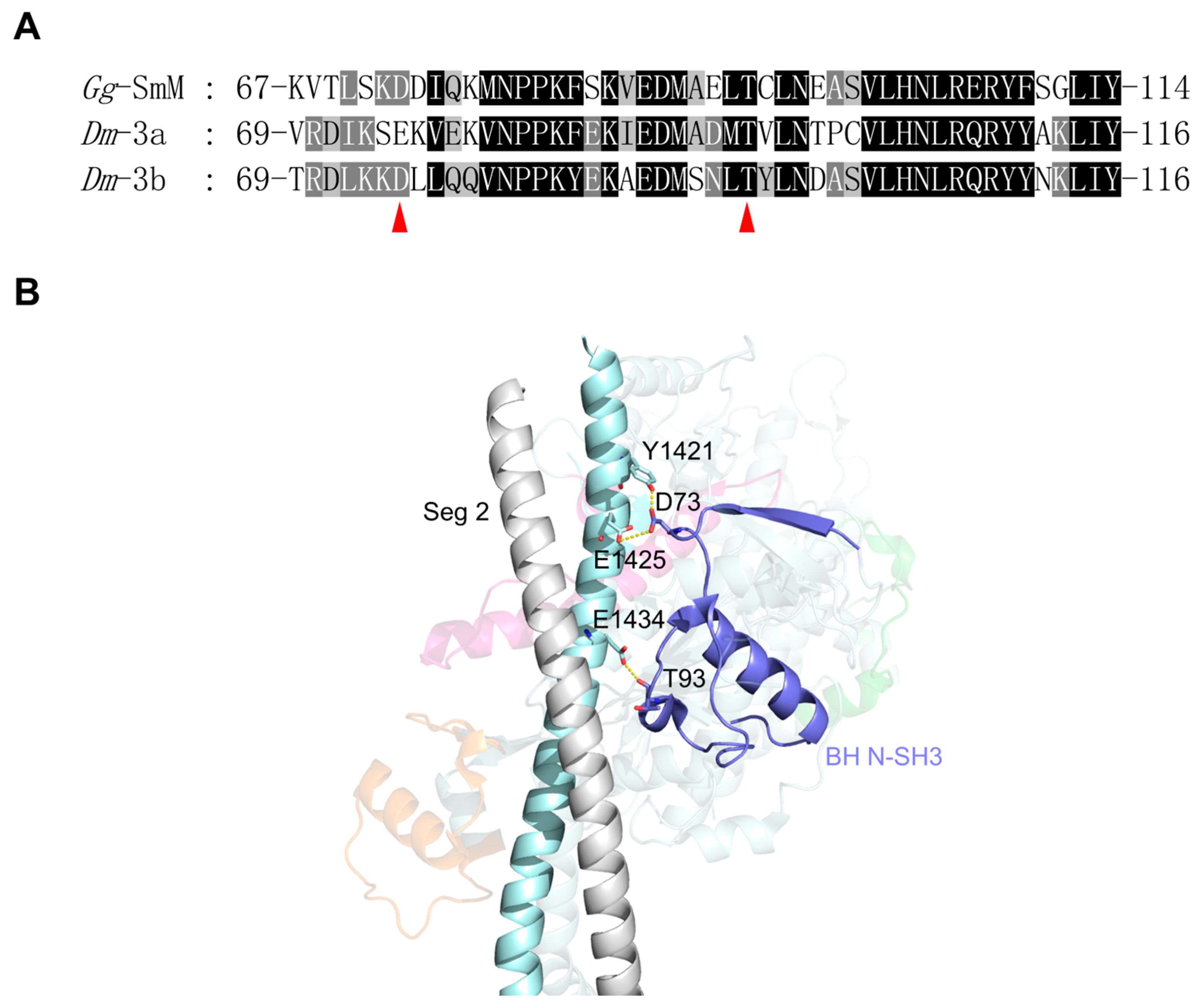
- Exon 3-encoded region
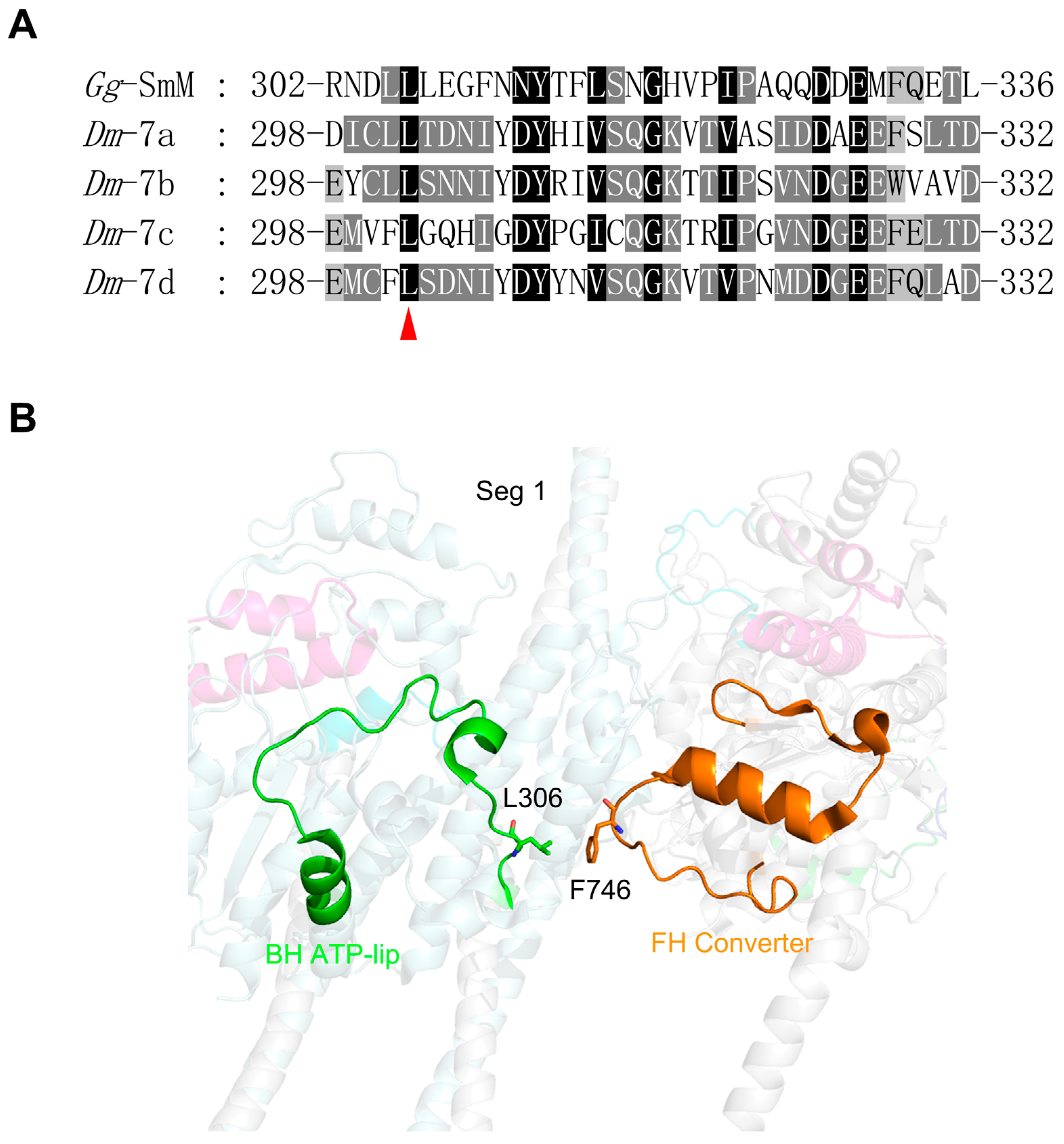
- Exon 7-encoded region
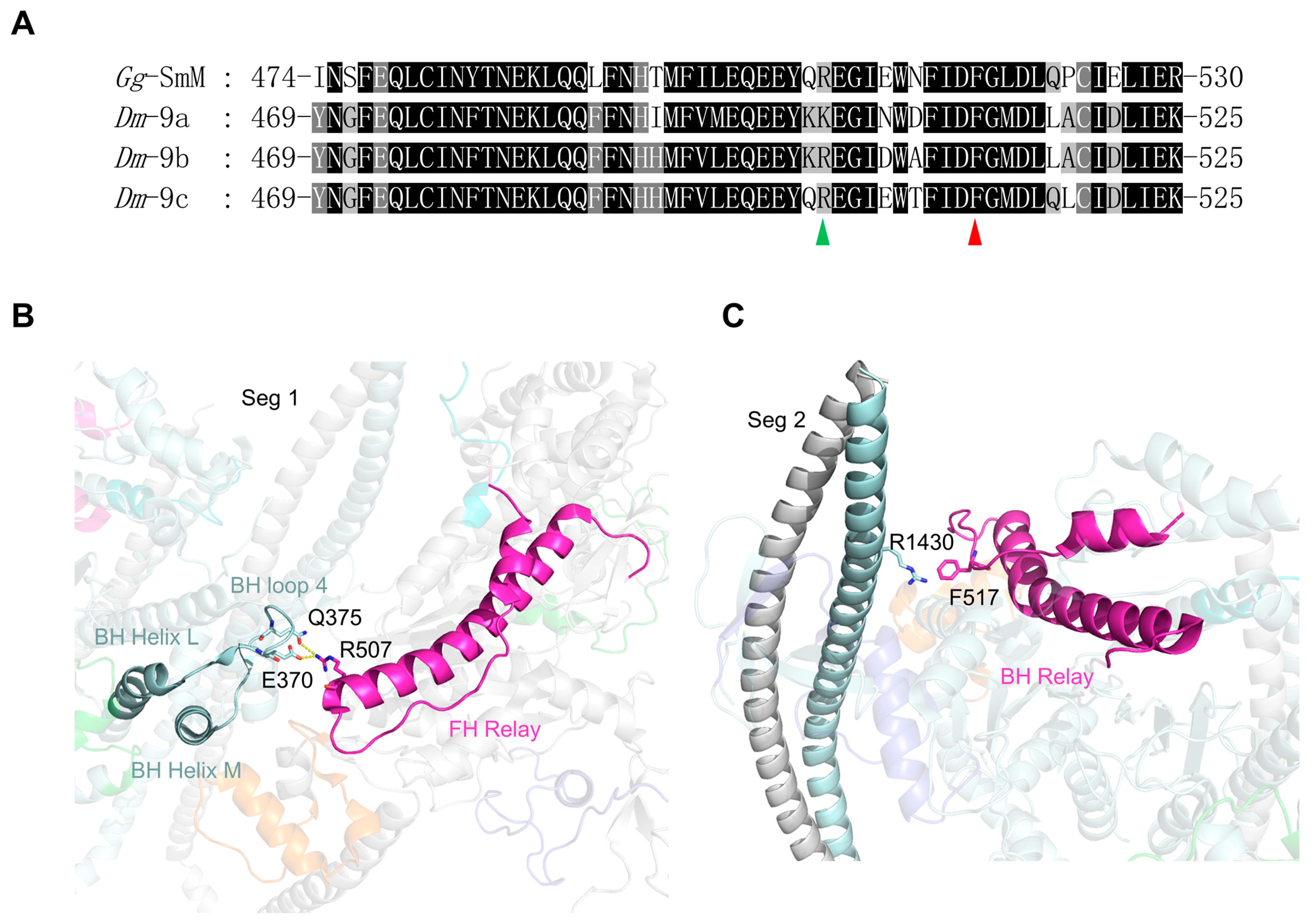
- Exon 9-encoded region
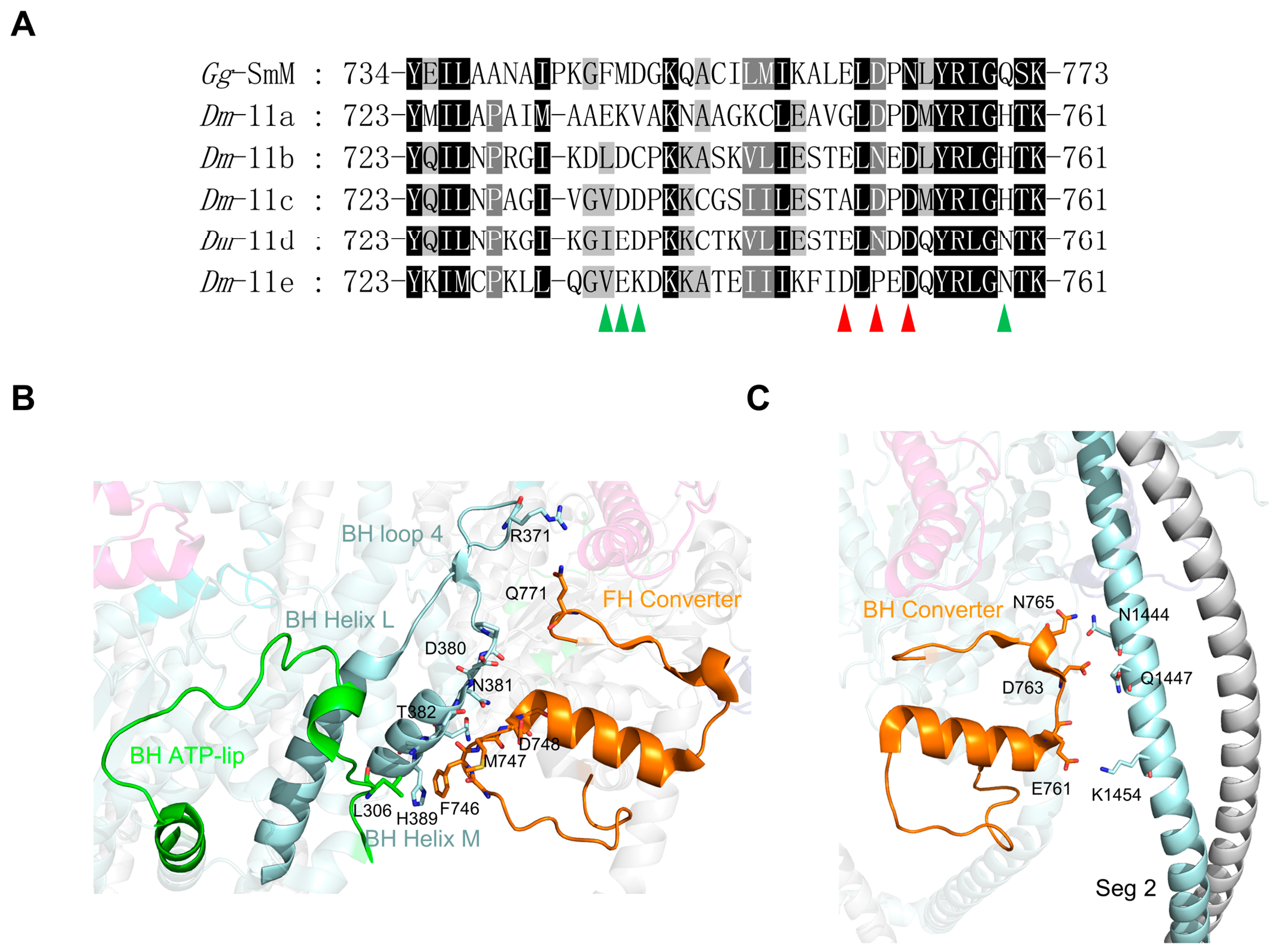
- Exon 11-encoded region
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Myosin-2 | Class II myosin |
| MHC | Myosin heavy chain |
| F-actin | Actin filament |
| SmM | Smooth muscle myosin |
| ELC | Essential light chain |
| RLC | Regulatory light chain |
| IHM | Interacting heads motif |
| BH | “Blocked” head |
| FH | “Free” head |
| MLCK | Myosin light chain kinase |
| EMB | Embryonic muscle |
| IFM | Indirect flight muscle |
| Gg | Gallus gallus |
| Dm | Drosophila melanogaster |
References
- Heissler, S.M.; Sellers, J.R. Kinetic adaptations of myosins for their diverse cellular functions. Traffic 2016, 17, 839–859. [Google Scholar] [CrossRef] [PubMed]
- Geeves, M.A.; Holmes, K.C. Structural mechanism of muscle contraction. Annu. Rev. Biochem. 1999, 68, 687–728. [Google Scholar] [CrossRef] [PubMed]
- Brito, C.; Sousa, S. Non-muscle myosin 2A (NM2A): Structure, regulation and function. Cells 2020, 9, 1590. [Google Scholar] [CrossRef] [PubMed]
- Sulbarán, G.; Alamo, L.; Pinto, A.; Márquez, G.; Méndez, F.; Padrón, R.; Craig, R. An invertebrate smooth muscle with striated muscle myosin filaments. Proc. Natl. Acad. Sci. USA 2015, 112, E5660–E5668. [Google Scholar] [CrossRef] [PubMed]
- Sellers, J.R. Myosins: A diverse superfamily. Biochim. Biophys. Acta 2000, 1496, 3–22. [Google Scholar] [CrossRef]
- Burgess, S.A.; Yu, S.; Walker, M.L.; Hawkins, R.J.; Chalovich, J.M.; Knight, P.J. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J. Mol. Biol. 2007, 372, 1165–1178. [Google Scholar] [CrossRef]
