Artificial Intelligence in Cardiovascular CT and MR Imaging
Abstract
1. Introduction
2. AI in Coronary Computed Tomography Angiography
2.1. Calcium Score
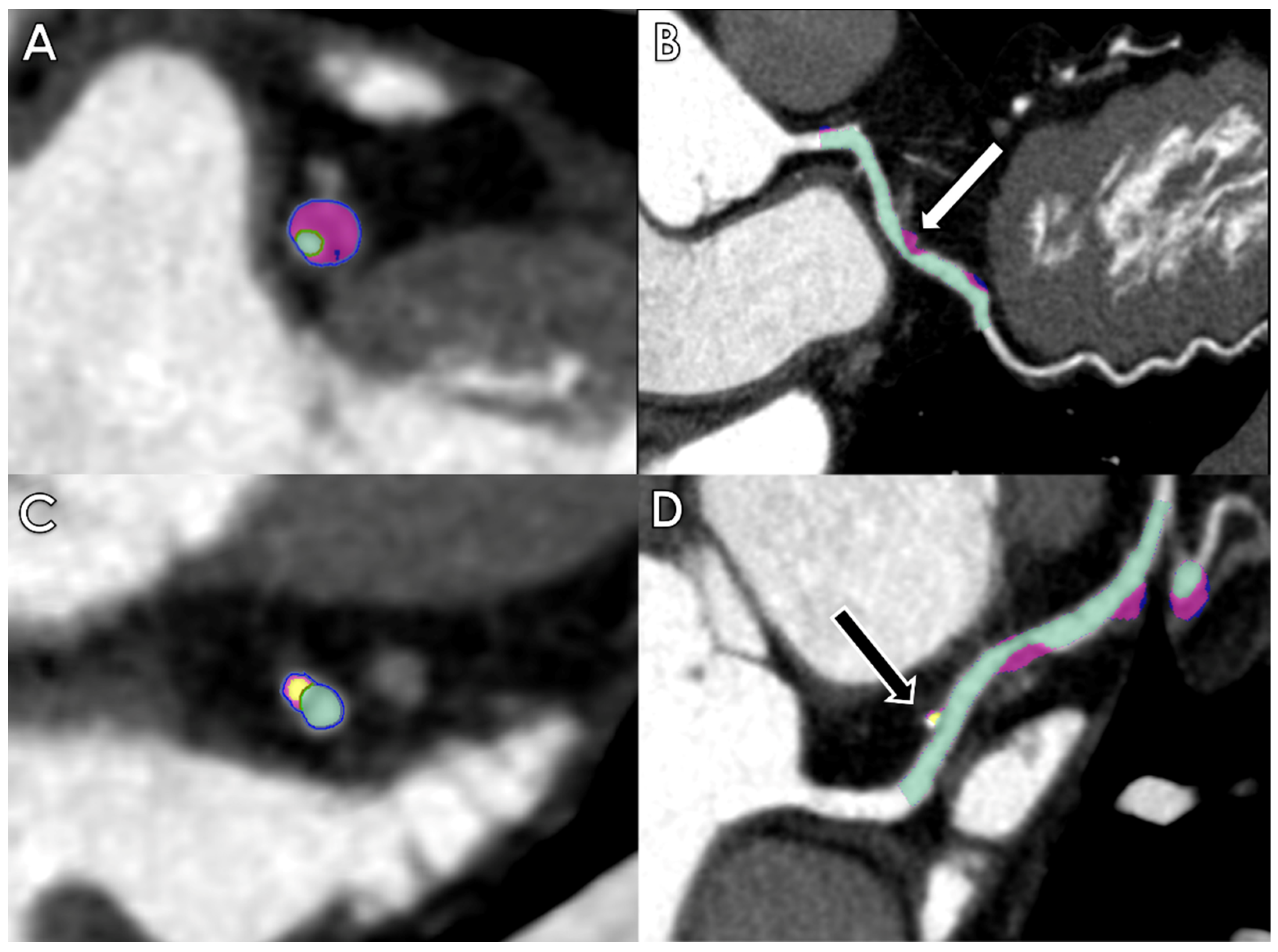
2.2. Coronary Stenosis and Plaque Analysis
2.3. FFRct and Myocardial Perfusion
3. AI in Cardiac Magnetic Resonance
3.1. Acquisition Phase
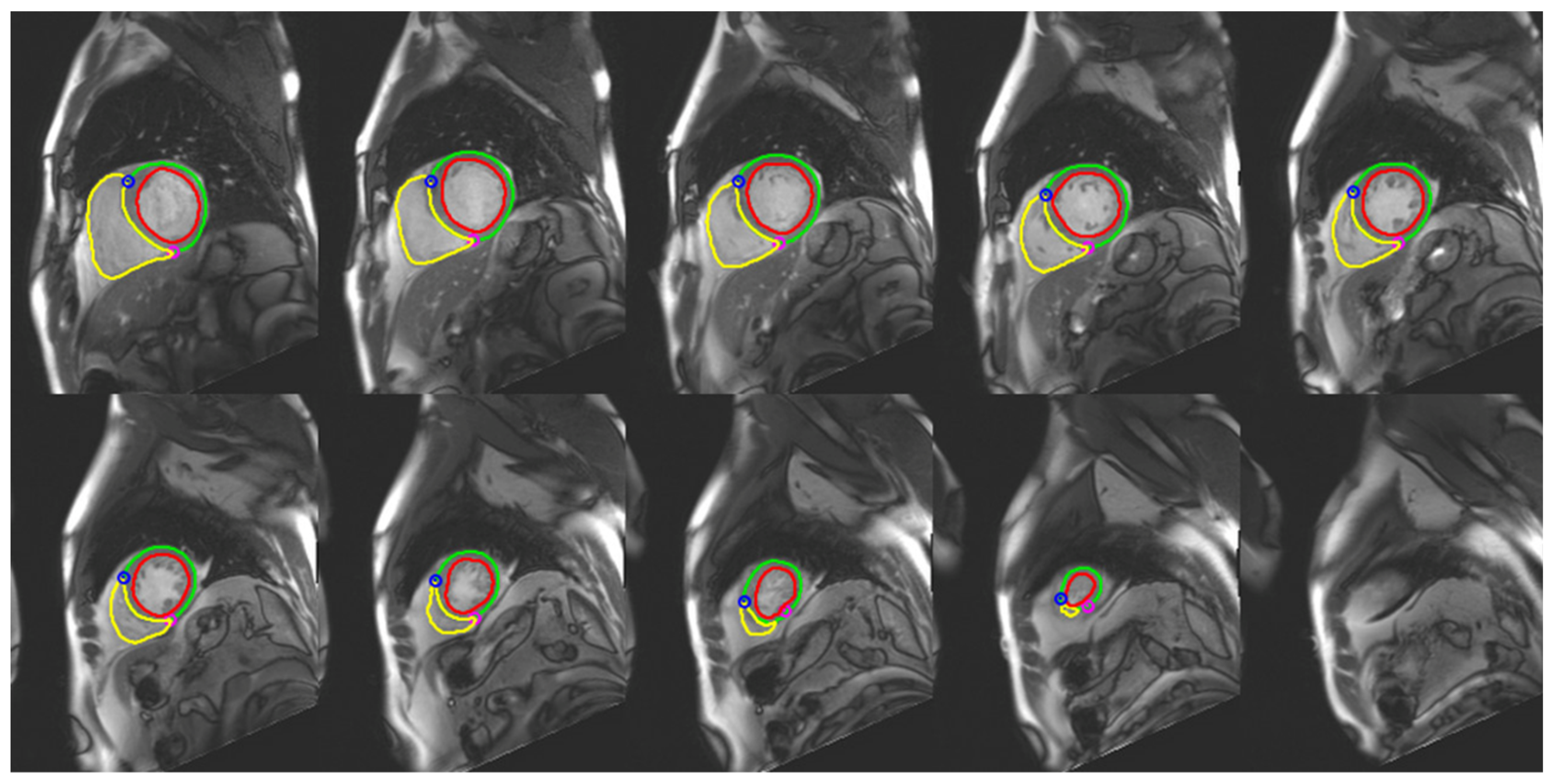
3.2. Image Segmentation
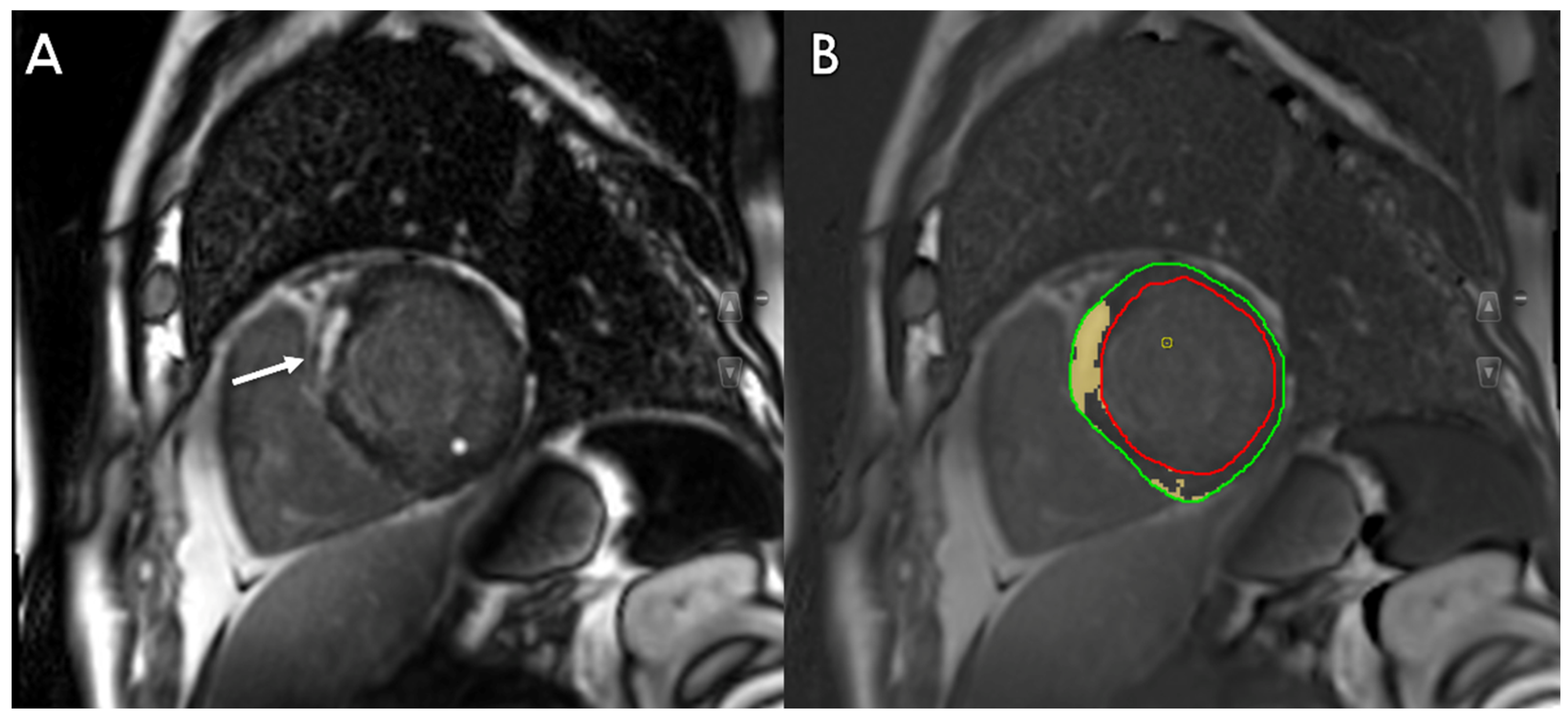
3.3. Tissue Characterization
3.4. Prognosis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mintz, Y.; Brodie, R. Introduction to artificial intelligence in medicine. Minim. Invasive Ther. Allied Technol. 2019, 28, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Kilic, A. Artificial Intelligence and Machine Learning in Cardiovascular Health Care. Ann. Thorac. Surg. 2020, 109, 1323–1329. [Google Scholar] [CrossRef]
- van Assen, M.; Lee, S.J.; De Cecco, C.N. Artificial intelligence from A to Z: From neural network to legal framework. Eur. J. Radiol. 2020, 129, 109083. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, T.; Caudo, D.; Blandino, A.; Albrecht, M.H.; Vogl, T.J.; Gruenewald, L.D.; Gaeta, M.; Yel, I.; Koch, V.; Martin, S.S.; et al. Artificial intelligence, machine learning and deep learning in musculoskeletal imaging: Current applications. J. Clin. Ultrasound. 2022, 50, 1414–1431. [Google Scholar] [CrossRef]
- Muscogiuri, G.; Chiesa, M.; Trotta, M.; Gatti, M.; Palmisano, V.; Dell’Aversana, S.; Baessato, F.; Cavaliere, A.; Cicala, G.; Loffreno, A.; et al. Performance of a deep learning algorithm for the evaluation of CAD-RADS classification with CCTA. Atherosclerosis 2020, 294, 25–32. [Google Scholar] [CrossRef]
