New Insights into HIV Life Cycle, Th1/Th2 Shift during HIV Infection and Preferential Virus Infection of Th2 Cells: Implications of Early HIV Treatment Initiation and Care
Abstract
1. Introduction
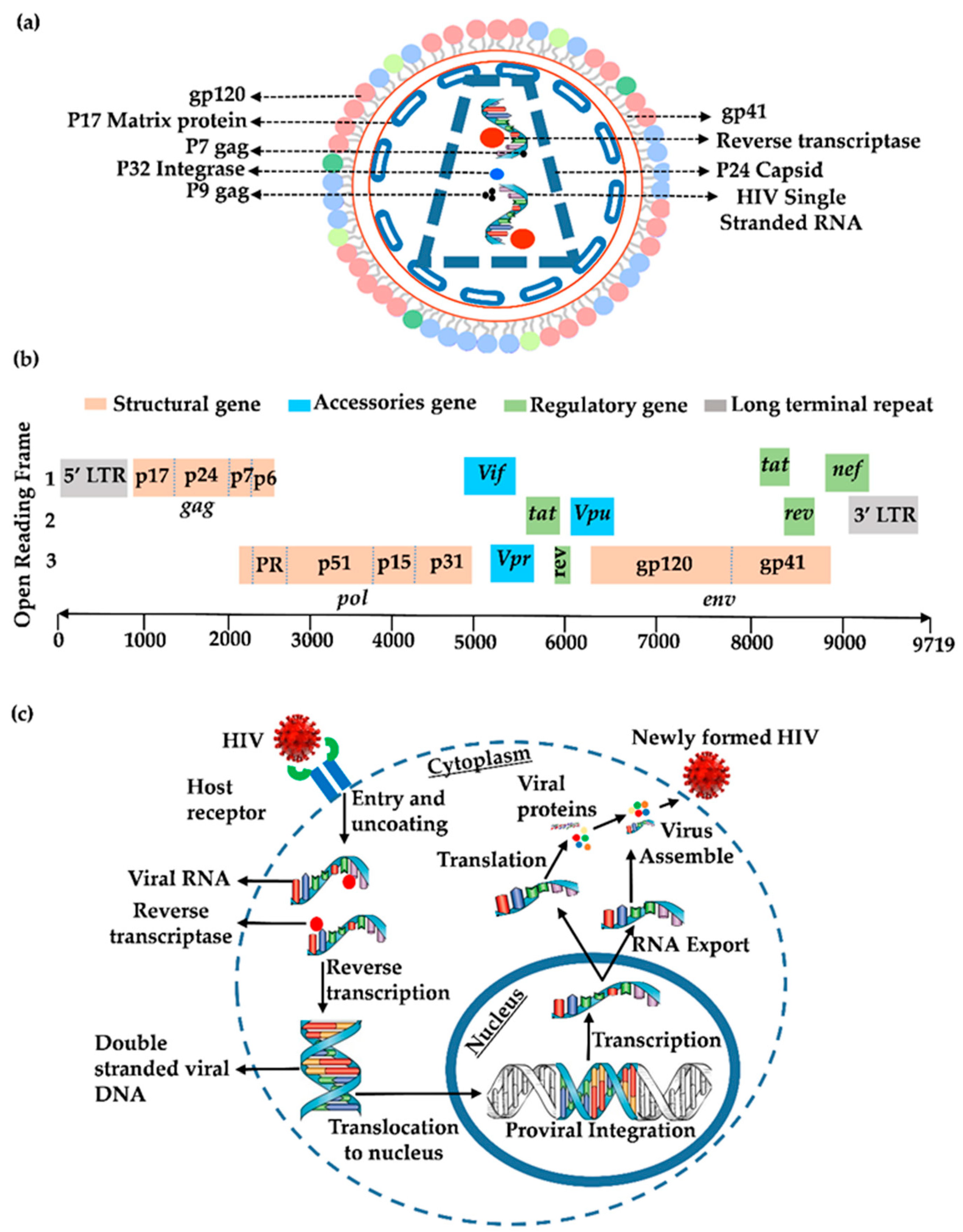
2. Genomics and Proteomics of HIV-1
3. HIV Lifecycle
3.1. Entry and Reverse Transcription
3.2. Integration
3.3. Transcription
3.4. Translation
3.5. New Viral Progeny
4. Overview of the Immune Response to HIV Infection and Th1 and Th2 Hypothesis
4.1. Mechanism of Th1 and Th2 Classification and Differentiation
4.2. Mechanism of Th1/Th2 Shift in HIV Infection and Preferential Virus Infection of Th2 Cells
4.3. The Th1/Th2 Shift in the Context of HIV and Co-Infections
4.4. Implications of Early HIV Treatment Initiation and Care
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gilbert, P.B.; McKeague, I.W.; Eisen, G.; Mullins, C.; Gueye, N.A.; Mboup, S.; Kanki, P.J. Comparison of HIV-1 and HIV-2 infectivity from a prospective cohort study in Senegal. Stat. Med. 2003, 22, 573–593. [Google Scholar] [CrossRef] [PubMed]
- Popper, S.J.; Sarr, A.D.; Travers, K.U.; Gueye-Ndiaye, A.; Mboup, S.; Essex, M.E.; Kanki, P.J. Lower human immunodeficiency virus (HIV) type 2 viral load reflects the difference in pathogenicity of HIV-1 and HIV-2. J. Infect. Dis. 1999, 180, 1116–1121. [Google Scholar] [CrossRef] [PubMed]
- Ruelle, J.; Mukadi, B.K.; Schutten, M.; Goubau, P. Quantitative real-time PCR on Lightcycler for the detection of human immunodeficiency virus type 2 (HIV-2). J. Virol. Methods 2004, 117, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Nyamweya, S.; Hegedus, A.; Jaye, A.; Rowland-Jones, S.; Flanagan, K.L.; Macallan, D.C. Comparing HIV-1 and HIV-2 infection: Lessons for viral immunopathogenesis. Rev. Med. Virol. 2013, 23, 221–240. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Hahn, B.H. The evolution of HIV-1 and the origin of AIDS. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 2487–2494. [Google Scholar] [CrossRef] [PubMed]
- Peeters, M.; Courgnaud, V. Overview of primate lentiviruses and their evolution in non-human primates in Africa. HIV Seq. Compend. 2002, 8, 451–457. [Google Scholar]
- Hokello, J.; Tyagi, P.; Dimri, S.; Sharma, A.L.; Tyagi, M. Comparison of the Biological Basis for Non-HIV Transmission to HIV-Exposed Seronegative Individuals, Disease Non-Progression in HIV Long-Term Non-Progressors and Elite Controllers. Viruses 2023, 15, 1362. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Herzig, E.; Doitsh, G.; Grimmett, Z.W.; Munoz-Arias, I.; Greene, W.C. HIV-2 Depletes CD4 T Cells through Pyroptosis despite Vpx-Dependent Degradation of SAMHD1. J. Virol. 2019, 93, e00666-19. [Google Scholar] [CrossRef]
- Baldauf, H.M.; Stegmann, L.; Schwarz, S.M.; Ambiel, I.; Trotard, M.; Martin, M.; Burggraf, M.; Lenzi, G.M.; Lejk, H.; Pan, X.; et al. Vpx overcomes a SAMHD1-independent block to HIV reverse transcription that is specific to resting CD4 T cells. Proc. Natl. Acad. Sci. USA 2017, 114, 2729–2734. [Google Scholar] [CrossRef]
- Sharp, P.M.; Hahn, B.H. Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 2011, 1, a006841. [Google Scholar] [CrossRef]
- Huet, T.; Cheynier, R.; Meyerhans, A.; Roelants, G.; Wain-Hobson, S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature 1990, 345, 356–359. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, V.M.; Olmsted, R.A.; Murphey-Corb, M.; Purcell, R.H.; Johnson, P.R. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 1989, 339, 389–392. [Google Scholar] [CrossRef] [PubMed]
- De Cock, K.M.; Adjorlolo, G.; Ekpini, E.; Sibailly, T.; Kouadio, J.; Maran, M.; Brattegaard, K.; Vetter, K.M.; Doorly, R.; Gayle, H.D. Epidemiology and transmission of HIV-2. Why there is no HIV-2 pandemic. JAMA 1993, 270, 2083–2086. [Google Scholar] [CrossRef] [PubMed]
- Clavel, F.; Guetard, D.; Brun-Vezinet, F.; Chamaret, S.; Rey, M.A.; Santos-Ferreira, M.O.; Laurent, A.G.; Dauguet, C.; Katlama, C.; Rouzioux, C.; et al. Isolation of a new human retrovirus from West African patients with AIDS. Science 1986, 233, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Esbjornsson, J.; Jansson, M.; Jespersen, S.; Mansson, F.; Honge, B.L.; Lindman, J.; Medina, C.; da Silva, Z.J.; Norrgren, H.; Medstrand, P.; et al. HIV-2 as a model to identify a functional HIV cure. AIDS Res. Ther. 2019, 16, 24. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Jetzt, A.E.; Sun, G.; Yu, H.; Klarmann, G.; Ron, Y.; Preston, B.D.; Dougherty, J.P. Human immunodeficiency virus type 1 recombination: Rate, fidelity, and putative hot spots. J. Virol. 2002, 76, 11273–11282. [Google Scholar] [CrossRef]
