Repurposing Castanea sativa Spiny Burr By-Products Extract as a Potentially Effective Anti-Inflammatory Agent for Novel Future Biotechnological Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of C. sativa Burr (CSB) Extract
2.3. Total Phenolic Content (TPC)
2.4. Total Flavonoid Content (TFC)
2.5. Determination of Reducing Power
2.6. ABTS Free-Radical Scavenging Activity
2.7. DPPH Free-Radical Scavenging Activity
2.8. UPLC-MS/MS
2.9. In Vitro Cytotoxicity and Anti-Inflammatory Activity
2.9.1. Cell Cultures
2.9.2. NIH3T3 Cytotoxicity
2.9.3. Cell Viability
2.9.4. Cell Stimulation
2.9.5. Quantification of Intracellular ROS Generation
2.9.6. Determination of NO Production
2.9.7. Protein Extraction
2.9.8. Western Blotting
2.9.9. Immunofluorescence Study
2.10. Mutagenicity Assay: Ames Test
2.11. Statistical Analysis
2.12. In Silico Studies
Structural Optimization and Resources
3. Results
3.1. Chemical Composition and Antioxidant Capacity of CSB
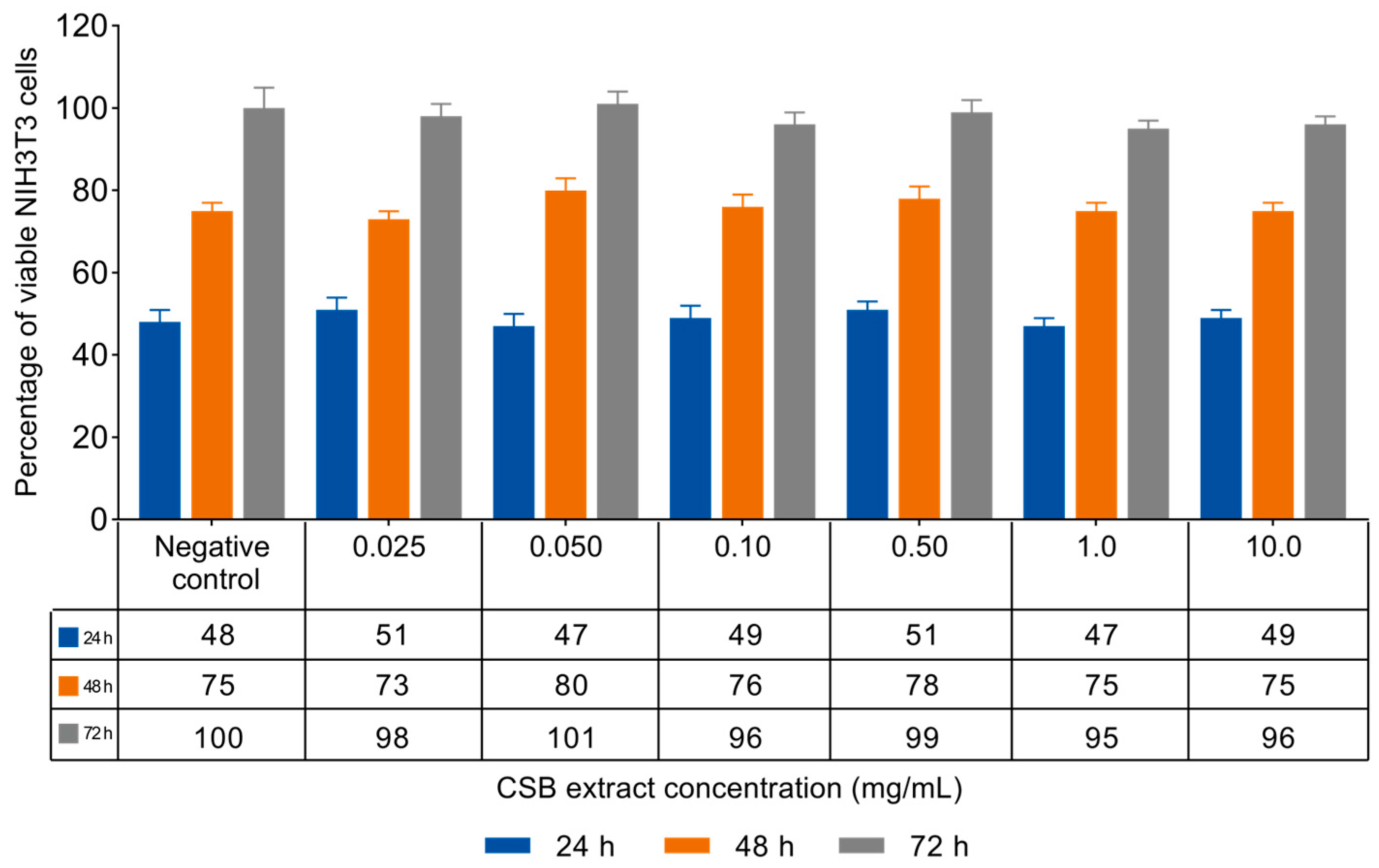
3.2. NIH3T3 Viability and Proliferation
3.3. CSB Inhibits LPS-Induced ROS Generation
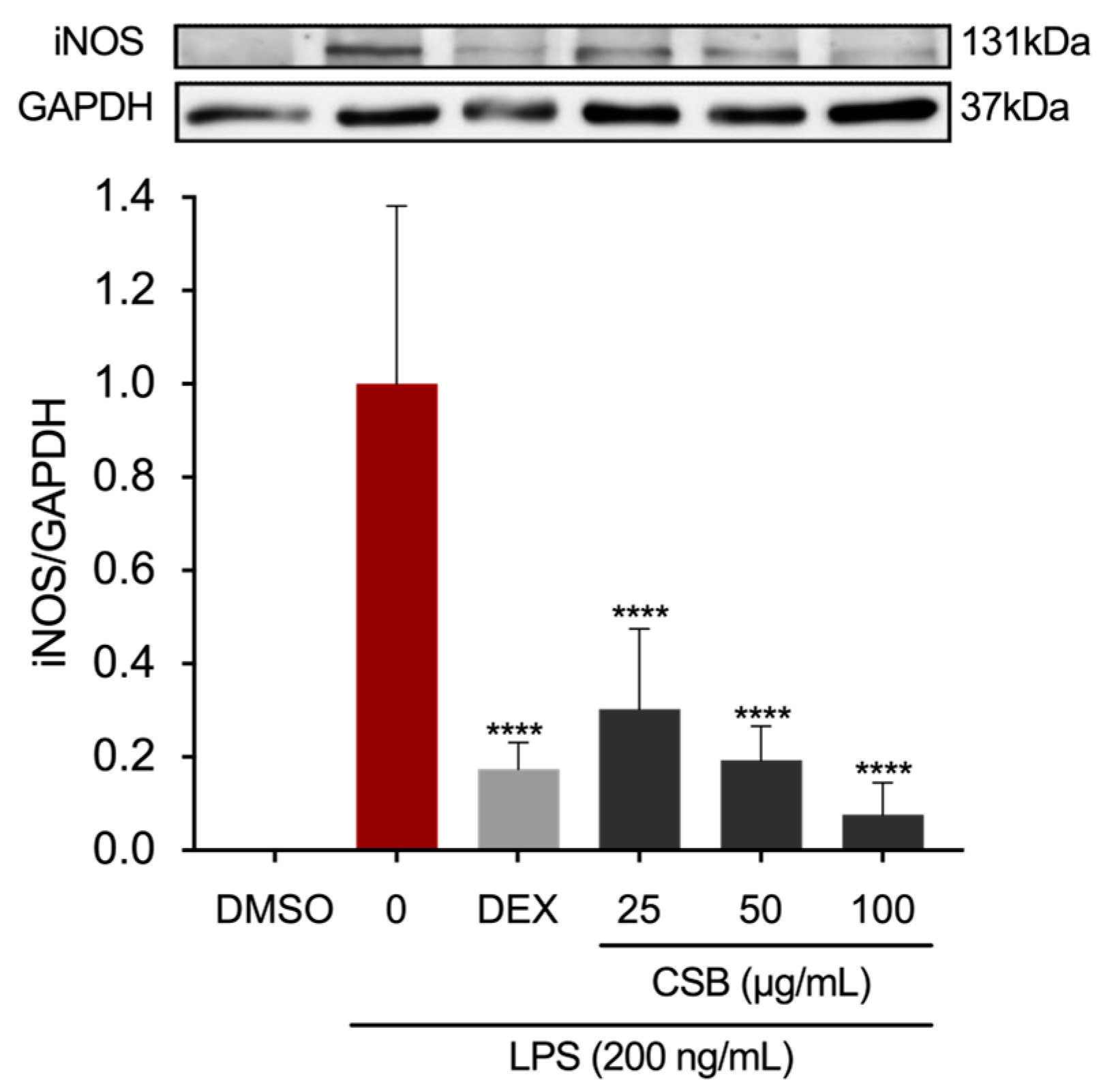
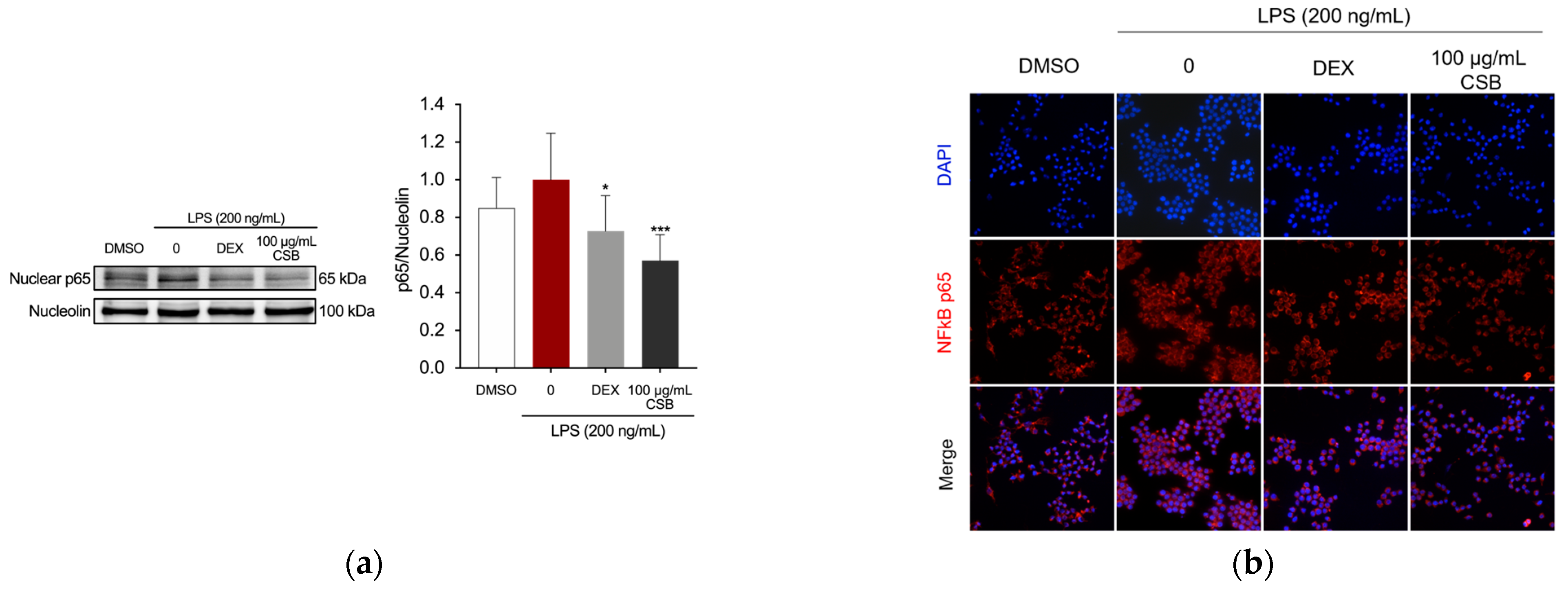
3.4. CSB Reduced LPS-Induced Inflammation in RAW 264.7 Cells
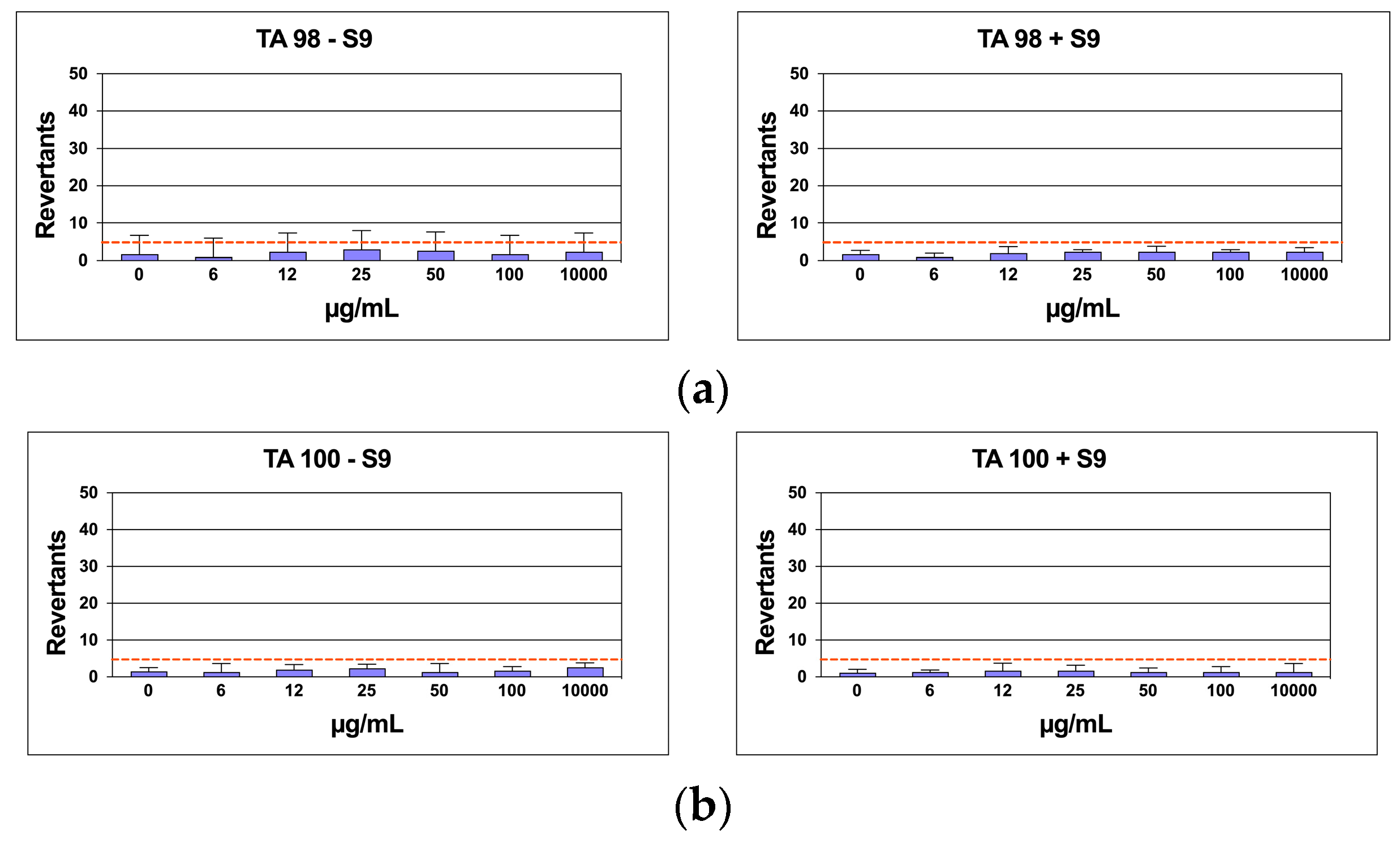
3.5. Mutagenicity Assay: Ames Test
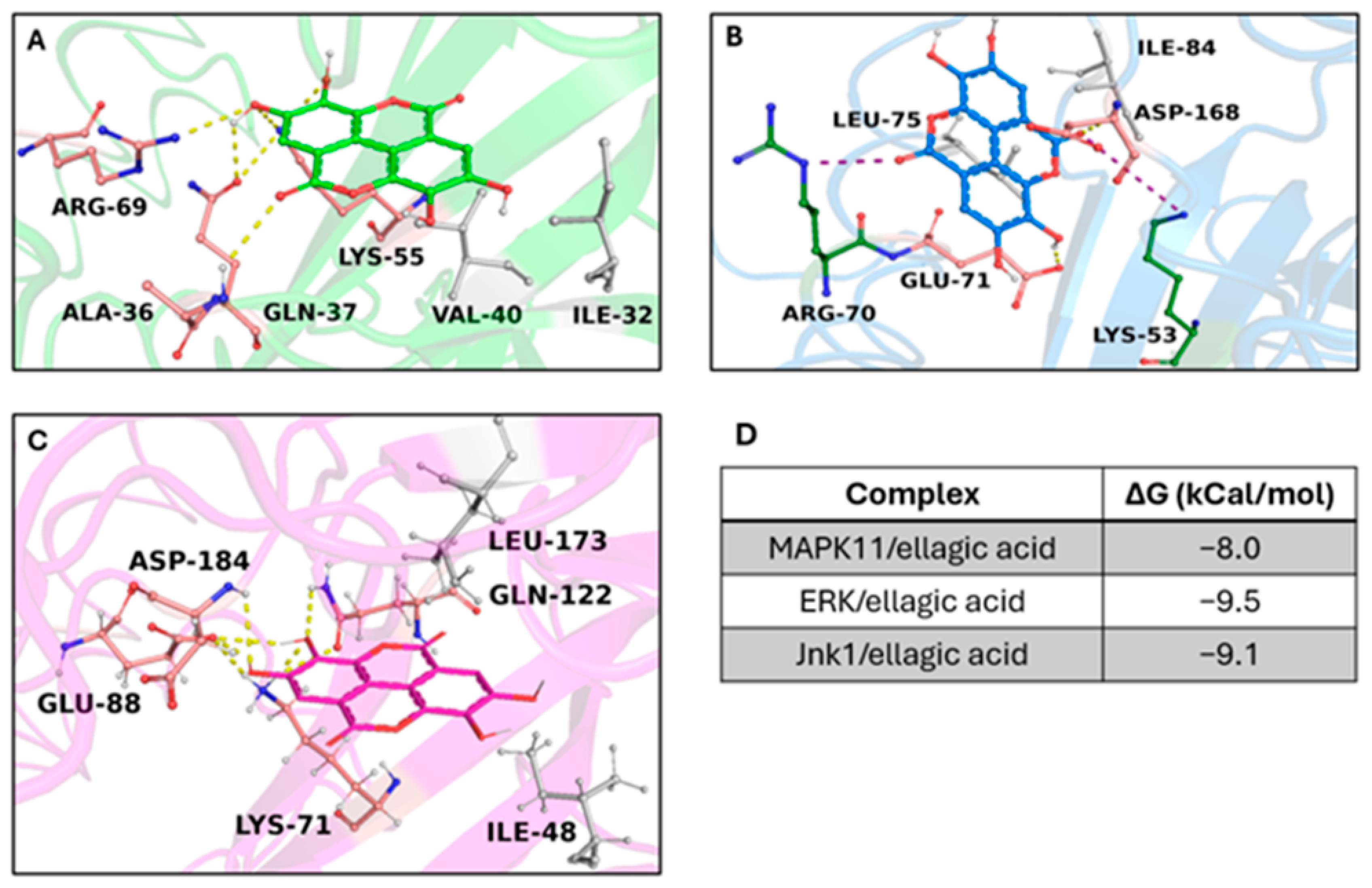
3.6. In Silico Results
Target/Compound Virtual Screening