- Muscogiuri, G.; Van Assen, M.; Tesche, C.; De Cecco, C.N.; Chiesa, M.; Scafuri, S.; Guglielmo, M.; Baggiano, A.; Fusini, L.; Guaricci, A.I.; et al. Artificial Intelligence in Coronary Computed Tomography Angiography: From Anatomy to Prognosis. Biomed. Res. Int. 2020, 2020, 6649410. [Google Scholar] [CrossRef] [PubMed]
- Hartaigh, B.O.; Valenti, V.; Cho, I.; Schulman-Marcus, J.; Gransar, H.; Knapper, J.; Kelkar, A.A.; Xie, J.X.; Chang, H.J.; Shaw, L.J.; et al. 15-Year prognostic utility of coronary artery calcium scoring for all-cause mortality in the elderly. Atherosclerosis 2016, 246, 361–366. [Google Scholar] [CrossRef]
- Tesche, C.; Duguay, T.M.; Schoepf, U.J.; van Assen, M.; De Cecco, C.N.; Albrecht, M.H.; Varga-Szemes, A.; Bayer, R.R. Current and future applications of CT coronary calcium assessment. Expert Rev. Cardiovasc. Ther. 2018, 16, 441–453. [Google Scholar] [CrossRef]
- Agatston, A.S.; Janowitz, W.R.; Hildner, F.J.; Zusmer, N.R.; Viamonte, M., Jr.; Detrano, R. Quantification of coronary artery calcium using ultrafast computed tomography. J. Am. Coll. Cardiol. 1990, 15, 827–832. [Google Scholar] [CrossRef]
- Isgum, I.; Rutten, A.; Prokop, M.; van Ginneken, B. Detection of coronary calcifications from computed tomography scans for automated risk assessment of coronary artery disease. Med. Phys. 2007, 34, 1450–1461. [Google Scholar] [CrossRef]
- Hecht, H.S.; Cronin, P.; Blaha, M.J.; Budoff, M.J.; Kazerooni, E.A.; Narula, J.; Yankelevitz, D.; Abbara, S. 2016 SCCT/STR guidelines for coronary artery calcium scoring of noncontrast noncardiac chest CT scans: A report of the Society of Cardiovascular Computed Tomography and Society of Thoracic Radiology. J. Thorac. Imaging 2017, 32, W54–W66. [Google Scholar] [CrossRef] [PubMed]
