The Intricate Interplay between Cancer Stem Cells and Oncogenic miRNAs in Breast Cancer Progression and Metastasis
Abstract
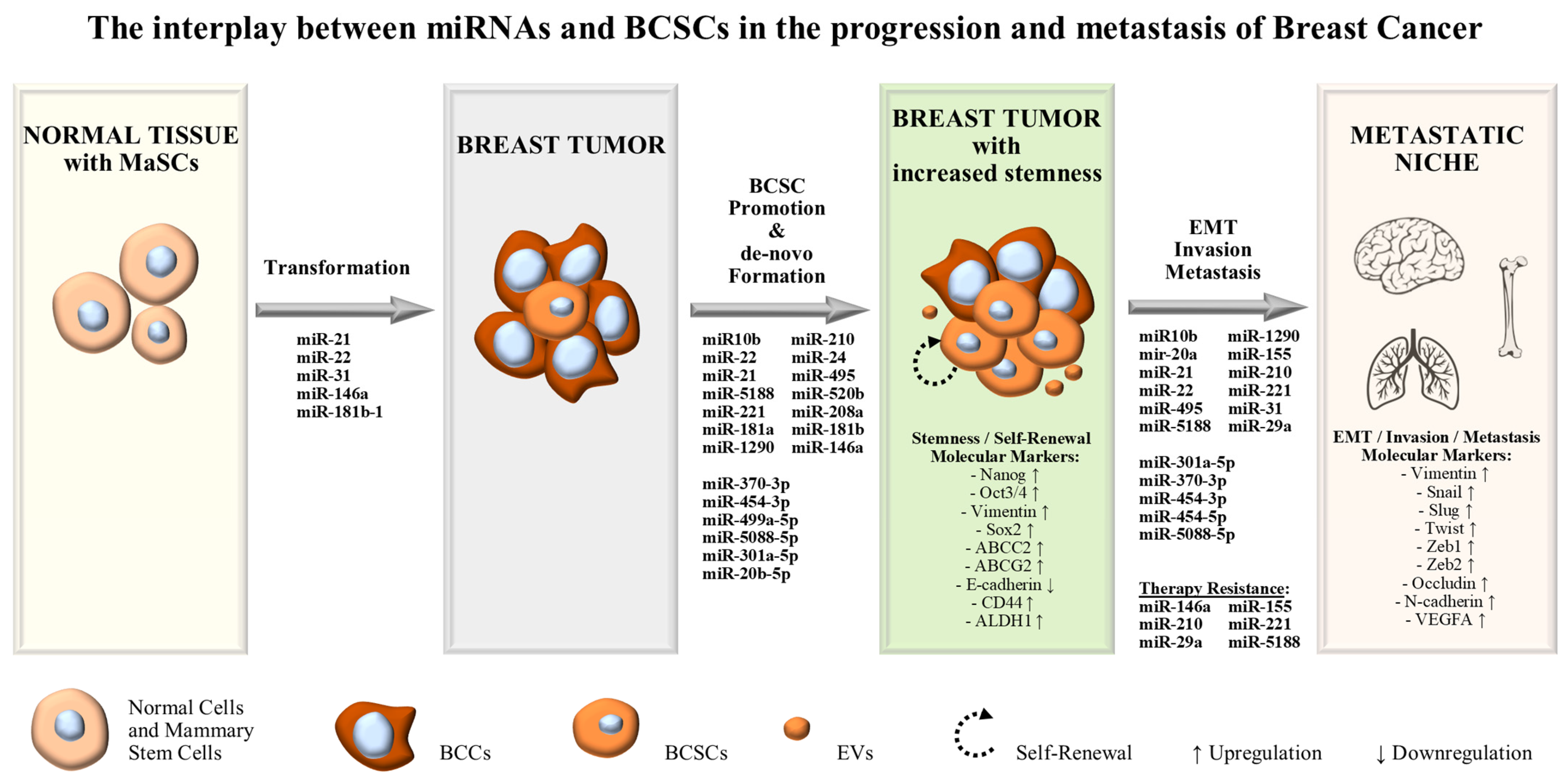
1. Introduction
2. MiRNAs Exerting Oncogenic Functions in BCSCs and Stem-Like BCCs
2.1. miR-155
2.2. miR-208a
2.3. miR-210
2.4. miR-24
2.5. miR-20a and miR-20b
2.6. miR-10b
2.7. miR-21
2.8. miR-495
2.9. miR-181a and miR-181b
2.10. miR-454-3p and -5p
2.11. miR-5188
2.12. miR-5088-5p
2.13. miR-370-3p
2.14. miR-499a-5p
2.15. miR-221
2.16. mir-301a-5p
2.17. miR-520b
2.18. miR-31
2.19. mir-1290
2.20. miR-29a
2.21. miR-22
2.22. miR-146a
3. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S.; et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast 2022, 66, 15–23. [Google Scholar] [CrossRef]
- Tomasetti, C.; Li, L.; Vogelstein, B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 2017, 355, 1330–1334. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhu, W.; Thompson, P.; Hannun, Y.A. Evaluating intrinsic and non-intrinsic cancer risk factors. Nat. Commun. 2018, 9, 3490. [Google Scholar] [CrossRef]
- Zhang, X.; Powell, K.; Li, L. Breast Cancer Stem Cells: Biomarkers, Identification and Isolation Methods, Regulating Mechanisms, Cellular Origin, and Beyond. Cancers 2020, 12, 3765. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar] [PubMed]
- Liu, Z.; Zhang, X.-S.; Zhang, S. Breast tumor subgroups reveal diverse clinical prognostic power. Sci. Rep. 2014, 4, 4002. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Shan, N.L.; Shin, Y.; Yang, G.; Furmanski, P.; Suh, N. Breast cancer stem cells: A review of their characteristics and the agents that affect them. Mol. Carcinog. 2021, 60, 73–100. [Google Scholar] [CrossRef]
- Velasco-Velázquez, M.A.; Popov, V.M.; Lisanti, M.P.; Pestell, R.G. The Role of Breast Cancer Stem Cells in Metastasis and Therapeutic Implications. Am. J. Pathol. 2011, 179, 2–11. [Google Scholar] [CrossRef]
- Ponti, D.; Costa, A.; Zaffaroni, N.; Pratesi, G.; Petrangolini, G.; Coradini, D.; Pilotti, S.; Pierotti, M.A.; Daidone, M.G. Isolation and In vitro Propagation of Tumorigenic Breast Cancer Cells with Stem/Progenitor Cell Properties. Cancer Res. 2005, 65, 5506–5511. [Google Scholar] [CrossRef]
- Wang, R.; Lv, Q.; Meng, W.; Tan, Q.; Zhang, S.; Mo, X.; Yang, X. Comparison of mammosphere formation from breast cancer cell lines and primary breast tumors. J. Thorac. Dis. 2014, 6, 829–837. [Google Scholar] [PubMed]
- Bhandary, L.; Bailey, P.C.; Chang, K.T.; Underwood, K.F.; Lee, C.J.; Whipple, R.A.; Jewell, C.M.; Ory, E.; Thompson, K.N.; Ju, J.A.; et al. Lipid tethering of breast tumor cells reduces cell aggregation during mammosphere formation. Sci. Rep. 2021, 11, 3214. [Google Scholar] [CrossRef] [PubMed]
- Yousefnia, S.; Ghaedi, K.; Forootan, F.S.; Esfahani, M.H.N. Characterization of the stemness potency of mammospheres isolated from the breast cancer cell lines. Tumor Biol. 2019, 41, 1010428319869101. [Google Scholar] [CrossRef]
- Chalfie, M.; Horvitz, H.R.; Sulston, J.E. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 1981, 24, 59–69. [Google Scholar] [CrossRef]
- Ambros, V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell 1989, 57, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef]
- Altuvia, Y.; Landgraf, P.; Lithwick, G.; Elefant, N.; Pfeffer, S.; Aravin, A.; Brownstein, M.J.; Tuschl, T.; Margalit, H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005, 33, 2697–2706. [Google Scholar] [CrossRef]