- Chang, A.N.; Mahajan, P.; Knapp, S.; Barton, H.; Sweeney, H.L.; Kamm, K.E.; Stull, J.T. Cardiac myosin light chain is phosphorylated by Ca2+/calmodulin-dependent and -independent kinase activities. Proc. Natl. Acad. Sci. USA 2016, 113, E3824–E3833. [Google Scholar] [CrossRef]
- Zoghbi, M.E.; Woodhead, J.L.; Moss, R.L.; Craig, R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc. Natl. Acad. Sci. USA 2008, 105, 2386–2390. [Google Scholar] [CrossRef]
- Woodhead, J.L.; Zhao, F.Q.; Craig, R.; Egelman, E.H.; Alamo, L.; Padrón, R. Atomic model of a myosin filament in the relaxed state. Nature 2005, 436, 1195–1199. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Nguyen, V.; Campbell, K.S.; Padrón, R.; Craig, R. Cryo-EM structure of the human cardiac myosin filament. Nature 2023, 623, 853–862. [Google Scholar] [CrossRef]
- Grinzato, A.; Auguin, D.; Kikuti, C.; Nandwani, N.; Moussaoui, D.; Pathak, D.; Kandiah, E.; Ruppel, K.M.; Spudich, J.A.; Houdusse, A.; et al. Cryo-EM structure of the folded-back state of human β-cardiac myosin. Nat. Commun. 2023, 14, 3166. [Google Scholar] [CrossRef]
- Alamo, L.; Qi, D.; Wriggers, W.; Pinto, A.; Zhu, J.; Bilbao, A.; Gillilan, R.E.; Hu, S.; Padrón, R. Conserved intramolecular interactions maintain myosin interacting-heads motifs explaining Tarantula muscle super-relaxed state structural basis. J. Mol. Biol. 2016, 428, 1142–1164. [Google Scholar] [CrossRef] [PubMed]
- Scarff, C.A.; Carrington, G.; Casas-Mao, D.; Chalovich, J.M.; Knight, P.J.; Ranson, N.A.; Peckham, M. Structure of the shutdown state of myosin-2. Nature 2020, 588, 515–520. [Google Scholar] [CrossRef]