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Squillaci, G.; Apone, F.; Sena, L.M.; Carola, A.; Tito, A.; Bimonte, M.; De Lucia, A.; Colucci, G.; La Cara, F.; Morana, A. Chestnut (Castanea sativa Mill.) Industrial Wastes as a Valued Bioresource for the Production of Active Ingredients. Process Biochem. 2018, 64, 228–236. [Google Scholar] [CrossRef]
- Pinto, D.; Braga, N.; Silva, A.M.; Costa, P.; Delerue-Matos, C.; Rodrigues, F. Chestnut. In Valorization of Fruit Processing by-Products; Elsevier: Amsterdam, The Netherlands, 2020; pp. 127–144. [Google Scholar]
- Battisti, A.; Benvegnù, I.; Colombari, F.; Haack, R.A. Invasion by the Chestnut Gall Wasp in Italy Causes Significant Yield Loss in Castanea sativa Nut Production. Agric. For. Entomol. 2014, 16, 75–79. [Google Scholar] [CrossRef]
- Torello Marinoni, D.; Akkak, A.; Beltramo, C.; Guaraldo, P.; Boccacci, P.; Bounous, G.; Ferrara, A.M.; Ebone, A.; Viotto, E.; Botta, R. Genetic and Morphological Characterization of Chestnut (Castanea sativa Mill.) Germplasm in Piedmont (North-Western Italy). Tree Genet. Genomes 2013, 9, 1017–1030. [Google Scholar] [CrossRef]
- Ventorino, V.; Parillo, R.; Testa, A.; Viscardi, S.; Espresso, F.; Pepe, O. Chestnut Green Waste Composting for Sustainable Forest Management: Microbiota Dynamics and Impact on Plant Disease Control. J. Environ. Manag. 2016, 166, 168–177. [Google Scholar] [CrossRef]
- Barreira, J.C.M.; Ferreira, I.C.F.R.; Oliveira, M.B.P.P.; Pereira, J.A. Antioxidant Potential of Chestnut (Castanea sativa L.) and Almond (Prunus dulcis L.) by-Products. Food Sci. Technol. Int. 2010, 16, 209–216. [Google Scholar] [CrossRef]