- Bentwich, I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005, 579, 5904–5910. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Baumann, V.; Winkler, J. miRNA-based therapies: Strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med. Chem. 2014, 6, 1967–1984. [Google Scholar] [CrossRef]
- Pan, G.; Liu, Y.; Shang, L.; Zhou, F.; Yang, S. EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun. 2021, 41, 199–217. [Google Scholar] [CrossRef]
- Majumder, M.; Dunn, L.; Liu, L.; Hasan, A.; Vincent, K.; Brackstone, M.; Hess, D.; Lala, P.K. COX-2 induces oncogenic micro RNA miR655 in human breast cancer. Sci. Rep. 2018, 8, 327. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Yu, Y.; Zhu, M.; Jing, W.; Yu, M.; Chai, H.; Liang, C.; Tu, J. Inhibition of miR-155, a therapeutic target for breast cancer, prevented in cancer stem cell formation. Cancer Biomark. 2018, 21, 383–392. [Google Scholar] [CrossRef]
- Santos, J.C.; da Silva Lima, N.; Sarian, L.O.; Matheu, A.; Ribeiro, M.L.; Derchain, S.F.M. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci. Rep. 2018, 8, 829. [Google Scholar] [CrossRef]
- Sun, X.; Jiang, S.; Liu, J.; Wang, H.; Zhang, Y.; Tang, S.C.; Wang, J.; Du, N.; Xu, C.; Wang, C.; et al. MiR-208a stimulates the cocktail of SOX2 and beta-catenin to inhibit the let-7 induction of self-renewal repression of breast cancer stem cells and formed miR208a/let-7 feedback loop via LIN28 and DICER1. Oncotarget 2015, 6, 32944–32954. [Google Scholar] [CrossRef]
- Camps, C.; Buffa, F.M.; Colella, S.; Moore, J.; Sotiriou, C.; Sheldon, H.; Harris, A.L.; Gleadle, J.M.; Ragoussis, J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin. Cancer Res. 2008, 14, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Griñán-Lisón, C.; Olivares-Urbano, M.A.; Jiménez, G.; López-Ruiz, E.; Del Val, C.; Morata-Tarifa, C.; Entrena, J.M.; González-Ramírez, A.R.; Boulaiz, H.; Herrera, M.Z.; et al. miRNAs as radio-response biomarkers for breast cancer stem cells. Mol. Oncol. 2020, 14, 556–570. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Yang, Z.; Zhu, Q.; Wu, Y.; Sun, K.; Alahdal, M.; Zhang, Y.; Xing, Y.; Shen, Y.; Xia, T.; et al. Up-regulation of miR-210 induced by a hypoxic microenvironment promotes breast cancer stem cells metastasis, proliferation, and self-renewal by targeting E-cadherin. FASEB J. 2018, 32, fj201801013R. [Google Scholar] [CrossRef] [PubMed]
- Khodadadi-Jamayran, A.; Akgol-Oksuz, B.; Afanasyeva, Y.; Heguy, A.; Thompson, M.; Ray, K.; Giro-Perafita, A.; Sánchez, I.; Wu, X.; Tripathy, D.; et al. Prognostic role of elevated mir-24-3p in breast cancer and its association with the metastatic process. Oncotarget 2018, 9, 12868–12878. [Google Scholar] [CrossRef] [PubMed]
- Roscigno, G.; Puoti, I.; Giordano, I.; Donnarumma, E.; Russo, V.; Affinito, A.; Adamo, A.; Quintavalle, C.; Todaro, M.; Vivanco, M.D.; et al. MiR-24 induces chemotherapy resistance and hypoxic advantage in breast cancer. Oncotarget 2017, 8, 19507–19521. [Google Scholar] [CrossRef] [PubMed]