- Jung, H.S.; Burgess, S.A.; Billington, N.; Colegrave, M.; Patel, H.; Chalovich, J.M.; Chantler, P.D.; Knight, P.J. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 6022–6026. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Sulbarán, G.; Yang, S.; Mun, J.Y.; Alamo, L.; Pinto, A.; Sato, O.; Ikebe, M.; Liu, X.; Korn, E.D.; et al. Interacting-heads motif has been conserved as a mechanism of myosin II inhibition since before the origin of animals. Proc. Natl. Acad. Sci. USA 2018, 115, E1991–E2000. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.S.; Komatsu, S.; Ikebe, M.; Craig, R. Head-head and head-tail interaction: A general mechanism for switching off myosin II activity in cells. Mol. Biol. Cell 2008, 19, 3234–3242. [Google Scholar] [CrossRef]
- Odronitz, F.; Kollmar, M. Comparative genomic analysis of the arthropod muscle myosin heavy chain genes allows ancestral gene reconstruction and reveals a new type of ’partially’ processed pseudogene. BMC Mol. Biol. 2008, 9, 21. [Google Scholar] [CrossRef]
- Swank, D.M.; Bartoo, M.L.; Knowles, A.F.; Iliffe, C.; Bernstein, S.I.; Molloy, J.E.; Sparrow, J.C. Alternative exon-encoded regions of Drosophila myosin heavy chain modulate ATPase rates and actin sliding velocity. J. Biol. Chem. 2001, 276, 15117–15124. [Google Scholar] [CrossRef]
- Li, J.; Lu, Z.; He, J.; Chen, Q.; Wang, X.; Kang, L.; Li, X.D. Alternative exon-encoding regions of Locusta migratoria muscle myosin modulate the pH dependence of ATPase activity. Insect Mol. Biol. 2016, 25, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Bernstein, S.I. Spatially and temporally regulated expression of myosin heavy chain alternative exons during Drosophila embryogenesis. Mech. Dev. 2001, 101, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Swank, D.M.; Knowles, A.F.; Kronert, W.A.; Suggs, J.A.; Morrill, G.E.; Nikkhoy, M.; Manipon, G.G.; Bernstein, S.I. Variable N-terminal regions of muscle myosin heavy chain modulate ATPase rate and actin sliding velocity. J. Biol. Chem. 2003, 278, 17475–17482. [Google Scholar] [CrossRef] [PubMed]