- Fernández-Agulló, A.; Freire, M.S.; Antorrena, G.; Pereira, J.A.; González-Álvarez, J. Effect of the Extraction Technique and Operational Conditions on the Recovery of Bioactive Compounds from Chestnut (Castanea sativa) Bur and Shell. Sep. Sci. Technol. 2014, 49, 267–277. [Google Scholar] [CrossRef]
- Silva, V.; Falco, V.; Dias, M.I.; Barros, L.; Silva, A.; Capita, R.; Alonso-Calleja, C.; Amaral, J.S.; Igrejas, G.; Ferreira, I.C.F.R.; et al. Evaluation of the Phenolic Profile of Castanea sativa Mill. by-Products and Their Antioxidant and Antimicrobial Activity against Multiresistant Bacteria. Antioxidants 2020, 9, 87. [Google Scholar] [CrossRef]
- Rodrigues, F.; Santos, J.; Pimentel, F.B.; Braga, N.; Palmeira-de-Oliveira, A.; Oliveira, M.B.P.P. Promising New Applications of Castanea sativa Shell: Nutritional Composition, Antioxidant Activity, Amino Acids and Vitamin E Profile. Food Funct. 2015, 6, 2854–2860. [Google Scholar] [CrossRef]
- Vella, F.M.; Laratta, B.; La Cara, F.; Morana, A. Recovery of Bioactive Molecules from Chestnut (Castanea sativa Mill.) by-Products through Extraction by Different Solvents. Nat. Prod. Res. 2018, 32, 1022–1032. [Google Scholar] [CrossRef]
- Braga, N.; Rodrigues, F.; Oliveira, M.B.P.P. Castanea sativa by-Products: A Review on Added Value and Sustainable Application. Nat. Prod. Res. 2015, 29, 1–18. [Google Scholar] [CrossRef]