- Nunes, D.N.; Dias-Neto, E.; Cardó-Vila, M.; Edwards, J.K.; Dobroff, A.S.; Giordano, R.J.; Mandelin, J.; Brentani, H.P.; Hasselgren, C.; Yao, V.J.; et al. Synchronous down-modulation of miR-17 family members is an early causative event in the retinal angiogenic switch. Proc. Natl. Acad. Sci. USA 2015, 112, 3770–3775. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-Y.; Zhang, Y.; Zhang, W.-H.; Jia, S.; Kang, Y.; Zhu, X.-Y. Differential Distribution of miR-20a and miR-20b may Underly Metastatic Heterogeneity of Breast Cancers. Asian Pac. J. Cancer Prev. 2012, 13, 1901–1906. [Google Scholar] [CrossRef]
- Guo, L.; Zhu, Y.; Li, L.; Zhou, S.; Yin, G.; Yu, G.; Cui, H. Breast cancer cell-derived exosomal miR-20a-5p promotes the proliferation and differentiation of osteoclasts by targeting SRCIN1. Cancer Med. 2019, 8, 5687–5701. [Google Scholar] [CrossRef]
- Wang, B.; Wang, Q.; Wang, Z.; Jiang, J.; Yu, S.-C.; Ping, Y.-F.; Yang, J.; Xu, S.-L.; Ye, X.-Z.; Xu, C.; et al. Metastatic consequences of immune escape from NK cell cytotoxicity by human breast cancer stem cells. Cancer Res. 2014, 74, 5746–5757. [Google Scholar] [CrossRef]
- Xia, L.; Li, F.; Qiu, J.; Feng, Z.; Xu, Z.; Chen, Z.; Sun, J. Oncogenic miR-20b-5p contributes to malignant behaviors of breast cancer stem cells by bidirectionally regulating CCND1 and E2F1. BMC Cancer 2020, 20, 949. [Google Scholar] [CrossRef]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef]
- Bahena-Ocampo, I.; Espinosa, M.; Ceballos-Cancino, G.; Lizarraga, F.; Campos-Arroyo, D.; Schwarz, A.; Maldonado, V.; Melendez-Zajgla, J.; Garcia-Lopez, P. miR-10b expression in breast cancer stem cells supports self-renewal through negative PTEN regulation and sustained AKT activation. EMBO Rep. 2016, 17, 648–658. [Google Scholar] [CrossRef]
- Han, M.; Liu, M.; Wang, Y.; Mo, Z.; Bi, X.; Liu, Z.; Fan, Y.; Chen, X.; Wu, C. Re-expression of miR-21 contributes to migration and invasion by inducing epithelial-mesenchymal transition consistent with cancer stem cell characteristics in MCF-7 cells. Mol. Cell Biochem. 2012, 363, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Wang, Y.; Liu, M.; Bi, X.; Bao, J.; Zeng, N.; Zhu, Z.; Mo, Z.; Wu, C.; Chen, X. MiR-21 regulates epithelial-mesenchymal transition phenotype and hypoxia-inducible factor-1α expression in third-sphere forming breast cancer stem cell-like cells. Cancer Sci. 2012, 103, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Bourguignon, L.Y.W. Hyaluronan-CD44 interaction promotes c-Jun signaling and miRNA21 expression leading to Bcl-2 expression and chemoresistance in breast cancer cells. Mol. Cancer 2014, 13, 52. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Jaeger, S.A.; Hirsch, H.A.; Bulyk, M.L.; Struhl, K. STAT3 Activation of miR-21 and miR-181b-1 via PTEN and CYLD Are Part of the Epigenetic Switch Linking Inflammation to Cancer. Mol. Cell 2010, 39, 493–506. [Google Scholar] [CrossRef]
- Chi, L.H.; Cross, R.S.N.; Redvers, R.P.; Davis, M.; Hediyeh-Zadeh, S.; Mathivanan, S.; Samuel, M.; Lucas, E.C.; Mouchemore, K.; Gregory, P.A.; et al. MicroRNA-21 is immunosuppressive and pro-metastatic via separate mechanisms. Oncogenesis 2022, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Cao, X.; Zhang, Y.; Xu, H.; Zhang, R.; Wu, Y.; Lu, P.; Jin, F. Co-expression of Oct-4 and Nestin in human breast cancers. Mol. Biol. Rep. 2012, 39, 5875–5881. [Google Scholar] [CrossRef]
- Chen, H.; Wang, X.; Bai, J.; He, A. Expression, regulation and function of miR-495 in healthy and tumor tissues. Oncol. Lett. 2017, 13, 2021–2026. [Google Scholar] [CrossRef]