- Kronert, W.A.; Dambacher, C.M.; Knowles, A.F.; Swank, D.M.; Bernstein, S.I. Alternative relay domains of Drosophila melanogaster myosin differentially affect ATPase activity, in vitro motility, myofibril structure and muscle function. J. Mol. Biol. 2008, 379, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Ikebe, M. Regulation of the function of mammalian myosin and its conformational change. Biochem. Biophys. Res. Commun. 2008, 369, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Onishi, H.; Takahashi, K.; Watanabe, S. Structure and function of chicken gizzard myosin. J. Biochem. 1978, 84, 1529–1542. [Google Scholar] [CrossRef] [PubMed]
- Wendt, T.; Taylor, D.; Trybus, K.M.; Taylor, K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc. Natl. Acad. Sci. USA 2001, 98, 4361–4366. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wendt, T.; Taylor, D.; Taylor, K. Refined model of the 10S conformation of smooth muscle myosin by cryo-electron microscopy 3D image reconstruction. J. Mol. Biol. 2003, 329, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Cross, R.A.; Cross, K.E.; Sobieszek, A. ATP-linked monomer-polymer equilibrium of smooth muscle myosin: The free folded monomer traps ADP.Pi. Embo J. 1986, 5, 2637–2641. [Google Scholar] [CrossRef]