- Flórez-Fernández, N.; Torres, M.D.; Gómez, S.; Couso, S.; Domínguez, H. Potential of Chestnut Wastes for Cosmetics and Pharmaceutical Applications. Waste Biomass Valorization 2020, 11, 4721–4730. [Google Scholar] [CrossRef]
- Pinto, D.; Braga, N.; Rodrigues, F.; Oliveira, M. Castanea sativa Bur: An Undervalued by-Product but a Promising Cosmetic Ingredient. Cosmetics 2017, 4, 50. [Google Scholar] [CrossRef]
- Pinto, D.; Rodrigues, F.; Braga, N.; Santos, J.; Pimentel, F.B.; Palmeira-de-Oliveira, A.; Oliveira, M.B.P.P. The Castanea sativa Bur as a New Potential Ingredient for Nutraceutical and Cosmetic Outcomes: Preliminary Studies. Food Funct. 2017, 8, 201–208. [Google Scholar] [CrossRef]
- Silva, A.M.; Costa, P.C.; Delerue-Matos, C.; Rodrigues, F. Assessment of a Formulation Containing a Castanea sativa Shells Extract on Skin Face Parameters: In Vivo Evaluation. Processes 2022, 10, 2230. [Google Scholar] [CrossRef]
- Cadau, S.; Gault, M.; Berthelemy, N.; Hsu, C.-Y.; Danoux, L.; Pelletier, N.; Goudounèche, D.; Pons, C.; Leprince, C.; André-Frei, V.; et al. An Inflamed and Infected Reconstructed Human Epidermis to Study Atopic Dermatitis and Skin Care Ingredients. Int. J. Mol. Sci. 2022, 23, 12880. [Google Scholar] [CrossRef]
- Savchenko, T.; Degtyaryov, E.; Radzyukevich, Y.; Buryak, V. Therapeutic Potential of Plant Oxylipins. Int. J. Mol. Sci. 2022, 23, 14627. [Google Scholar] [CrossRef]
- Available online: https://www.marketgrowthreports.com/Global-Biomedical-Materials-Market-21051012 (accessed on 15 May 2024).