- Takx, R.A.; de Jong, P.A.; Leiner, T.; Oudkerk, M.; de Koning, H.J.; Mol, C.P.; Viergever, M.A.; Isgum, I. Automated coronary artery calcification scoring in non-gated chest CT: Agreement and reliability. PLoS ONE 2014, 9, e91239. [Google Scholar] [CrossRef]
- Cano-Espinosa, C.; Gonzalez, G.; Washko, G.R.; Cazorla, M.; Estepar, R.S.J. Automated Agatston Score Computation in non-ECG Gated CT Scans Using Deep Learning. Proc. SPIE Int. Soc. Opt. Eng. 2018, 10574, 673–678. [Google Scholar] [CrossRef]
- Sandstedt, M.; Henriksson, L.; Janzon, M.; Nyberg, G.; Engvall, J.; De Geer, J.; Alfredsson, J.; Persson, A. Evaluation of an AI-based, automatic coronary artery calcium scoring software. Eur. Radiol. 2020, 30, 1671–1678. [Google Scholar] [CrossRef] [PubMed]
- Winkelmann, M.T.; Jacoby, J.; Schwemmer, C.; Faby, S.; Krumm, P.; Artzner, C.; Bongers, M.N. Fully Automated Artery-Specific Calcium Scoring Based on Machine Learning in Low-Dose Computed Tomography Screening. Rofo 2022, 194, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.S.; van Assen, M.; Rapaka, S.; Hudson, H.T., Jr.; Fischer, A.M.; Varga-Szemes, A.; Sahbaee, P.; Schwemmer, C.; Gulsun, M.A.; Cimen, S.; et al. Evaluation of a Deep Learning-Based Automated CT Coronary Artery Calcium Scoring Algorithm. JACC Cardiovasc. Imaging 2020, 13 Pt 1, 524–526. [Google Scholar] [CrossRef]
- Lee, J.G.; Kim, H.; Kang, H.; Koo, H.J.; Kang, J.W.; Kim, Y.H.; Yang, D.H. Fully Automatic Coronary Calcium Score Software Empowered by Artificial Intelligence Technology: Validation Study Using Three CT Cohorts. Korean J. Radiol. 2021, 22, 1764–1776. [Google Scholar] [CrossRef] [PubMed]