- Yang, S.; Lee, K.H.; Woodhead, J.L.; Sato, O.; Ikebe, M.; Craig, R. The central role of the tail in switching off 10S myosin II activity. J. Gen. Physiol. 2019, 151, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Kiboku, T.; Katoh, T.; Nakamura, A.; Kitamura, A.; Kinjo, M.; Murakami, Y.; Takahashi, M. Nonmuscle myosin II folds into a 10S form via two portions of tail for dynamic subcellular localization. Genes Cells 2013, 18, 90–109. [Google Scholar] [CrossRef]
- Milton, D.L.; Schneck, A.N.; Ziech, D.A.; Ba, M.; Facemyer, K.C.; Halayko, A.J.; Baker, J.E.; Gerthoffer, W.T.; Cremo, C.R. Direct evidence for functional smooth muscle myosin II in the 10S self-inhibited monomeric conformation in airway smooth muscle cells. Proc. Natl. Acad. Sci. USA 2011, 108, 1421–1426. [Google Scholar] [CrossRef] [PubMed]
- Heissler, S.M.; Arora, A.S.; Billington, N.; Sellers, J.R.; Chinthalapudi, K. Cryo-EM structure of the autoinhibited state of myosin-2. Sci. Adv. 2021, 7, eabk3273. [Google Scholar] [CrossRef]
- Yang, S.; Tiwari, P.; Lee, K.H.; Sato, O.; Ikebe, M.; Padron, R.; Craig, R. Cryo-EM structure of the inhibited (10S) form of myosin II. Nature 2020, 588, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Trybus, K.M.; Lowey, S. Mechanism of smooth muscle myosin phosphorylation. J. Biol. Chem. 1985, 260, 15988–15995. [Google Scholar] [CrossRef] [PubMed]
- Persechini, A.; Hartshorne, D.J. Phosphorylation of smooth muscle myosin: Evidence for cooperativity between the myosin heads. Science 1981, 213, 1383–1385. [Google Scholar] [CrossRef] [PubMed]
- Alamo, L.; Pinto, A.; Sulbarán, G.; Mavárez, J.; Padrón, R. Lessons from a tarantula: New insights into myosin interacting-heads motif evolution and its implications on disease. Biophys. Rev. 2018, 10, 1465–1477. [Google Scholar] [CrossRef] [PubMed]
- Alamo, L.; Koubassova, N.; Pinto, A.; Gillilan, R.; Tsaturyan, A.; Padrón, R. Lessons from a tarantula: New insights into muscle thick filament and myosin interacting-heads motif structure and function. Biophys. Rev. 2017, 9, 461–480. [Google Scholar] [CrossRef]