- Li, L.; Peng, P.; Ding, N.; Jia, W.; Huang, C.; Tang, Y. Oxidative Stress, Inflammation, Gut Dysbiosis: What Can Polyphenols Do in Inflammatory Bowel Disease? Antioxidants 2023, 12, 967. [Google Scholar] [CrossRef]
- Chen, G.Y. Regulation of the Gut Microbiome by Inflammasomes. Free Radic. Biol. Med. 2017, 105, 35–40. [Google Scholar] [CrossRef][Green Version]
- Lillo, S.; Saleh, M. Inflammasomes in Cancer Progression and Anti-Tumor Immunity. Front. Cell Dev. Biol. 2022, 10, 839041. [Google Scholar] [CrossRef]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic Inflammation in the Etiology of Disease across the Life Span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef]
- Sawada, Y.; Saito-Sasaki, N.; Mashima, E.; Nakamura, M. Daily Lifestyle and Inflammatory Skin Diseases. Int. J. Mol. Sci. 2021, 22, 5204. [Google Scholar] [CrossRef]
- Nema, N.; Rajan, N.; Babu, M.; Sabu, S.; Khamborkar, S.; Sarojam, S.; Sajan, L.; Peter, A.; Chacko, B.; Jacob, V. Plant Polyphenols as Nutraceuticals and Their Antioxidant Potentials. In Polyphenols; Wiley: Hoboken, NJ, USA, 2023; pp. 21–44. [Google Scholar]
- Chiocchio, I.; Prata, C.; Mandrone, M.; Ricciardiello, F.; Marrazzo, P.; Tomasi, P.; Angeloni, C.; Fiorentini, D.; Malaguti, M.; Poli, F.; et al. Leaves and Spiny Burs of Castanea sativa from an Experimental Chestnut Grove: Metabolomic Analysis and Anti-Neuroinflammatory Activity. Metabolites 2020, 10, 408. [Google Scholar] [CrossRef]
- Fernando, A.L.; Duarte, M.P.; Vatsanidou, A.; Alexopoulou, E. Environmental Aspects of Fiber Crops Cultivation and Use. Ind. Crops Prod. 2015, 68, 105–115. [Google Scholar] [CrossRef]
- Durán-Zuazo, V.H.; Rodríguez, B.C.; García-Tejero, I.F.; Ruiz, B.G. Suitability and Opportunities for Cannabis sativa L. as an Alternative Crop for Mediterranean Environments. In Current Applications, Approaches, and Potential Perspectives for Hemp; Elsevier: Amsterdam, The Netherlands, 2023; pp. 3–47. [Google Scholar]
- Song, F.L.; Gan, R.Y.; Zhang, Y.; Xiao, Q.; Kuang, L.; Li, H.B. Total Phenolic Contents and Antioxidant Capacities of Selected Chinese Medicinal Plants. Int. J. Mol. Sci. 2010, 11, 2362–2372. [Google Scholar] [CrossRef]
- Chang, C.-C.; Yang, M.-H.; Wen, H.-M.; Chern, J.-C. Estimation of Total Flavonoid Content in Propolis by Two Complementary Colometric Methods. J. Food Drug Anal. 2020, 10, 3. [Google Scholar] [CrossRef]
- Jayaprakasha, G.K.; Singh, R.P.; Sakariah, K.K. Antioxidant Activity of Grape Seed (Vitis vinifera) Extracts on Peroxidation Models in vitro. Food Chem. 2001, 73, 285–290. [Google Scholar] [CrossRef]
- Ilyasov, I.R.; Beloborodov, V.L.; Selivanova, I.A.; Terekhov, R.P. ABTS/PP Decolorization Assay of Antioxidant Capacity Reaction Pathways. Int. J. Mol. Sci. 2020, 21, 1131. [Google Scholar] [CrossRef]
- Yen, G.-C.; Chen, H.-Y. Antioxidant Activity of Various Tea Extracts in Relation to Their Antimutagenicity. J. Agric. Food Chem. 1995, 43, 27–32. [Google Scholar] [CrossRef]
- Frusciante, L.; Geminiani, M.; Trezza, A.; Olmastroni, T.; Mastroeni, P.; Salvini, L.; Lamponi, S.; Bernini, A.; Grasso, D.; Dreassi, E.; et al. Phytochemical Composition, Anti-Inflammatory Property, and Anti-Atopic Effect of Chaetomorpha linum Extract. Mar. Drugs 2024, 22, 226. [Google Scholar] [CrossRef]
- ISO 10993-5; Biological Evaluation of Medical Devices. Part 5: Tests for In vitro Cytotoxicity. ISO: Geneva, Switzerland, 2009.
- Lamponi, S. Preliminary In Vitro Cytotoxicity, Mutagenicity and Antitumoral Activity Evaluation of Graphene Flake and Aqueous Graphene Paste. Life 2022, 12, 242. [Google Scholar] [CrossRef]
- Ng, N.; Ooi, L. A Simple Microplate Assay for Reactive Oxygen Species Generation and Rapid Cellular Protein Normalization. Bio-Protocol 2021, 11, e3877. [Google Scholar] [CrossRef]
- Feoktistova, M.; Geserick, P.; Leverkus, M. Crystal Violet Assay for Determining Viability of Cultured Cells. Cold Spring Harb. Protoc. 2016, 2016, pdb.prot087379. [Google Scholar] [CrossRef]
- Mortelmans, K.; Zeiger, E. The Ames Salmonella/Microsome Mutagenicity Assay. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2000, 455, 29–60. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The Harmonizome: A Collection of Processed Datasets Gathered to Serve and Mine Knowledge about Genes and Proteins. Database 2016, 2016, baw100. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; O’Donovan, C.; Magrane, M.; Alpi, E.; Antunes, R.; Bely, B.; Bingley, M.; Bonilla, C.; Britto, R.; et al. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Trezza, A.; Geminiani, M.; Cutrera, G.; Dreassi, E.; Frusciante, L.; Lamponi, S.; Spiga, O.; Santucci, A. A Drug Discovery Approach to a Reveal Novel Antioxidant Natural Source: The Case of Chestnut Burr Biomass. Int. J. Mol. Sci. 2024, 25, 2517. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 Update: Improved Access to Chemical Data. Nucleic Acids Res 2019, 47, D1102–D1109. [Google Scholar] [CrossRef]