- Ramirez, B.C.; Simon-Loriere, E.; Galetto, R.; Negroni, M. Implications of recombination for HIV diversity. Virus Res. 2008, 134, 64–73. [Google Scholar] [CrossRef]
- Gao, F.; Yue, L.; Robertson, D.L.; Hill, S.C.; Hui, H.; Biggar, R.J.; Neequaye, A.E.; Whelan, T.M.; Ho, D.D.; Shaw, G.M.; et al. Genetic diversity of human immunodeficiency virus type 2: Evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 1994, 68, 7433–7447. [Google Scholar] [CrossRef]
- Kanki, P.J.; Travers, K.U.; Marlink, R.G.; Essex, M.E.; MBoup, S.; Gueye-Ndiaye, A.; Siby, T.; Thior, I.; Sankalé, J.-L.; Hsieh, C.-C.; et al. Slower heterosexual spread of HIV-2 than HIV-1. Lancet 1994, 343, 943–946. [Google Scholar] [CrossRef]
- Adjorlolo-Johnson, G.; De Cock, K.M.; Ekpini, E.; Vetter, K.M.; Sibailly, T.; Brattegaard, K.; Yavo, D.; Doorly, R.; Whitaker, J.P.; Kestens, L.; et al. Prospective comparison of mother-to-child transmission of HIV-1 and HIV-2 in Abidjan, Ivory Coast. JAMA 1994, 272, 462–466. [Google Scholar] [CrossRef]
- O’Donovan, D.; Ariyoshi, K.; Milligan, P.; Ota, M.; Yamuah, L.; Sarge-Njie, R.; Whittle, H. Maternal plasma viral RNA levels determine marked differences in mother-to-child transmission rates of HIV-1 and HIV-2 in The Gambia. MRC/Gambia Government/University College London Medical School working group on mother-child transmission of HIV. AIDS 2000, 14, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Steele, E.; Awasana, A.A.; Corrah, T.; Sabally, S.; van der Sande, M.; Jaye, A.; Togun, T.; Sarge-Njie, R.; McConkey, S.J.; Whittle, H.; et al. Is HIV-2- induced AIDS different from HIV-1-associated AIDS? Data from a West African clinic. AIDS 2007, 21, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Barre-Sinoussi, F.; Chermann, J.C.; Rey, F.; Nugeyre, M.T.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axler-Blin, C.; Vezinet-Brun, F.; Rouzioux, C.; et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 1983, 220, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Gallo, R.C.; Salahuddin, S.Z.; Popovic, M.; Shearer, G.M.; Kaplan, M.; Haynes, B.F.; Palker, T.J.; Redfield, R.; Oleske, J.; Safai, B.; et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 1984, 224, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Siekevitz, M.; Josephs, S.F.; Dukovich, M.; Peffer, N.; Wong-Staal, F.; Greene, W.C. Activation of the HIV-1 LTR by T cell mitogens and the trans-activator protein of HTLV-I. Science 1987, 238, 1575–1578. [Google Scholar] [CrossRef]
- Frankel, A.D.; Young, J.A. HIV-1: Fifteen proteins and an RNA. Annu. Rev. Biochem. 1998, 67, 1–25. [Google Scholar] [CrossRef]
- Freed, E.O. HIV-1 gag proteins: Diverse functions in the virus life cycle. Virology 1998, 251, 1–15. [Google Scholar] [CrossRef]
- Chatterji, U.; Bobardt, M.D.; Gaskill, P.; Sheeter, D.; Fox, H.; Gallay, P.A. Trim5α accelerates degradation of cytosolic capsid associated with productive HIV-1 entry. J. Biol. Chem. 2006, 281, 37025–37033. [Google Scholar] [CrossRef]
- Binley, J.M.; Sanders, R.W.; Master, A.; Cayanan, C.S.; Wiley, C.L.; Schiffner, L.; Travis, B.; Kuhmann, S.; Burton, D.R.; Hu, S.L.; et al. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 2002, 76, 2606–2616. [Google Scholar] [CrossRef]
- Doms, R.W.; Peiper, S.C. Unwelcomed guests with master keys: How HIV uses chemokine receptors for cellular entry. Virology 1997, 235, 179–190. [Google Scholar] [CrossRef]
- Mao, Y.; Wang, L.; Gu, C.; Herschhorn, A.; Xiang, S.H.; Haim, H.; Yang, X.; Sodroski, J. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat. Struct. Mol. Biol. 2012, 19, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Summers, M.F.; Henderson, L.E.; Chance, M.R.; Bess, J.W., Jr.; South, T.L.; Blake, P.R.; Sagi, I.; Perez-Alvarado, G.; Sowder, R.C., 3rd; Hare, D.R.; et al. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992, 1, 563–574. [Google Scholar] [CrossRef]
- King, S.R. HIV: Virology and mechanisms of disease. Ann. Emerg. Med. 1994, 24, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Craigie, R.; Bushman, F.D. HIV DNA integration. Cold Spring Harb. Perspect. Med. 2012, 2, a006890. [Google Scholar] [CrossRef] [PubMed]