- Pinto, A.; Sánchez, F.; Alamo, L.; Padrón, R. The myosin interacting-heads motif is present in the relaxed thick filament of the striated muscle of scorpion. J. Struct. Biol. 2012, 180, 469–478. [Google Scholar] [CrossRef]
- Hu, Z.; Taylor, D.W.; Reedy, M.K.; Edwards, R.J.; Taylor, K.A. Structure of myosin filaments from relaxed Lethocerus flight muscle by cryo-EM at 6 Å resolution. Sci. Adv. 2016, 30, e1600058. [Google Scholar] [CrossRef]
- Hess, N.K.; Singer, P.A.; Trinh, K.; Nikkhoy, M.; Bernstein, S.I. Transcriptional regulation of the Drosophila melanogaster muscle myosin heavy-chain gene. Gene Expr. Patterns 2007, 7, 413–422. [Google Scholar] [CrossRef]
- Bernstein, S.I.; Milligan, R.A. Fine tuning a molecular motor: The location of alternative domains in the Drosophila myosin head. J. Mol. Biol. 1997, 271, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, J.T.; Mermelstein, D.J.; Walker, R.C.; Bernstein, S.I.; Huxford, T. X-ray crystallographic and molecular dynamic analyses of Drosophila melanogaster embryonic muscle myosin define domains responsible for isoform-specific properties. J. Mol. Biol. 2020, 432, 427–447. [Google Scholar] [CrossRef] [PubMed]
- Swank, D.M.; Braddock, J.; Brown, W.; Lesage, H.; Bernstein, S.I.; Maughan, D.W. An alternative domain near the ATP binding pocket of Drosophila myosin affects muscle fiber kinetics. Biophys. J. 2006, 90, 2427–2435. [Google Scholar] [CrossRef]
- Miller, B.M.; Bloemink, M.J.; Nyitrai, M.; Bernstein, S.I.; Geeves, M.A. A variable domain near the ATP-binding site in Drosophila muscle myosin is part of the communication pathway between the nucleotide and actin-binding sites. J. Mol. Biol. 2007, 368, 1051–1066. [Google Scholar] [CrossRef][Green Version]