- Fusi, F.; Trezza, A.; Spiga, O.; Sgaragli, G.; Bova, S. Ca v 1.2 Channel Current Block by the PKA Inhibitor H-89 in Rat Tail Artery Myocytes via a PKA-Independent Mechanism: Electrophysiological, Functional, and Molecular Docking Studies. Biochem. Pharmacol. 2017, 140, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Fusi, F.; Durante, M.; Spiga, O.; Trezza, A.; Frosini, M.; Floriddia, E.; Teodori, E.; Dei, S.; Saponara, S. In Vitro and In Silico Analysis of the Vascular Effects of Asymmetrical N,N-Bis(Alkanol)Amine Aryl Esters, Novel Multidrug Resistance-Reverting Agents. Naunyn Schmiedebergs Arch. Pharmacol. 2016, 389, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the Scope of the Protein–Ligand Interaction Profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Sharif, O.; Bolshakov, V.N.; Raines, S.; Newham, P.; Perkins, N.D. Transcriptional Profiling of the LPS Induced NF-ΚB Response in Macrophages. BMC Immunol. 2007, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Murthy, H.N.; Bapat, V.A. (Eds.) Bioactive Compounds in Underutilized Fruits and Nuts; Springer International Publishing: Cham, Switzerland, 2020; ISBN 978-3-030-30181-1. [Google Scholar]
- Wen, C.; Zhang, J.; Zhang, H.; Dzah, C.S.; Zandile, M.; Duan, Y.; Ma, H.; Luo, X. Advances in Ultrasound Assisted Extraction of Bioactive Compounds from Cash Crops—A Review. Ultrason. Sonochem. 2018, 48, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Chemat, F.; Rombaut, N.; Sicaire, A.-G.; Meullemiestre, A.; Fabiano-Tixier, A.-S.; Abert-Vian, M. Ultrasound Assisted Extraction of Food and Natural Products. Mechanisms, Techniques, Combinations, Protocols and Applications. A Review. Ultrason. Sonochem. 2017, 34, 540–560. [Google Scholar] [CrossRef] [PubMed]
- Vilkhu, K.; Mawson, R.; Simons, L.; Bates, D. Applications and Opportunities for Ultrasound Assisted Extraction in the Food Industry—A Review. Innov. Food Sci. Emerg. Technol. 2008, 9, 161–169. [Google Scholar] [CrossRef]
- Kumar, K.; Srivastav, S.; Sharanagat, V.S. Ultrasound Assisted Extraction (UAE) of Bioactive Compounds from Fruit and Vegetable Processing by-Products: A Review. Ultrason. Sonochem. 2021, 70, 105325. [Google Scholar] [CrossRef]
- Cerulli, A.; Napolitano, A.; Hošek, J.; Masullo, M.; Pizza, C.; Piacente, S. Antioxidant and In Vitro Preliminary Anti-Inflammatory Activity of Castanea sativa (Italian Cultivar “Marrone Di Roccadaspide” PGI) Burs, Leaves, and Chestnuts Extracts and Their Metabolite Profiles by LC-ESI/LTQOrbitrap/MS/MS. Antioxidants 2021, 10, 278. [Google Scholar] [CrossRef]
- Seibert, K.; Zhang, Y.; Leahy, K.; Hauser, S.; Masferrer, J.; Perkins, W.; Lee, L.; Isakson, P. Pharmacological and Biochemical Demonstration of the Role of Cyclooxygenase 2 in Inflammation and Pain. Proc. Natl. Acad. Sci. USA 1994, 91, 12013–12017. [Google Scholar] [CrossRef] [PubMed]
- Zamora, R.; Vodovotz, Y.; Billiar, T.R. Inducible Nitric Oxide Synthase and Inflammatory Diseases. Mol. Med. 2000, 6, 347–373. [Google Scholar] [CrossRef] [PubMed]
- Facchin, B.M.; dos Reis, G.O.; Vieira, G.N.; Mohr, E.T.B.; da Rosa, J.S.; Kretzer, I.F.; Demarchi, I.G.; Dalmarco, E.M. Inflammatory Biomarkers on an LPS-Induced RAW 264.7 Cell Model: A Systematic Review and Meta-Analysis. Inflamm. Res. 2022, 71, 741–758. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.-H.; Shiao, H.-Y.; Tu, C.-H.; Liu, P.-M.; Hsu, J.T.-A.; Amancha, P.K.; Wu, J.-S.; Coumar, M.S.; Chen, C.-H.; Wang, S.-Y.; et al. Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors. J. Med. Chem. 2013, 56, 3889–3903. [Google Scholar] [CrossRef] [PubMed]
- Arter, C.; Trask, L.; Ward, S.; Yeoh, S.; Bayliss, R. Structural Features of the Protein Kinase Domain and Targeted Binding by Small-Molecule Inhibitors. J. Biol. Chem. 2022, 298, 102247. [Google Scholar] [CrossRef] [PubMed]
- Evtyugin, D.D.; Magina, S.; Evtuguin, D.V. Recent Advances in the Production and Applications of Ellagic Acid and Its Derivatives. A Review. Molecules 2020, 25, 2745. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Zhang, Y.; Tang, C.; Hou, Y.; Ai, X.; Chen, X.; Zhang, Y.; Wang, X.; Meng, X. Gallic Acid: Pharmacological Activities and Molecular Mechanisms Involved in Inflammation-Related Diseases. Biomed. Pharmacother. 2021, 133, 110985. [Google Scholar] [CrossRef] [PubMed]
- Mard, S.A.; Mojadami, S.; Farbood, Y.; Kazem, M.; Naseri, G.; Ali, S.; Phd, M. The Anti-Inflammatory and Anti-Apoptotic Effects of Gallic Acid against Mucosal Inflammation- and Erosions-Induced by Gastric Ischemia-Reperfusion in Rats. Vet. Res. Forum 2015, 6, 305–311. [Google Scholar]
- Chagas, M.d.S.S.; Behrens, M.D.; Moragas-Tellis, C.J.; Penedo, G.X.M.; Silva, A.R.; Gonçalves-de-Albuquerque, C.F. Flavonols and Flavones as Potential Anti-Inflammatory, Antioxidant, and Antibacterial Compounds. Oxidative Med. Cell. Longev. 2022, 2022, 9966750. [Google Scholar] [CrossRef]