- Hwang-Verslues, W.W.; Chang, P.-H.; Wei, P.-C.; Yang, C.-Y.; Huang, C.-K.; Kuo, W.-H.; Shew, J.-Y.; Chang, K.-J.; Lee, E.Y.-H.P.; Lee, W.-H. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene 2011, 30, 2463–2474. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Tsuyada, A.; Ren, X.; Wu, X.; Stubblefield, K.; Rankin-Gee, E.K.; Wang, S.E. Transforming growth factor-beta regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene 2011, 30, 1470–1480. [Google Scholar] [CrossRef]
- Bisso, A.; Faleschini, M.; Zampa, F.; Capaci, V.; De Santa, J.; Santarpia, L.; Piazza, S.; Cappelletti, V.; Daidone, M.G.; Agami, R.; et al. Oncogenic miR-181a/b affect the DNA damage response in aggressive breast cancer. Cell Cycle 2013, 12, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.P.; Yu, Y.Z.; Zhao, H.; Xie, L.J.; Wang, Q.T.; Wang, Y.; Mu, Q. MicroRNA-454-5p promotes breast cancer progression by inducing epithelial-mesenchymal transition via targeting the FoxJ2/E-cadherin axis. Oncol. Rep. 2021, 46, 127. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Chen, H.; Song, J.; Chen, X.; Lin, C.; Zhang, X.; Hou, N.; Pan, J.; Zhou, Z.; Wang, L.; et al. MiR-454-3p-Mediated Wnt/beta-catenin Signaling Antagonists Suppression Promotes Breast Cancer Metastasis. Theranostics 2019, 9, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Lin, X.; Bu, J.; Lin, Z.; Chen, Y.; Qiu, Y.; Mo, H.; Tang, Y.; Fang, W.; Wu, Z. Timeless-Stimulated miR-5188-FOXO1/beta-Catenin-c-Jun Feedback Loop Promotes Stemness via Ubiquitination of beta-Catenin in Breast Cancer. Mol. Ther. 2020, 28, 313–327. [Google Scholar] [CrossRef]
- Xie, X.; Kaoud, T.S.; Edupuganti, R.; Zhang, T.; Kogawa, T.; Zhao, Y.; Chauhan, G.B.; Giannoukos, D.N.; Qi, Y.; Tripathy, D.; et al. c-Jun N-terminal kinase promotes stem cell phenotype in triple-negative breast cancer through upregulation of Notch1 via activation of c-Jun. Oncogene 2017, 36, 2599–2608. [Google Scholar] [CrossRef]
- Seok, H.J.; Choi, Y.E.; Choi, J.Y.; Yi, J.M.; Kim, E.J.; Choi, M.Y.; Lee, S.-J.; Bae, I.H. Novel miR-5088-5p promotes malignancy of breast cancer by inhibiting DBC2. Mol. Ther. Nucleic Acids 2021, 25, 127–142. [Google Scholar] [CrossRef]
- Mao, J.; Wang, L.; Wu, J.; Wang, Y.; Wen, H.; Zhu, X.; Wang, B.; Yang, H. miR-370-3p as a Novel Biomarker Promotes Breast Cancer Progression by Targeting FBLN5. Stem Cells Int. 2021, 2021, 4649890. [Google Scholar] [CrossRef]
- Ren, Z.; Lv, M.; Yu, Q.; Bao, J.; Lou, K.; Li, X. MicroRNA-370-3p shuttled by breast cancer cell-derived extracellular vesicles induces fibroblast activation through the CYLD/Nf-κB axis to promote breast cancer progression. FASEB J. 2021, 35, e21383. [Google Scholar] [CrossRef]
- Mandal, S.; Gamit, N.; Varier, L.; Dharmarajan, A.; Warrier, S. Inhibition of breast cancer stem-like cells by a triterpenoid, ursolic acid, via activation of Wnt antagonist, sFRP4 and suppression of miRNA-499a-5p. Life Sci. 2021, 265, 118854. [Google Scholar] [CrossRef]
- Ke, J.; Zhao, Z.; Hong, S.-H.; Bai, S.; He, Z.; Malik, F.; Xu, J.; Zhou, L.; Chen, W.; Martin-Trevino, R.; et al. Role of microRNA221 in regulating normal mammary epithelial hierarchy and breast cancer stem-like cells. Oncotarget 2015, 6, 3709–3721. [Google Scholar] [CrossRef]