- Houdusse, A.; Kalabokis, V.N.; Himmel, D.; Szent-Györgyi, A.G.; Cohen, C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: A novel conformation of the myosin head. Cell 1999, 97, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ramanath, S.; Kronert, W.A.; Bernstein, S.I.; Maughan, D.W.; Swank, D.M. Alternative versions of the myosin relay domain differentially respond to load to influence Drosophila muscle kinetics. Biophys. J. 2008, 95, 5228–5237. [Google Scholar] [CrossRef] [PubMed]
- Bloemink, M.J.; Dambacher, C.M.; Knowles, A.F.; Melkani, G.C.; Geeves, M.A.; Bernstein, S.I. Alternative exon 9-encoded relay domains affect more than one communication pathway in the Drosophila myosin head. J. Mol. Biol. 2009, 389, 707–721. [Google Scholar] [CrossRef][Green Version]
- Ramanath, S.; Wang, Q.; Bernstein, S.I.; Swank, D.M. Disrupting the myosin converter-relay interface impairs Drosophila indirect flight muscle performance. Biophys. J. 2011, 101, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Kronert, W.A.; Melkani, G.C.; Melkani, A.; Bernstein, S.I. Alternative relay and converter domains tune native muscle myosin isoform function in Drosophila. J. Mol. Biol. 2012, 416, 543–557. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kronert, W.A.; Melkani, G.C.; Melkani, A.; Bernstein, S.I. Mapping interactions between myosin relay and converter domains that power muscle function. J. Biol. Chem. 2014, 289, 12779–12790. [Google Scholar] [CrossRef] [PubMed]
- Bloemink, M.J.; Melkani, G.C.; Bernstein, S.I.; Geeves, M.A. The relay/converter interface influences hydrolysis of ATP by skeletal muscle myosin II. J. Biol. Chem. 2016, 291, 1763–1773. [Google Scholar] [CrossRef]