- Lim, H.; Heo, M.Y.; Kim, H.P. Flavonoids: Broad Spectrum Agents on Chronic Inflammation. Biomol. Ther. 2019, 27, 241–253. [Google Scholar] [CrossRef]
- Ahmed, M.; Ji, M.; Qin, P.; Gu, Z.; Liu, Y.; Sikandar, A.; Iqbal, M.F.; Javeed, A. Phytochemical screening, total phenolic and flavonoids contents and antioxidant activities of Citrullus colocynthis L. and Cannabis sativa L. Appl. Ecol. Environ. Res. 2019, 17, 6961–6979. [Google Scholar] [CrossRef]
- Mkpenie, V.N.; Essien, E.E.; Udoh, I.I. Effect of Extraction Conditions on Total Polyphenol Contents, Antioxidant and Antimicrobial Activities of Cannabis sativa L. Electron. J. Environ. Agric. Food Chem. 2012, 11, 300–307. [Google Scholar]
- Liu, H.-M.; Cheng, M.-Y.; Xun, M.-H.; Zhao, Z.-W.; Zhang, Y.; Tang, W.; Cheng, J.; Ni, J.; Wang, W. Possible Mechanisms of Oxidative Stress-Induced Skin Cellular Senescence, Inflammation, and Cancer and the Therapeutic Potential of Plant Polyphenols. Int. J. Mol. Sci. 2023, 24, 3755. [Google Scholar] [CrossRef]
- Vernarelli, J.A.; Lambert, J.D. Flavonoid Intake Is Inversely Associated with Obesity and C-Reactive Protein, a Marker for Inflammation, in US Adults. Nutr. Diabetes 2017, 7, e276. [Google Scholar] [CrossRef]
- Lefèvre-Arbogast, S.; Gaudout, D.; Bensalem, J.; Letenneur, L.; Dartigues, J.F.; Hejblum, B.P.; Féart, C.; Delcourt, C.; Samieri, C. Pattern of Polyphenol Intake and the Long-Term Risk of Dementia in Older Persons. Neurology 2018, 90, e1979–e1988. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; He, F.; Wu, C.; Li, P.; Li, N.; Deng, J.; Zhu, G.; Ren, W.; Peng, Y. Betaine in Inflammation: Mechanistic Aspects and Applications. Front. Immunol. 2018, 9, 1070. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.K.; Paal, M.C.; Donohue, T.M.; Ganesan, M.; Osna, N.A.; Kharbanda, K.K. Beneficial Effects of Betaine: A Comprehensive Review. Biology 2021, 10, 456. [Google Scholar] [CrossRef]
- Go, E.K.; Jung, K.J.; Kim, J.Y.; Yu, B.P.; Chung, H.Y. Betaine Suppresses Proinflammatory Signaling During Aging: The Involvement of Nuclear Factor- B via Nuclear Factor-Inducing Kinase/I B Kinase and Mitogen-Activated Protein Kinases. J. Gerontol. A Biol. Sci. Med. Sci. 2005, 60, 1252–1264. [Google Scholar] [CrossRef]



| Antioxidant Capacity | |||||
|---|---|---|---|---|---|
| TPC (mg GAE/g) | TFC (mg QE/g) | TRP (mg AAE/g) | ABTS (IC50 µg/mL) | DPPH (IC50 µg/mL) | |
| CSB | 243.98 ± 17.77 | 27.54 ± 0.60 | 272.12 ± 4.64 | 8.16 ± 1.11 | 29.57 ± 0.57 |
| No. | Name | Retention Time (min) | Formula | Calculated MW | Theoretical m/z | Reference Ion | Mass Error (ppm) | Peak Area (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | Ellagic acid | 15.388 | C14H6O8 | 302.00617 | 300.9988 | [M − H]−1 | −0.33 | 51.7 |
| 2 | Betaine | 1.863 | C5H11NO2 | 117.07923 | 118.0865 | [M + H]+1 | 2.17 | 22.0 |
| 3 | 5,7-dihydroxy-3.8-dimethoxy-2-phenyl-4h-chromen-4-one | 29.172 | C17H14O6 | 314.07985 | 315.0871 | [M + H]+1 | 2.59 | 15.8 |
| 4 | Mollioside | 25.285 | C26H40O10 | 512.26256 | 513.2698 | [M + H]+1 | 0.81 | 1.7 |
| 5 | (±)-(2e)-abscisic acid | 11.733 | C15H20O4 | 264.13625 | 265.1435 | [M + H]+1 | 0.34 | 1.6 |
| 6 | 3,8-di-o-methylellagic acid | 21.233 | C16H10O8 | 330.03838 | 331.0457 | [M + H]+1 | 2.46 | 1.4 |
| 7 | 12-hydroxyjasmonic acid | 18.885 | C12H18O4 | 226.12108 | 227.1284 | [M + H]+1 | 2.53 | 0.9 |
| 8 | Epi-jasmonic acid | 15.442 | C12H18O3 | 210.12618 | 211.1335 | [M + H]+1 | 2.77 | 0.7 |
| 9 | Protocatechuic aldehyde | 6.164 | C7H6O3 | 138.03177 | 139.0391 | [M + H]+1 | 0.56 | 0.4 |
| 10 | Gibberellin A2 o-beta-d-glucoside | 23.076 | C25H36O11 | 512.22551 | 513.2328 | [M + H]+1 | −0.49 | 0.3 |
| 11 | Sinapaldehyde | 21.782 | C11H12O4 | 208.07372 | 209.081 | [M + H]+1 | 0.76 | 0.3 |
| 12 | 12-hydroxyjasmonic acid 12-o-beta-d-glucoside | 19.078 | C19H30O8 | 386.1931 | 387.2004 | [M + H]+1 | −2.5 | 0.3 |
| 13 | 5,7-dihydroxy-3′,4′,5′-trimethoxyflavanone | 18.774 | C18H18O7 | 346.10628 | 347.1136 | [M + H]+1 | 2.97 | 0.2 |
| 14 | (+)-Gibberellic acid | 16.236 | C19H22O6 | 346.14242 | 347.1497 | [M + H]+1 | 2.25 | 0.2 |
| 15 | N-propyl galiate | 10.343 | C10H12O5 | 212.06876 | 235.058 | [M + Na]+1 | 1.35 | 0.2 |
| 16 | Syringaldehyde | 12.269 | C9H10O4 | 182.05821 | 183.0655 | [M + H]+1 | 1.68 | 0.2 |
| 17 | Retusin (flavonol) | 18.648 | C19H18O7 | 358.10628 | 359.1136 | [M + H]+1 | 2.87 | 0.2 |
| 18 | Acaciin | 11.453 | C28H32O14 | 592.17833 | 593.1856 | [M + H]+1 | −1.48 | 0.2 |
| 19 | Kaempferol | 17.111 | C15H10O6 | 286.04805 | 287.0553 | [M + H]+1 | 1.1 | 0.2 |
| 20 | Scopoletin | 16.019 | C10H8O4 | 192.04282 | 193.0501 | [M + H]+1 | 2.91 | 0.2 |
| 21 | Isorhamnetin 3-rhamnosyl-(1->2)-gentiobiosyl-(1->6)-glucoside | 28.24 | C40H52O26 | 948.27548 | 949.2828 | [M + H]+1 | 0.84 | 0.1 |
| 22 | 5,7-methoxyflavanone | 12.653 | C17H16O4 | 284.10549 | 285.1128 | [M + H]+1 | 2.21 | 0.1 |
| 23 | 4′.5.7-trimethoxyflavone | 20.486 | C18H16O5 | 312.10053 | 313.1079 | [M + H]+1 | 2.44 | 0.1 |