- Apfaltrer, P.; Schoepf, U.J.; Vliegenthart, R.; Rowe, G.W.; Spears, J.R.; Fink, C.; Nance, J.W., Jr. Coronary computed tomography--present status and future directions. Int. J. Clin. Pract. Suppl. 2011, 65, 3–13. [Google Scholar] [CrossRef]
- Investigators, S.-H. CT coronary angiography in patients with suspected angina due to coronary heart disease (SCOT-HEART): An open-label, parallel-group, multicentre trial. Lancet 2015, 385, 2383–2391. [Google Scholar] [CrossRef]
- Williams, M.C.; Moss, A.J.; Dweck, M.; Adamson, P.D.; Alam, S.; Hunter, A.; Shah, A.S.V.; Pawade, T.; Weir-McCall, J.R.; Roditi, G.; et al. Coronary Artery Plaque Characteristics Associated With Adverse Outcomes in the SCOT-HEART Study. J. Am. Coll. Cardiol. 2019, 73, 291–301. [Google Scholar] [CrossRef]
- Jonas, R.A.; Weerakoon, S.; Fisher, R.; Griffin, W.F.; Kumar, V.; Rahban, H.; Marques, H.; Karlsberg, R.P.; Jennings, R.S.; Crabtree, T.R.; et al. Interobserver variability among expert readers quantifying plaque volume and plaque characteristics on coronary CT angiography: A CLARIFY trial sub-study. Clin. Imaging 2022, 91, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Cury, R.C.; Leipsic, J.; Abbara, S.; Achenbach, S.; Berman, D.; Bittencourt, M.; Budoff, M.; Chinnaiyan, K.; Choi, A.D.; Ghoshhajra, B.; et al. CAD-RADS 2.0—2022 Coronary Artery Disease—Reporting and Data System.: An expert consensus document of the Society of Cardiovascular Computed Tomography (SCCT), the American College of Cardiology (ACC), the American College of Radiology (ACR) and the North America Society of Cardiovascular Imaging (NASCI). J. Am. Coll. Radiol. 