- Littlefield, K.P.; Swank, D.M.; Sanchez, B.M.; Knowles, A.F.; Warshaw, D.M.; Bernstein, S.I. The converter domain modulates kinetic properties of Drosophila myosin. Am. J. Physiol. Cell Physiol. 2003, 284, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Hao, J.; Yao, L.L.; Wei, M.; Chen, W.; Yang, Q.; Li, X.D. Insect Sf9 cells are suitable for functional expression of insect, but not vertebrate, striated muscle myosin. Biochem. Biophys. Res. Commun. 2022, 635, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Liu, C.; Zhang, N.; Li, J.; Ni, T.; Qu, M.; Li, X.D. Alternative relay regulates the adenosine triphosphatase activity of Locusta migratoria striated muscle myosin. Insect Sci. 2024, 31, 435–447. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, S.; Lu, Y.-N.; Li, X.-d. Structure of the Inhibited Smooth Muscle Myosin and Its Implications on the Regulation of Insect Striated Muscle Myosin. Life 2025, 15, 379. https://doi.org/10.3390/life15030379
Sun S, Lu Y-N, Li X-d. Structure of the Inhibited Smooth Muscle Myosin and Its Implications on the Regulation of Insect Striated Muscle Myosin. Life. 2025; 15(3):379. https://doi.org/10.3390/life15030379
Chicago/Turabian StyleSun, Shaopeng, Yi-Ning Lu, and Xiang-dong Li. 2025. "Structure of the Inhibited Smooth Muscle Myosin and Its Implications on the Regulation of Insect Striated Muscle Myosin" Life 15, no. 3: 379. https://doi.org/10.3390/life15030379
APA StyleSun, S., Lu, Y.-N., & Li, X.-d. (2025). Structure of the Inhibited Smooth Muscle Myosin and Its Implications on the Regulation of Insect Striated Muscle Myosin. Life, 15(3), 379. https://doi.org/10.3390/life15030379