- Ye, X.; Bai, W.; Zhu, H.; Zhang, X.; Chen, Y.; Wang, L.; Yang, A.; Zhao, J.; Jia, L. MiR-221 promotes trastuzumab-resistance and metastasis in HER2-positive breast cancers by targeting PTEN. BMB Rep. 2014, 47, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Paul, S.; Singh, A.; Ghosh, A.; Roy, A.; Ansari, S.A.; Prasad, R.; Mukherjee, A.; Sen, P. Triple-negative breast cancer-derived microvesicles transfer microRNA221 to the recipient cells and thereby promote epithelial-to-mesenchymal transition. J. Biol. Chem. 2019, 294, 13681–13696. [Google Scholar] [CrossRef]
- Roscigno, G.; Quintavalle, C.; Donnarumma, E.; Puoti, I.; Lagares, A.D.; Iaboni, M.; Fiore, D.; Russo, V.; Todaro, M.; Romano, G.; et al. MiR-221 promotes stemness of breast cancer cells by targeting DNMT3b. Oncotarget 2016, 7, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Lettlova, S.; Brynychova, V.; Blecha, J.; Vrana, D.; Vondrusova, M.; Soucek, P.; Truksa, J. MiR-301a-3p Suppresses Estrogen Signaling by Directly Inhibiting ESR1 in ERalpha Positive Breast Cancer. Cell Physiol. Biochem. 2018, 46, 2601–2615. [Google Scholar] [CrossRef]
- Zheng, J.-Z.; Huang, Y.-N.; Yao, L.; Liu, Y.-R.; Liu, S.; Hu, X.; Liu, Z.-B.; Shao, Z.-M. Elevated miR-301a expression indicates a poor prognosis for breast cancer patients. Sci. Rep. 2018, 8, 2225. [Google Scholar] [CrossRef]
- Yu, H.; Li, H.; Qian, H.; Jiao, X.; Zhu, X.; Jiang, X.; Dai, G.; Huang, J. Upregulation of miR-301a correlates with poor prognosis in triple-negative breast cancer. Med. Oncol. 2014, 31, 283. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lang, T.-Y.; Zou, D.-L.; Zhou, L.; Lou, M.; Liu, J.-S.; Li, Y.-Z.; Ding, D.-Y.; Li, Y.-C.; Zhang, N.; et al. miR-520b Promotes Breast Cancer Stemness Through Hippo/YAP Signaling Pathway. OncoTargets Ther. 2019, 12, 11691–11700. [Google Scholar] [CrossRef]
- Lv, C.; Li, F.; Li, X.; Tian, Y.; Zhang, Y.; Sheng, X.; Song, Y.; Meng, Q.; Yuan, S.; Luan, L.; et al. MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nat. Commun. 2017, 8, 1036. [Google Scholar] [CrossRef] [PubMed]
- Sirkisoon, S.R.; Wong, G.L.; Aguayo, N.R.; Doheny, D.L.; Zhu, D.; Regua, A.T.; Arrigo, A.; Manore, S.G.; Wagner, C.; Thomas, A.; et al. Breast cancer extracellular vesicles-derived miR-1290 activates astrocytes in the brain metastatic microenvironment via the FOXA2-->CNTF axis to promote progression of brain metastases. Cancer Lett. 2022, 540, 215726. [Google Scholar] [CrossRef]
- Wong, G.L.; Najjar, M.; Esquenazi, Y.; Tandon, N.; Regua, A.T.; Lo, H.-W. Abstract 3769: Intracellular miR-1290 promotes breast cancer stemness in HER2-enriched and triple-negative breast cancer. Cancer Res. 2023, 83, 3769. [Google Scholar] [CrossRef]
- Chou, J.; Lin, J.H.; Brenot, A.; Kim, J.-W.; Provot, S.; Werb, Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat. Cell Biol. 2013, 15, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Singh, H.; Rajapakshe, K.; Tachibana, K.; Ganesan, N.; Pan, Y.; Gunaratne, P.H.; Coarfa, C.; Bedrosian, I. Regulation of miRNA-29c and its downstream pathways in preneoplastic progression of triple-negative breast cancer. Oncotarget 2017, 8, 19645–19660. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shi, W.; Tang, T.; Wang, Y.; Yin, X.; Chen, Y.; Zhang, Y.; Xing, Y.; Shen, Y.; Xia, T.; et al. miR-29a contributes to breast cancer cells epithelial–mesenchymal transition, migration, and invasion via down-regulating histone H4K20 trimethylation through directly targeting SUV420H2. Cell Death Dis. 2019, 10, 176. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Ding, M.; Zhang, H.; Xu, X.; Tang, J. Molecular mechanisms and clinical applications of miR-22 in regulating malignant progression in human cancer (Review). Int. J. Oncol. 2017, 50, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.K.; Zhang, Y.; Zhang, S.; Li, Y.; Tucker-Kellogg, G.; Yang, H.; Jha, S. TIP60-miR-22 axis as a prognostic marker of breast cancer progression. Oncotarget 2015, 6, 41290–41306. [Google Scholar] [CrossRef]