- Pettit, S.C.; Everitt, L.E.; Choudhury, S.; Dunn, B.M.; Kaplan, A.H. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. J. Virol. 2004, 78, 8477–8485. [Google Scholar] [CrossRef] [PubMed]
- Ju, S.M.; Song, H.Y.; Lee, J.A.; Lee, S.J.; Choi, S.Y.; Park, J. Extracellular HIV-1 Tat up-regulates expression of matrix metalloproteinase-9 via a MAPK-NF-κB dependent pathway in human astrocytes. Exp. Mol. Med. 2009, 41, 86–93. [Google Scholar] [CrossRef]
- Mahlknecht, U.; Dichamp, I.; Varin, A.; Van Lint, C.; Herbein, G. NF-κB-dependent control of HIV-1 transcription by the second coding exon of Tat in T cells. J. Leucoc. Biol. 2008, 83, 718–727. [Google Scholar] [CrossRef]
- Nekhai, S.; Jerebtsova, M.; Jackson, A.; Southerland, W. Regulation of HIV-1 transcription by protein phosphatase 1. Curr. HIV Res. 2007, 5, 3–9. [Google Scholar] [CrossRef]
- Piguet, V.; Schwartz, O.; Le Gall, S.; Trono, D. The downregulation of CD4 and MHC-I by primate lentiviruses: A paradigm for the modulation of cell surface receptors. Immunol. Rev. 1999, 168, 51–63. [Google Scholar] [CrossRef]
- Popov, S.; Rexach, M.; Zybarth, G.; Reiling, N.; Lee, M.A.; Ratner, L.; Lane, C.M.; Moore, M.S.; Blobel, G.; Bukrinsky, M. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998, 17, 909–917. [Google Scholar] [CrossRef]
- Schubert, U.; Anton, L.C.; Bacik, I.; Cox, J.H.; Bour, S.; Bennink, J.R.; Orlowski, M.; Strebel, K.; Yewdell, J.W. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J. Virol. 1998, 72, 2280–2288. [Google Scholar] [CrossRef] [PubMed]
- Simmons, A.; Aluvihare, V.; McMichael, A. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity 2001, 14, 763–777. [Google Scholar] [CrossRef] [PubMed]
- Varin, A.; Manna, S.K.; Quivy, V.; Decrion, A.Z.; Van Lint, C.; Herbein, G.; Aggarwal, B.B. Exogenous Nef protein activates NF-κB, AP-1, and c-Jun N-terminal kinase and stimulates HIV transcription in promonocytic cells. Role in AIDS pathogenesis. J. Biol. Chem. 2003, 278, 2219–2227. [Google Scholar] [CrossRef] [PubMed]
- Leonhardt, S.A.; Purdy, M.D.; Grover, J.R.; Yang, Z.; Poulos, S.; McIntire, W.E.; Tatham, E.A.; Erramilli, S.K.; Nosol, K.; Lai, K.K. Antiviral HIV-1 SERINC restriction factors disrupt virus membrane asymmetry. Nat. Commun. 2023, 14, 4368. [Google Scholar] [CrossRef] [PubMed]
- Trono, D. HIV accessory proteins: Leading roles for the supporting cast. Cell 1995, 82, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Nabel, G.; Baltimore, D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 1987, 326, 711–713. [Google Scholar] [CrossRef] [PubMed]
- West, M.J.; Lowe, A.D.; Karn, J. Activation of human immunodeficiency virus transcription in T cells revisited: NF-κB p65 stimulates transcriptional elongation. J. Virol. 2001, 75, 8524–8537. [Google Scholar] [CrossRef]
- Kinoshita, S.; Su, L.; Amano, M.; Timmerman, L.A.; Kaneshima, H.; Nolan, G.P. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity 1997, 6, 235–244. [Google Scholar] [CrossRef]
- Karn, J. Tackling Tat. J. Mol. Biol. 1999, 293, 235–254. [Google Scholar] [CrossRef]
- Kim, Y.K.; Bourgeois, C.F.; Isel, C.; Churcher, M.J.; Karn, J. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol. Cell. Biol. 2002, 22, 4622–4637. [Google Scholar] [CrossRef]
- Marciniak, R.A.; Sharp, P.A. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991, 10, 4189–4196. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, M.; Rusnati, M.; Presta, M.; Giacca, M. Internalization of HIV-1 tat requires cell surface heparan sulfate proteoglycans. J. Biol. Chem. 2001, 276, 3254–3261. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, M.; Bukrinsky, M. Human immunodeficiency virus (HIV) latency: The major hurdle in HIV eradication. Mol. Med. 2012, 18, 1096–1108. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, M.; Romerio, F. Models of HIV-1 persistence in the CD4+ T cell compartment: Past, present and future. Curr. HIV Res. 2011, 9, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Marzio, G.; Tyagi, M.; Gutierrez, M.I.; Giacca, M. HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc. Natl. Acad. Sci. USA 1998, 95, 13519–13524. [Google Scholar] [CrossRef]
- Silhol, M.; Tyagi, M.; Giacca, M.; Lebleu, B.; Vives, E. Different mechanisms for cellular internalization of the HIV-1 Tat-derived cell penetrating peptide and recombinant proteins fused to Tat. Eur. J. Biochem. 2002, 269, 494–501. [Google Scholar] [CrossRef]