| 24 | Ethyl gallate | 13.29 | C9H10O5 | 198.05319 | 199.0605 | [M + H]+1 | 1.87 | 0.1 |
| 25 | 2-(2,6-dimethoxyphenyl)-5,6-dimethoxy-4h-chromen-4-one (zapotin) | 16.541 | C19H18O6 | 342.11131 | 343.1186 | [M + H]+1 | 2.85 | 0.1 |
| 26 | Helichrysoside | 20.13 | C30H26O14 | 610.13403 | 633.1237 | [M + Na]+1 | 2.91 | 0.1 |
| 27 | 1,4-dihydro-4-oxo-3-(2-pyrrolidinyl)-2-quinolinecarboxylic acid | 17.834 | C14H14N2O3 | 258.10066 | 259.1079 | [M + H]+1 | 0.86 | 0.1 |
| 28 | Afrormosin | 16.195 | C17H14O5 | 298.08489 | 299.0922 | [M + H]+1 | 2.56 | 0.1 |
| 29 | Gibberellin A17 | 6.043 | C20H26O7 | 378.16813 | 377.1609 | [M − H]−1 | 0.75 | 0.1 |
| 30 | Quercetin | 17.258 | C15H10O7 | 302.04329 | 303.0506 | [M + H]+1 | 2.11 | 0.1 |
| 31 | 1,3-bis-(5-carboxypentyl)-urea | 11.099 | C13H24N2O5 | 288.16887 | 289.1762 | [M + H]+1 | 1.21 | 0.1 |
| 32 | 5-carboxyvanillic acid | 13.026 | C9H8O6 | 212.0318 | 211.0245 | [M − H]−1 | −1.37 | tr. |
| 33 | 3-hydroxyflavone | 18.639 | C15H10O3 | 238.06363 | 239.0709 | [M + H]+1 | 2.68 | tr. |
| 34 | Coniferaldehyde | 19.593 | C10H10O3 | 178.0632 | 179.0705 | [M + H]+1 | 1.18 | tr. |
| 35 | Gibberellin A1/A34 | 39.565 | C19H24O6 | 348.15815 | 349.1654 | [M + H]+1 | 2.47 | tr. |
| 36 | Isorhamnetin 3-o-alpha-l-[6′-p-coumaroyl-beta-d-glucopyranosyl-(1->2)-rhamnopyranoside] | 20.825 | C37H38O18 | 770.20801 | 793.1978 | [M + Na]+1 | 2.85 | tr. |
| 37 | Gibberellin A53 | 38.444 | C20H28O5 | 348.19405 | 349.2013 | [M + H]+1 | 1.07 | tr. |
| 38 | 2′,5-digalloylhamamelofuranose | 12.064 | C20H20O14 | 484.08397 | 485.0913 | [M + H]+1 | −2.76 | tr. |
| 39 | (+)-Catechin 7-o-beta-d-xyloside | 18.011 | C20H22O10 | 422.12248 | 445.1117 | [M + Na]+1 | 2.81 | tr. |
| 40 | Digallic acid | 2.807 | C14H10O9 | 322.03294 | 321.0257 | [M − H]−1 | 1.43 | tr. |
| 41 | (E)-ferulic acid | 29.676 | C10H10O4 | 194.05788 | 195.0652 | [M + H]+1 | −0.16 | tr. |
| 42 | 1,3-dibutyl-1,3-dimethylurea | 13.625 | C11H24N2O | 200.18829 | 199.181 | [M−H]−1 | −2.86 | tr. |
| 43 | Kaempferol-3-o-(6′-trans-p-coumaroyl-2′-glucosyl)rhamnoside | 21.66 | C36H36O17 | 740.19518 | 741.2025 | [M + H]+1 | −0.1 | tr. |
| 44 | Tomentosin | 15.583 | C15H20O3 | 248.14121 | 249.1487 | [M + H]+1 | −0.15 | tr. |
| 45 | Catechin gallate. (-)- | 18.618 | C22H18O10 | 442.08926 | 443.0965 | [M + H]+1 | −1.68 | tr. |
| 46 | Coniferyl aldehyde | 24.95 | C10H10O3 | 178.0632 | 179.0705 | [M + H]+1 | 1.18 | tr. |
| 47 | Quercetin-3-o-(6′-trans-p-coumaroyl-2′-glucosyl)rhamnoside | 18.932 | C36H36O18 | 756.19158 | 779.1812 | [M + Na]+1 | 1.87 | tr. |
| 48 | Gibberellin A24 | 37.98 | C20H26O5 | 346.17818 | 347.1855 | [M + H]+1 | 0.45 | tr. |
| 49 | Glucogallin | 3.747 | C13H16O10 | 332.07488 | 331.0676 | [M − H]−1 | 1.61 | tr. |
| 50 | 5′-desgalloylstachyurin | 11.878 | C34H24O22 | 784.0757 | 783.0684 | [M − H]−1 | −0.29 | tr. |
| 51 | Gibberellin A12 | 44.209 | C20H28O4 | 332.19787 | 333.2052 | [M + H]+1 | −2.67 | tr. |
| 52 | (+)-Gallocatechin | 2.929 | C15H14O7 | 306.07341 | 305.0661 | [M − H]−1 | −1.78 | tr. |
| 53 | Isorhamnetin | 23.872 | C16H12O7 | 316.05908 | 315.0518 | [M − H]−1 | 2.47 | tr. |
| 54 | Myricetin-3-o-glucoside | 8.465 | C21H20O13 | 480.09172 | 479.0844 | [M − H]−1 | 2.77 | tr. |
| 55 | 1,6-bis-o-galloyl-beta-d-glucose | 8.974 | C20H20O14 | 484.0862 | 483.0789 | [M − H]−1 | 1.84 | tr. |
| 56 | Castalagin/vescalagin | 15.21 | C41H26O26 | 934.06952 | 933.0623 | [M − H]−1 | −1.83 | tr. |
| Tot | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frusciante, L.; Geminiani, M.; Olmastroni, T.; Mastroeni, P.; Trezza, A.; Salvini, L.; Lamponi, S.; Spiga, O.; Santucci, A. Repurposing Castanea sativa Spiny Burr By-Products Extract as a Potentially Effective Anti-Inflammatory Agent for Novel Future Biotechnological Applications. Life 2024, 14, 763. https://doi.org/10.3390/life14060763
Frusciante L, Geminiani M, Olmastroni T, Mastroeni P, Trezza A, Salvini L, Lamponi S, Spiga O, Santucci A. Repurposing Castanea sativa Spiny Burr By-Products Extract as a Potentially Effective Anti-Inflammatory Agent for Novel Future Biotechnological Applications. Life. 2024; 14(6):763. https://doi.org/10.3390/life14060763
Chicago/Turabian StyleFrusciante, Luisa, Michela Geminiani, Tommaso Olmastroni, Pierfrancesco Mastroeni, Alfonso Trezza, Laura Salvini, Stefania Lamponi, Ottavia Spiga, and Annalisa Santucci. 2024. "Repurposing Castanea sativa Spiny Burr By-Products Extract as a Potentially Effective Anti-Inflammatory Agent for Novel Future Biotechnological Applications" Life 14, no. 6: 763. https://doi.org/10.3390/life14060763
APA StyleFrusciante, L., Geminiani, M., Olmastroni, T., Mastroeni, P., Trezza, A., Salvini, L., Lamponi, S., Spiga, O., & Santucci, A. (2024). Repurposing Castanea sativa Spiny Burr By-Products Extract as a Potentially Effective Anti-Inflammatory Agent for Novel Future Biotechnological Applications. Life, 14(6), 763. https://doi.org/10.3390/life14060763