- Song, S.J.; Poliseno, L.; Song, M.S.; Ala, U.; Webster, K.; Ng, C.; Beringer, G.; Brikbak, N.J.; Yuan, X.; Cantley, L.C.; et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 2013, 154, 311–324. [Google Scholar] [CrossRef]
- Shimono, Y.; Zabala, M.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009, 138, 592–603. [Google Scholar] [CrossRef]
- Vares, G.; Cui, X.; Wang, B.; Nakajima, T.; Nenoi, M. Generation of Breast Cancer Stem Cells by Steroid Hormones in Irradiated Human Mammary Cell Lines. PLoS ONE 2013, 8, e77124. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Wang, D.; Wei, X. miR-22 suppresses tumorigenesis and improves radiosensitivity of breast cancer cells by targeting Sirt1. Biol. Res. 2017, 50, 27. [Google Scholar] [CrossRef]
- Pogribny, I.P.; Filkowski, J.N.; Tryndyak, V.P.; Golubov, A.; Shpyleva, S.I.; Kovalchuk, O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int. J. Cancer 2010, 127, 1785–1794. [Google Scholar] [CrossRef]
- Wang, X.; Lu, H.; Li, T.; Yu, L.; Liu, G.; Peng, X.; Zhao, J. Krüppel-like factor 8 promotes tumorigenic mammary stem cell induction by targeting miR-146a. Am. J. Cancer Res. 2013, 3, 356–373. [Google Scholar] [PubMed]
- Tordonato, C.; Marzi, M.J.; Giangreco, G.; Freddi, S.; Bonetti, P.; Tosoni, D.; Di Fiore, P.P.; Nicassio, F. miR-146 connects stem cell identity with metabolism and pharmacological resistance in breast cancer. J. Cell Biol. 2021, 220, e202009053. [Google Scholar] [CrossRef] [PubMed]
- Sin, W.C.; Lim, C.L. Breast cancer stem cells—From origins to targeted therapy. Stem Cell Investig. 2017, 4, 96. [Google Scholar] [CrossRef] [PubMed]
- Chaffer, C.L.; Marjanovic, N.D.; Lee, T.; Bell, G.; Kleer, C.G.; Reinhardt, F.; D’Alessio, A.C.; Young, R.A.; Weinberg, R.A. Poised Chromatin at the ZEB1 Promoter Enables Breast Cancer Cell Plasticity and Enhances Tumorigenicity. Cell 2013, 154, 61–74. [Google Scholar] [CrossRef]
- Lagadec, C.; Vlashi, E.; Della Donna, L.; Dekmezian, C.; Pajonk, F. Radiation-Induced Reprogramming of Breast Cancer Cells. Stem Cells 2012, 30, 833–844. [Google Scholar] [CrossRef]
- Li, Y.; Laterra, J. Cancer stem cells: Distinct entities or dynamically regulated phenotypes? Cancer Res. 2012, 72, 576–580. [Google Scholar] [CrossRef]
- Rich, J.N. Cancer stem cells: Understanding tumor hierarchy and heterogeneity. Medicine 2016, 95 (Suppl. 1), S2–S7. [Google Scholar] [CrossRef]
- Soysal, S.D.; Tzankov, A.; Muenst, S.E. Role of the Tumor Microenvironment in Breast Cancer. Pathobiology 2015, 82, 142–152. [Google Scholar] [CrossRef]
- Fouad, Y.A.; Aanei, C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017, 7, 1016–1036. [Google Scholar]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Wu, W.-S.; You, R.-I.; Cheng, C.-C.; Lee, M.-C.; Lin, T.-Y.; Hu, C.-T. Snail collaborates with EGR-1 and SP-1 to directly activate transcription of MMP 9 and ZEB1. Sci. Rep. 2017, 7, 17753. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhong, J.; Zhao, Z.; Sheng, J.; Wang, J.; Liu, J.; Cui, K.; Chang, J.; Zhao, H.; Wong, S. Epithelial derived CTGF promotes breast tumor progression via inducing EMT and collagen I fibers deposition. Oncotarget 2015, 6, 25320–25338. [Google Scholar] [CrossRef]