- Harris, R.S.; Bishop, K.N.; Sheehy, A.M.; Craig, H.M.; Petersen-Mahrt, S.K.; Watt, I.N.; Neuberger, M.S.; Malim, M.H. DNA deamination mediates innate immunity to retroviral infection. Cell 2003, 113, 803–809. [Google Scholar] [CrossRef]
- Yang, X.; Goncalves, J.; Gabuzda, D. Phosphorylation of Vif and its role in HIV-1 replication. J. Biol. Chem. 1996, 271, 10121–10129. [Google Scholar] [CrossRef]
- Emerman, M. HIV-1, Vpr and the cell cycle. Curr. Biol. 1996, 6, 1096–1103. [Google Scholar] [CrossRef][Green Version]
- Mansky, L.M. The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology 1996, 222, 391–400. [Google Scholar] [CrossRef]
- Neil, S.J.; Zang, T.; Bieniasz, P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 2008, 451, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Cohen, E.A.; Subbramanian, R.A.; Gottlinger, H.G. Role of auxiliary proteins in retroviral morphogenesis. Morphog. Matur. Retroviruses 1996, 214, 219–235. [Google Scholar] [CrossRef]
- Dalgleish, A.G.; Beverley, P.C.; Clapham, P.R.; Crawford, D.H.; Greaves, M.F.; Weiss, R.A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 1984, 312, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Liu, R.; Ellmeier, W.; Choe, S.; Unutmaz, D.; Burkhart, M.; Di Marzio, P.; Marmon, S.; Sutton, R.E.; Hill, C.M.; et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature 1996, 381, 661–666. [Google Scholar] [CrossRef]
- Feng, Y.; Broder, C.C.; Kennedy, P.E.; Berger, E.A. HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 1996, 272, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Zaitseva, M.; Peden, K.; Golding, H. HIV coreceptors: Role of structure, posttranslational modifications, and internalization in viral-cell fusion and as targets for entry inhibitors. Biochim. Biophys. Acta 2003, 1614, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.K.; Chen, M.Y.; Chuang, C.Y.; Jeang, K.T.; Huang, L.M. Molecular biology of human immunodeficiency virus type 1. J. Microbiol. Immunol. Infect. = Wei Mian Yu Gan Ran Za Zhi 2000, 33, 131–140. [Google Scholar] [PubMed]
- Gifford, L.B.; Melikian, G. Human Immunodeficiency Virus 1 Capsid Uncoating in the Nucleus Progresses Through Defect Formation in the Capsid Lattice. bioRxiv 2023. [Google Scholar] [CrossRef]
- Müller, T.G.; Zila, V.; Müller, B.; Kräusslich, H.-G. Nuclear capsid uncoating and reverse transcription of HIV-1. Annu. Rev. Virol. 2022, 9, 261–284. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.; van Kooyk, Y. DC-SIGN: A novel HIV receptor on DCs that mediates HIV-1 transmission. Curr. Top. Microbiol. Immunol. 2003, 276, 31–54. [Google Scholar] [CrossRef]
- Hoorelbeke, B.; Xue, J.; LiWang, P.J.; Balzarini, J. Role of the carbohydrate-binding sites of griffithsin in the prevention of DC-SIGN-mediated capture and transmission of HIV-1. PLoS ONE 2013, 8, e64132. [Google Scholar] [CrossRef] [PubMed]
- de Witte, L.; Nabatov, A.; Pion, M.; Fluitsma, D.; de Jong, M.A.; de Gruijl, T.; Piguet, V.; van Kooyk, Y.; Geijtenbeek, T.B. Langerin is a natural barrier to HIV-1 transmission by Langerhans cells. Nat. Med. 2007, 13, 367–371. [Google Scholar] [CrossRef] [PubMed]
- de Witte, L.; Nabatov, A.; Geijtenbeek, T.B. Distinct roles for DC-SIGN+-dendritic cells and Langerhans cells in HIV-1 transmission. Trends Mol. Med. 2008, 14, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Cicala, C.; Arthos, J.; Fauci, A.S. HIV-1 envelope, integrins and co-receptor use in mucosal transmission of HIV. J. Transl. Med. 2011, 9 (Suppl. S1), S2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Freed, E.O. HIV-1 replication. Somat. Cell Mol. Genet. 2001, 26, 13–33. [Google Scholar] [CrossRef]
- Francis, A.C.; Melikyan, G.B. Single HIV-1 Imaging Reveals Progression of Infection through CA-Dependent Steps of Docking at the Nuclear Pore, Uncoating, and Nuclear Transport. Cell Host Microbe 2018, 23, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Rankovic, S.; Varadarajan, J.; Ramalho, R.; Aiken, C.; Rousso, I. Reverse Transcription Mechanically Initiates HIV-1 Capsid Disassembly. J. Virol. 2017, 91, e00289-17. [Google Scholar] [CrossRef]
- Cosnefroy, O.; Murray, P.J.; Bishop, K.N. HIV-1 capsid uncoating initiates after the first strand transfer of reverse transcription. Retrovirology 2016, 13, 58. [Google Scholar] [CrossRef]
- Stevenson, M. HIV-1 pathogenesis. Nat. Med. 2003, 9, 853–860. [Google Scholar] [CrossRef]