2022, 19, 1185–1212. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.F.; Rohnean, A.; Giroussens, H.; Pressat-Laffouilhere, T.; Wong, T. Evaluation of a deep learning model on coronary CT angiography for automatic stenosis detection. Diagn. Interv. Imaging 2022, 103, 316–323. [Google Scholar] [CrossRef]
- Choi, A.D.; Marques, H.; Kumar, V.; Griffin, W.F.; Rahban, H.; Karlsberg, R.P.; Zeman, R.K.; Katz, R.J.; Earls, J.P. CT Evaluation by Artificial Intelligence for Atherosclerosis, Stenosis and Vascular Morphology (CLARIFY): A Multi-center, international study. J. Cardiovasc. Comput. Tomogr. 2021, 15, 470–476. [Google Scholar] [CrossRef]
- Hell, M.M.; Dey, D.; Marwan, M.; Achenbach, S.; Schmid, J.; Schuhbaeck, A. Non-invasive prediction of hemodynamically significant coronary artery stenoses by contrast density difference in coronary CT angiography. Eur. J. Radiol. 2015, 84, 1502–1508. [Google Scholar] [CrossRef]
- Griffin, W.F.; Choi, A.D.; Riess, J.S.; Marques, H.; Chang, H.J.; Choi, J.H.; Doh, J.H.; Her, A.Y.; Koo, B.K.; Nam, C.W.; et al. AI Evaluation of Stenosis on Coronary CT Angiography, Comparison With Quantitative Coronary Angiography and Fractional Flow Reserve: A CREDENCE Trial Substudy. JACC Cardiovasc. Imaging 2022, 16, 193–205. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, T.; Cicero, G.; Mazziotti, S.; Ascenti, G.; Albrecht, M.H.; Martin, S.S.; Othman, A.E.; Vogl, T.J.; Wichmann, J.L. Dual energy computed tomography virtual monoenergetic imaging: Technique and clinical applications. Br. J. Radiol. 2019, 92, 20180546. [Google Scholar] [CrossRef]