- Ocaña, O.H.; Córcoles, R.; Fabra, Á.; Moreno-Bueno, G.; Acloque, H.; Vega, S.; Barrallo-Gimeno, A.; Cano, A.; Nieto, M.A. Metastatic Colonization Requires the Repression of the Epithelial-Mesenchymal Transition Inducer Prrx1. Cancer Cell 2012, 22, 709–724. [Google Scholar] [CrossRef]
- Manzo, G. Similarities Between Embryo Development and Cancer Process Suggest New Strategies for Research and Therapy of Tumors: A New Point of View. Front. Cell Dev. Biol. 2019, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zeng, Z.; Wang, J.; Wu, Y.; Chen, W.; Zheng, L.; Xi, T.; Wang, A.; Lu, Y. MicroRNA-9 and breast cancer. Biomed Pharm. 2020, 122, 109687. [Google Scholar] [CrossRef]
- Luo, Q.; Wei, C.; Li, X.; Li, J.; Chen, L.; Huang, Y.; Song, H.; Li, D.; Fang, L. MicroRNA-195-5p is a potential diagnostic and therapeutic target for breast cancer. Oncol. Rep. 2014, 31, 1096–1102. [Google Scholar] [CrossRef]
- Shen, M.; Dong, C.; Ruan, X.; Yan, W.; Cao, M.; Pizzo, D.; Wu, X.; Yang, L.; Liu, L.; Ren, X.; et al. Chemotherapy-Induced Extracellular Vesicle miRNAs Promote Breast Cancer Stemness by Targeting ONECUT2. Cancer Res. 2019, 79, 3608–3621. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat. Rev. 2018, 69, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; Van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.G.; Lowery, A.J.; Miller, N.; Kerin, M.J. MicroRNA Expression Profiles and Breast Cancer Chemotherapy. Int. J. Mol. Sci. 2021, 22, 10812. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.M.; Salvatore, M.; Incoronato, M. miRNA-Based Therapeutics in Breast Cancer: A Systematic Review. Front. Oncol. 2021, 11, 668464. [Google Scholar] [CrossRef] [PubMed]
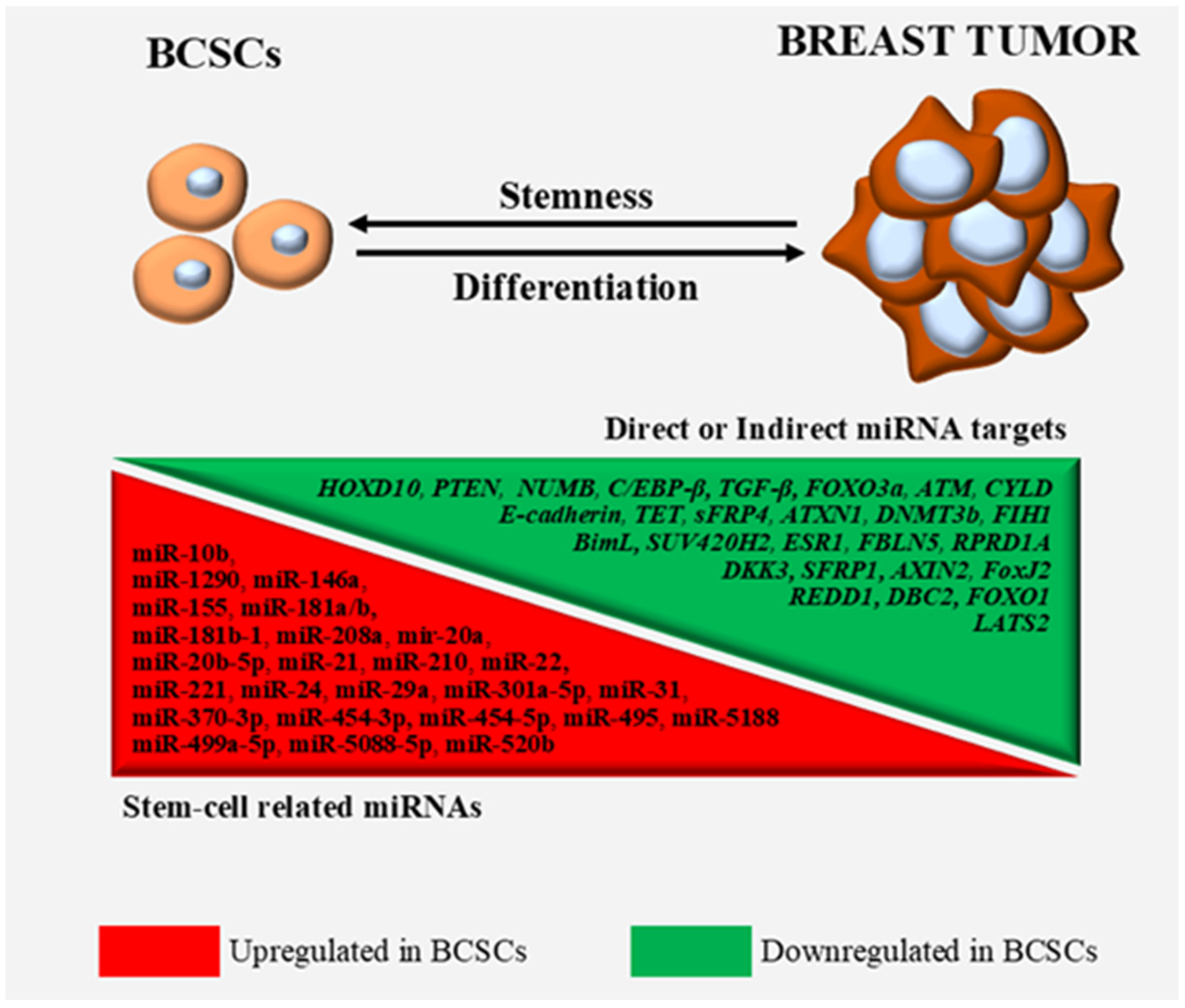
| miRNA | Effect | Targets | Implicated Cells & Cell Lines | References |
|---|---|---|---|---|
| miR-10b | Invasion, Migration | HOXD10 | MDA-MB-231, SUM149, SUM159 | [40] |
| Self-renewal, Stemness, EMT | PTEN | MCF-7, SKBR-3, MDA-MB-231 | [41] | |
| miR-1290 | Mammosphere formation, TME modulation, Brain metastasis, | - | SKBR3 | [69,70] |
| miR-146a | Malignant transformation, Pro-tumorigenic mammary stem cell formation | NUMB | MCF-10A | [81] |
| Self-renewal, Mammosphere formation, Tumor initiating cells frequency, Drug resistance | - | SUM159 | [82] | |
| miR-155 | Sphere growth, Stem-cell formation, Doxorubicinol resistance | - | MDA-MB-231 | [27] |
| Stem-like characteristics, EMT-related resistance, Migration | C/EBP-β, TGF-β, FOXO3a | MCF-7, MDA-MB-231 | [28] | |
| miR-181a/b | DDR impairment | ATM | MDA-MB-231, SUM159PT | [51] |
| Mammosphere formation, Stem-like characteristics | ATM | BT474, MDA-MB-361, MCF7 | [50] | |
| miR-181b-1 | Malignant transformation, BCSC formation | CYLD | MCF-10A | [45] |
| miR-208a | BCSC phenotype, Increased BCSC proportion | - | MDA-MB-231, ER+ ZR75-1 | [29] |
| mir-20a | BCSC resistance to NK cell cytotoxicity, Lung metastasis | MICA, MICB | Primary BCSCs | [38] |
| miR-20b-5p | BCSC proliferation, BCSCs inhibition of apoptosis | CCND1, E2F1 | T47D-BCSCs | [39] |