- Kingsman, S.M.; Kingsman, A.J. The regulation of human immunodeficiency virus type-1 gene expression. Eur. J. Biochem. 1996, 240, 491–507. [Google Scholar] [CrossRef]
- Pearson, R.; Kim, Y.K.; Hokello, J.; Lassen, K.; Friedman, J.; Tyagi, M.; Karn, J. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 2008, 82, 12291–12303. [Google Scholar] [CrossRef] [PubMed]
- Hokello, J.; Sharma, A.L.; Tyagi, M. AP-1 and NF-κB synergize to transcriptionally activate latent HIV upon T-cell receptor activation. FEBS Lett. 2021, 559, 577–594. [Google Scholar] [CrossRef] [PubMed]
- Hokello, J.; Sharma, A.; Tyagi, M. Combinatorial Use of Both Epigenetic and Non-Epigenetic Mechanisms to Efficiently Reactivate HIV Latency. Int. J. Mol. Sci. 2021, 22, 3697. [Google Scholar] [CrossRef] [PubMed]
- Hokello, J.; Sharma, A.L.; Tyagi, M. Efficient Non-Epigenetic Activation of HIV Latency through the T-Cell Receptor Signalosome. Viruses 2020, 12, 868. [Google Scholar] [CrossRef] [PubMed]
- Hokello, J.; Sharma, A.L.; Tyagi, P.; Bhushan, A.; Tyagi, M. Human immunodeficiency virus type-1 (HIV-1) transcriptional regulation, latency and therapy in the central nervous system. Vaccines 2021, 9, 1272. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.L.; Shafer, D.; Netting, D.; Tyagi, M. Cocaine sensitizes the CD4+ T cells for HIV infection by co-stimulating NFAT and AP-1. iScience 2022, 25, 105651. [Google Scholar] [CrossRef]
- Sharma, A.L.; Hokello, J.; Sonti, S.; Zicari, S.; Sun, L.; Alqatawni, A.; Bukrinsky, M.; Simon, G.; Chauhan, A.; Daniel, R.; et al. CBF-1 Promotes the Establishment and Maintenance of HIV Latency by Recruiting Polycomb Repressive Complexes, PRC1 and PRC2, at HIV LTR. Viruses 2020, 12, 1040. [Google Scholar] [CrossRef]
- Zicari, S.; Sharma, A.L.; Sahu, G.; Dubrovsky, L.; Sun, L.; Yue, H.; Jada, T.; Ochem, A.; Simon, G.; Bukrinsky, M.; et al. DNA dependent protein kinase (DNA-PK) enhances HIV transcription by promoting RNA polymerase II activity and recruitment of transcription machinery at HIV LTR. Oncotarget 2020, 11, 699–726. [Google Scholar] [CrossRef]
- Sonti, S.; Tyagi, K.; Pande, A.; Daniel, R.; Sharma, A.L.; Tyagi, M. Crossroads of Drug Abuse and HIV Infection: Neurotoxicity and CNS Reservoir. Vaccines 2022, 10, 202. [Google Scholar] [CrossRef]
- Sonti, S.; Sharma, A.L.; Tyagi, M. HIV-1 persistence in the CNS: Mechanisms of latency, pathogenesis and an update on eradication strategies. Virus Res. 2021, 303, 198523. [Google Scholar] [CrossRef]
- Kim, Y.K.; Bourgeois, C.F.; Pearson, R.; Tyagi, M.; West, M.J.; Wong, J.; Wu, S.Y.; Chiang, C.M.; Karn, J. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J. 2006, 25, 3596–3604. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.P.; Akoulitchev, S.; Reinberg, D. Promoter-proximal stalling results from the inability to recruit transcription factor IIH to the transcription complex and is a regulated event. Proc. Natl. Acad. Sci. USA 1998, 95, 9767–9772. [Google Scholar] [CrossRef] [PubMed]
- Pantaleo, G.; Graziosi, C.; Butini, L.; Pizzo, P.A.; Schnittman, S.M.; Kotler, D.P.; Fauci, A.S. Lymphoid organs function as major reservoirs for human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 1991, 88, 9838–9842. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S. Immunopathogenesis of HIV infection. J. Acquir. Immune Defic. Syndr. 1993, 6, 655–662. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Poli, G.; Pantaleo, G.; Fauci, A.S. Immunopathogenesis of human immunodeficiency virus infection. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 1993, 17 (Suppl. S1), S224–S229. [Google Scholar]
- Fauci, A.S. Multifactorial nature of human immunodeficiency virus disease: Implications for therapy. Science 1993, 262, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Pantaleo, G.; Graziosi, C.; Demarest, J.F.; Butini, L.; Montroni, M.; Fox, C.H.; Orenstein, J.M.; Kotler, D.P.; Fauci, A.S. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature 1993, 362, 355–358. [Google Scholar] [CrossRef]
- Pantaleo, G.; Graziosi, C.; Fauci, A.S. The role of lymphoid organs in the immunopathogenesis of HIV infection. AIDS 1993, 7 (Suppl. 1), S19–S23. [Google Scholar] [CrossRef]
- Weber, J. The pathogenesis of HIV-1 infection. Br. Med. Bull. 2001, 58, 61–72. [Google Scholar] [CrossRef]
- Mosmann, T.R.; Cherwinski, H.; Bond, M.W.; Giedlin, M.A.; Coffman, R.L. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986, 136, 2348–2357. [Google Scholar] [CrossRef]
- Mosmann, T.R.; Coffman, R.L. TH1 and TH2 cells: Different patterns of lymphokine secretion lead to different functional properties. Annu. Rev. Immunol. 1989, 7, 145–173. [Google Scholar] [CrossRef] [PubMed]
- Romagnani, S. Biology of human TH1 and TH2 cells. J. Clin. Immunol. 1995, 15, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Fishman, M.A.; Perelson, A.S. Th1/Th2 cross regulation. J. Theor. Biol. 1994, 170, 25–56. [Google Scholar] [CrossRef]
- Orciani, M.; Campanati, A.; Caffarini, M.; Ganzetti, G.; Consales, V.; Lucarini, G.; Offidani, A.; Di Primio, R. T helper (Th)1, Th17 and Th2 imbalance in mesenchymal stem cells of adult patients with atopic dermatitis: At the origin of the problem. Br. J. Dermatol. 2017, 176, 1569–1576. [Google Scholar] [CrossRef] [PubMed]
- Pinto, R.A.; Arredondo, S.M.; Bono, M.R.; Gaggero, A.A.; Diaz, P.V. T helper 1/T helper 2 cytokine imbalance in respiratory syncytial virus infection is associated with increased endogenous plasma cortisol. Pediatrics 2006, 117, e878–e886. [Google Scholar] [CrossRef] [PubMed]
- Jenner, R.G.; Townsend, M.J.; Jackson, I.; Sun, K.; Bouwman, R.D.; Young, R.A.; Glimcher, L.H.; Lord, G.M. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc. Natl. Acad. Sci. USA 2009, 106, 17876–17881. [Google Scholar] [CrossRef] [PubMed]
- Hertweck, A.; Vila de Mucha, M.; Barber, P.R.; Dagil, R.; Porter, H.; Ramos, A.; Lord, G.M.; Jenner, R.G. The TH1 cell lineage-determining transcription factor T-bet suppresses TH2 gene expression by redistributing GATA3 away from TH2 genes. Nucleic Acids Res. 2022, 50, 4557–4573. [Google Scholar] [CrossRef]
- Djuretic, I.M.; Levanon, D.; Negreanu, V.; Groner, Y.; Rao, A.; Ansel, K.M. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 2007, 8, 145–153. [Google Scholar] [CrossRef]
- Clerici, M.; Shearer, G.M. A TH1→ TH2 switch is a critical step in the etiology of HIV infection. Immunol. Today 1993, 14, 107–111. [Google Scholar] [CrossRef]
- Becker, Y. The changes in the T helper 1 (Th1) and T helper 2 (Th2) cytokine balance during HIV-1 infection are indicative of an allergic response to viral proteins that may be reversed by Th2 cytokine inhibitors and immune response modifiers—A review and hypothesis. Virus Genes 2004, 28, 5–18. [Google Scholar] [CrossRef]
- Gorry, P.R.; Ancuta, P. Coreceptors and HIV-1 pathogenesis. Curr. HIV/AIDS Rep. 2011, 8, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Connor, R.I.; Sheridan, K.E.; Ceradini, D.; Choe, S.; Landau, N.R. Change in coreceptor use correlates with disease progression in HIV-1—Infected individuals. J. Exp. Med. 1997, 185, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Di Marzio, P.; Tse, J.; Landau, N.R. Chemokine receptor regulation and HIV type 1 tropism in monocyte-macrophages. AIDS Res. Hum. Retroviruses 1998, 14, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Tersmette, M.; de Goede, R.E.; Al, B.J.; Winkel, I.N.; Gruters, R.A.; Cuypers, H.T.; Huisman, H.G.; Miedema, F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: Frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J. Virol. 1988, 62, 2026–2032. [Google Scholar] [CrossRef]
- Tersmette, M.; Gruters, R.A.; de Wolf, F.; de Goede, R.E.; Lange, J.M.; Schellekens, P.T.; Goudsmit, J.; Huisman, H.G.; Miedema, F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: Studies on sequential HIV isolates. J. Virol. 1989, 63, 2118–2125. [Google Scholar] [CrossRef]
- Connor, R.I.; Ho, D.D. Human immunodeficiency virus type 1 variants with increased replicative capacity develop during the asymptomatic stage before disease progression. J. Virol. 1994, 68, 4400–4408. [Google Scholar] [CrossRef]
- Richman, D.D.; Bozzette, S.A. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J. Infect. Dis. 1994, 169, 968–974. [Google Scholar] [CrossRef]
- Syrbe, U.; Siveke, J.; Hamann, A. Th1/Th2 subsets: Distinct differences in homing and chemokine receptor expression? Springer Semin. Immunopathol. 1999, 21, 263–285. [Google Scholar] [CrossRef]
- Sallusto, F.; Lanzavecchia, A.; Mackay, C.R. Chemokines and chemokine receptors in T-cell priming and Th1/Th2-mediated responses. Immunol. Today 1998, 19, 568–574. [Google Scholar] [CrossRef]
- Orlova-Fink, N.; Chowdhury, F.Z.; Xiaoming, S.; Harrington, S.; Rosenberg, E.S.; Lichterfeld, M. Preferential susceptibility of Th9 and Th2 CD4 T cells to X4-tropic HIV-1 infection. AIDS 2017, 31, 2211. [Google Scholar] [CrossRef]