| miR-21 | Chemoresistance | - | MDA-MB-468 | [44] |
| BCSC-like characteristics, Colony and mammosphere formation, Motility, Invasiveness, Tumor formation capabilities | PTEN | MCF-10A | [45] | |
| Cell growth, Self-renewal, Clonogenicity, Invasion, Migration, EMT | - | MCF-7 | [42,43] | |
| miR-210 | Radioresistance | - | MCF-7, MDA-MB-231 | [31] |
| Proliferation, Self-renewal, Invasion, Migration | E-cadherin | MCF-7 | [32] | |
| miR-22 | BCSC promotion, Tumor formation, Stemness, EMT, Metastasis | TET | MCF-7, MCF-10A | [76] |
| miR-221 | BCSC promotion, EMT, | ATXN1 | MCF-7 | [60] |
| Invasion, Metastasis, Chemoresistance | PTEN | SK-BR-3 | [61] | |
| Invasion, Metastasis, Chemoresistance | PTEN | MCF-7, MDA-MB-231 | [62] | |
| BCSC phenotype, Mammosphere formation | DNMT3b | T47D | [63] | |
| miR-24 | Mammosphere formation, Stemness, Adaptive hypoxia response, Cisplatin resistance | FIH1, bimL | MCF-7, T47D, MDA-MB-231 | [34] |
| miR-29a | Invasion, Migration, EMT | SUV420H2 | MCF-7, MDA-MB-231 | [73] |
| miR-301a-5p | BCSC phenotype, EMT, Metastasis | ESR1 | MCF-7 | [64] |
| miR-31 | Tumor formation, BCSC promotion, Lung metastasis | Dkk1, Axin1, Gsk3β, Smad3, Smad4 | Mammary epithelial cells, CD24+/CD90+ BCSCs from PyVT tumors | [68] |
| miR-370-3p | Stemness, EMT | FBLN5 | MDA-MB-231 | [57] |
| Stemness, Migration, Invasion, EMT, Mammosphere formation | CYLD | MCF-7, MDA-MB-231, MDA-MB-436, Primary NFs | [58] | |
| miR-454-3p | Stemness, Metastasis | RPRD1A, DKK3, SFRP1, AXIN2 | MCF-7, MDA-MB-231 | [53] |
| miR-454-5p | EMT | FoxJ2 | MDA-MB-231 | [52] |
| miR-495 | Stemness, Invasion, Metastasis, Early relapse | E-cadherin, REDD1 | MDA-MB-231, SKBR3 | [49] |
| miR-499a-5p | BCSC phenotype, Mammosphere formation, Proliferation | sFRP4 | MDA-MB-231 | [59] |
| miR-5088-5p | BCSC phenotype, Mammosphere formation, EMT, Invasion, Metastasis | DBC2 | MCF-7, MDA-MB-231 | [56] |
| miR-5188 | Stemness, Mammosphere formation, Proliferation, Metastasis, Chemoresistance | FOXO1 | MCF-7, MDA-MB-468 | [54] |
| miR-520b | Stemness, Sphere formation, Migration | LATS2 | MCF-7, MDA-MB-231 | [67] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsintarakis, A.; Papalouka, C.; Kontarini, C.; Zoumpourlis, P.; Karakostis, K.; Adamaki, M.; Zoumpourlis, V. The Intricate Interplay between Cancer Stem Cells and Oncogenic miRNAs in Breast Cancer Progression and Metastasis. Life 2023, 13, 1361. https://doi.org/10.3390/life13061361
Tsintarakis A, Papalouka C, Kontarini C, Zoumpourlis P, Karakostis K, Adamaki M, Zoumpourlis V. The Intricate Interplay between Cancer Stem Cells and Oncogenic miRNAs in Breast Cancer Progression and Metastasis. Life. 2023; 13(6):1361. https://doi.org/10.3390/life13061361
Chicago/Turabian StyleTsintarakis, Antonis, Chara Papalouka, Christina Kontarini, Panagiotis Zoumpourlis, Konstantinos Karakostis, Maria Adamaki, and Vassilis Zoumpourlis. 2023. "The Intricate Interplay between Cancer Stem Cells and Oncogenic miRNAs in Breast Cancer Progression and Metastasis" Life 13, no. 6: 1361. https://doi.org/10.3390/life13061361
APA StyleTsintarakis, A., Papalouka, C., Kontarini, C., Zoumpourlis, P., Karakostis, K., Adamaki, M., & Zoumpourlis, V. (2023). The Intricate Interplay between Cancer Stem Cells and Oncogenic miRNAs in Breast Cancer Progression and Metastasis. Life, 13(6), 1361. https://doi.org/10.3390/life13061361