- Romagnani, S.; Maggi, E.; Del Prete, G. An alternative view of the Th1/Th2 switch hypothesis in HIV infection. AIDS Res. Hum. Retroviruses 1994, 10, iii. [Google Scholar] [CrossRef] [PubMed]
- Abrahem, R.; Chiang, E.; Haquang, J.; Nham, A.; Ting, Y.-S.; Venketaraman, V. The Role of Dendritic Cells in TB and HIV Infection. J. Clin. Med. 2020, 9, 2661. [Google Scholar] [CrossRef] [PubMed]
- Yandrapally, S.; Agarwal, A.; Chatterjee, A.; Sarkar, S.; Mohareer, K.; Banerjee, S. Mycobacterium tuberculosis EspR modulates Th1-Th2 shift by transcriptionally regulating IL-4, steering increased mycobacterial persistence and HIV propagation during co-infection. Front. Immunol. 2023, 14, 1276817. [Google Scholar] [CrossRef]
- Rodrigues, D.B.R.; Correia, D.; Marra, M.D.; Giraldo, L.E.R.; Lages-Silva, E.; Silva-Vergara, M.L.; Barata, C.H.; Rodrigues Junior, V. Cytokine serum levels in patients infected by human immunodeficiency virus with and without Trypanosoma cruzi coinfection. Rev. Soc. Bras. Med. Trop. 2005, 38, 483–487. [Google Scholar] [CrossRef] [PubMed]
- Obuku, A.E.; Asiki, G.; Abaasa, A.; Ssonko, I.; Harari, A.; van Dam, G.J.; Corstjens, P.L.; Joloba, M.; Ding, S.; Mpendo, J. Effect of Schistosoma mansoni infection on innate and HIV-1-specific T-cell immune responses in HIV-1-infected Ugandan fisher folk. AIDS Res. Hum. Retroviruses 2016, 32, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Hokello, J.; Sharma, A.L.; Dimri, M.; Tyagi, M. Insights into the HIV Latency and the Role of Cytokines. Pathogens 2019, 8, 137. [Google Scholar] [CrossRef] [PubMed]
- Eholié, S.P.; Badje, A.; Kouame, G.M.; N’takpe, J.-B.; Moh, R.; Danel, C.; Anglaret, X. Antiretroviral treatment regardless of CD4 count: The universal answer to a contextual question. AIDS Res. Ther. 2016, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Harrington, M.; Carpenter, C.C. Hit HIV-1 hard, but only when necessary. Lancet 2000, 355, 2147–2152. [Google Scholar] [CrossRef]
- Grinsztejn, B.; Hosseinipour, M.C.; Ribaudo, H.J.; Swindells, S.; Eron, J.; Chen, Y.Q.; Wang, L.; Ou, S.-S.; Anderson, M.; McCauley, M. Effects of early versus delayed initiation of antiretroviral treatment on clinical outcomes of HIV-1 infection: Results from the phase 3 HPTN 052 randomised controlled trial. Lancet Infect. Dis. 2014, 14, 281–290. [Google Scholar] [CrossRef]
- Vujkovic-Cvijin, I.; Dunham, R.M.; Iwai, S.; Maher, M.C.; Albright, R.G.; Broadhurst, M.J.; Hernandez, R.D.; Lederman, M.M.; Huang, Y.; Somsouk, M. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 2013, 5, ra191–ra193. [Google Scholar] [CrossRef]
- Xu, H.; Wang, X.; Veazey, R.S. Mucosal immunology of HIV infection. Immunol. Rev. 2013, 254, 10–33. [Google Scholar] [CrossRef] [PubMed]
- Zevin, A.S.; McKinnon, L.; Burgener, A.; Klatt, N.R. Microbial translocation and microbiome dsybiosis in HIV-associated immune activation. Curr. Opin. HIV AIDS 2016, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, J.; Hussain, T.; Simar, D.; Palchaudhuri, R.; Abdel-Mohsen, M.; Crowe, S.M.; Mbogo, G.W.; Palmer, C.S. Inflammatory and immunometabolic consequences of gut dysfunction in HIV: Parallels with IBD and implications for reservoir persistence and non-AIDS comorbidities. eBioMedicine 2019, 46, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Tugizov, S. Human immunodeficiency virus-associated disruption of mucosal barriers and its role in HIV transmission and pathogenesis of HIV/AIDS disease. Tissue Barriers 2016, 4, e1159276. [Google Scholar] [CrossRef] [PubMed]
- Estes, J.D.; Harris, L.D.; Klatt, N.R.; Tabb, B.; Pittaluga, S.; Paiardini, M.; Barclay, G.R.; Smedley, J.; Pung, R.; Oliveira, K.M. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 2010, 6, e1001052. [Google Scholar] [CrossRef] [PubMed]
- Nazli, A.; Chan, O.; Dobson-Belaire, W.N.; Ouellet, M.; Tremblay, M.J.; Gray-Owen, S.D.; Arsenault, A.L.; Kaushic, C. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation. PLoS Pathog. 2010, 6, e1000852. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, G.; Tincati, C.; Silvestri, G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin. Microbiol. Rev. 2013, 26, 2–18. [Google Scholar] [CrossRef]
- Rajasuriar, R.; Wright, E.; Lewin, S.R. Impact of antiretroviral therapy (ART) timing on chronic immune activation/inflammation and end-organ damage. Curr. Opin. HIV AIDS 2015, 10, 35. [Google Scholar] [CrossRef]
- Krebs, S.J.; Ananworanich, J. Immune activation during acute HIV infection and the impact of early antiretroviral therapy. Curr. Opin. HIV AIDS 2016, 11, 163–172. [Google Scholar] [CrossRef]
- Planchais, C.; Hocqueloux, L.; Ibanez, C.; Gallien, S.; Copie, C.; Surenaud, M.; Kök, A.; Lorin, V.; Fusaro, M.; Delfau-Larue, M.-H. Early antiretroviral therapy preserves functional follicular helper T and HIV-specific B cells in the gut mucosa of HIV-1–infected individuals. J. Immunol. 2018, 200, 3519–3529. [Google Scholar] [CrossRef]
- Kök, A.; Hocqueloux, L.; Hocini, H.; Carrière, M.; Lefrou, L.; Guguin, A.; Tisserand, P.; Bonnabau, H.; Avettand-Fenoel, V.; Prazuck, T. Early initiation of combined antiretroviral therapy preserves immune function in the gut of HIV-infected patients. Mucosal Immunol. 2015, 8, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Brenchley, J.M.; Price, D.A.; Schacker, T.W.; Asher, T.E.; Silvestri, G.; Rao, S.; Kazzaz, Z.; Bornstein, E.; Lambotte, O.; Altmann, D.; et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 2006, 12, 1365–1371. [Google Scholar] [CrossRef] [PubMed]
- Planchais, C.; Molinos-Albert, L.M.; Rosenbaum, P.; Hieu, T.; Kanyavuz, A.; Clermont, D.; Prazuck, T.; Lefrou, L.; Dimitrov, J.D.; Hüe, S. HIV-1 treatment timing shapes the human intestinal memory B-cell repertoire to commensal bacteria. Nat. Commun. 2023, 14, 6326. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | HIV-1 | HIV-2 | References |
|---|---|---|---|
| Origin | Chimpanzee | Sooty mangabey | [10,11,12] |
| Strain | Predominant | Rare | [13,14] |
| Pathogenicity and infectivity | High | Low | [4,15] |
| Viral replicative fitness | 100-fold more fit | Less fit | |
| Genetic diversity | Diverse | Low | [16,17,18] |
| Transmission | High | Low | [19,20,21] |
| MTCT | High | Rare | [20,21] |
| Prevalence | Worldwide | West Africa | |
| Blood plasma viral load | High | Low | [2] |
| Mortality rate | High (87%) | Average (52%) | [22] |
| CD4 count at time of AIDS | Below 100 cells/µL | Above 100 cells/µL | [22] |
| Type | Gene | HIV Protein | Main Function | Refs. |
|---|---|---|---|---|
| Structural | env | gp120 gp41 | Binds to HIV receptor, CD4 molecule and CCR5 and CXCR4 co-receptors of host Promotes virion fusion to host cell | [30] [31] |
| gag | P24 P17 P7 p6 | Capsid protein Matrix protein Capsid protein Capsid protein | [32] [33] [33] | |
| pol | RT IN PR | Converts viral RNA into dsDNA Integration of dsDNA into host Cleavage of gag and pol precursors | [34] [35] | |
| Regulatory | tat | Tat | Promotes proviral DNA transcription | [36,37,38] |
| rev | Rev | Exports unspliced viral RNA to cytoplasm | ||
| nef | Nef | Downregulation of CD4, MHC-1 and other receptors | [39] | |
| Accessory | vpr | Vpr | Regulation of PIC nuclear import | [40] |
| vif | Vif | Promotes virion infectivity | ||
| vpu | vpu | Intracellular CD4 degradation | [41] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hokello, J.; Tyagi, K.; Owor, R.O.; Sharma, A.L.; Bhushan, A.; Daniel, R.; Tyagi, M. New Insights into HIV Life Cycle, Th1/Th2 Shift during HIV Infection and Preferential Virus Infection of Th2 Cells: Implications of Early HIV Treatment Initiation and Care. Life 2024, 14, 104. https://doi.org/10.3390/life14010104
Hokello J, Tyagi K, Owor RO, Sharma AL, Bhushan A, Daniel R, Tyagi M. New Insights into HIV Life Cycle, Th1/Th2 Shift during HIV Infection and Preferential Virus Infection of Th2 Cells: Implications of Early HIV Treatment Initiation and Care. Life. 2024; 14(1):104. https://doi.org/10.3390/life14010104
Chicago/Turabian StyleHokello, Joseph, Kratika Tyagi, Richard Oriko Owor, Adhikarimayum Lakhikumar Sharma, Alok Bhushan, Rene Daniel, and Mudit Tyagi. 2024. "New Insights into HIV Life Cycle, Th1/Th2 Shift during HIV Infection and Preferential Virus Infection of Th2 Cells: Implications of Early HIV Treatment Initiation and Care" Life 14, no. 1: 104. https://doi.org/10.3390/life14010104
APA StyleHokello, J., Tyagi, K., Owor, R. O., Sharma, A. L., Bhushan, A., Daniel, R., & Tyagi, M. (2024). New Insights into HIV Life Cycle, Th1/Th2 Shift during HIV Infection and Preferential Virus Infection of Th2 Cells: Implications of Early HIV Treatment Initiation and Care. Life, 14(1), 104. https://doi.org/10.3390/life14010104