- Arendt, C.T.; Czwikla, R.; Lenga, L.; Wichmann, J.L.; Albrecht, M.H.; Booz, C.; Martin, S.S.; Leithner, D.; Tischendorf, P.; Blandino, A.; et al. Improved coronary artery contrast enhancement using noise-optimised virtual monoenergetic imaging from dual-source dual-energy computed tomography. Eur. J. Radiol. 2020, 122, 108666. [Google Scholar] [CrossRef]
- Lenga, L.; Albrecht, M.H.; Othman, A.E.; Martin, S.S.; Leithner, D.; D’Angelo, T.; Arendt, C.; Scholtz, J.E.; De Cecco, C.N.; Schoepf, U.J.; et al. Monoenergetic Dual-energy Computed Tomographic Imaging: Cardiothoracic Applications. J. Thorac. Imaging 2017, 32, 151–158. [Google Scholar] [CrossRef]
- Cicero, G.; Ascenti, G.; Albrecht, M.H.; Blandino, A.; Cavallaro, M.; D’Angelo, T.; Carerj, M.L.; Vogl, T.J.; Mazziotti, S. Extra-abdominal dual-energy CT applications: A comprehensive overview. Radiol. Med. 2020, 125, 384–397. [Google Scholar] [CrossRef]
- Yi, Y.; Xu, C.; Guo, N.; Sun, J.; Lu, X.; Yu, S.; Wang, Y.; Vembar, M.; Jin, Z.; Wang, Y. Performance of an Artificial Intelligence-based Application for the Detection of Plaque-based Stenosis on Monoenergetic Coronary CT Angiography: Validation by Invasive Coronary Angiography. Acad. Radiol. 2022, 29 (Suppl. 4), S49–S58. [Google Scholar] [CrossRef] [PubMed]
- Tesche, C.; De Cecco, C.N.; Albrecht, M.H.; Duguay, T.M.; Bayer, R.R., 2nd; Litwin, S.E.; Steinberg, D.H.; Schoepf, U.J. Coronary CT Angiography-derived Fractional Flow Reserve. Radiology 2017, 285, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Coenen, A.; Kim, Y.H.; Kruk, M.; Tesche, C.; De Geer, J.; Kurata, A.; Lubbers, M.L.; Daemen, J.; Itu, L.; Rapaka, S.; et al. Diagnostic Accuracy of a Machine-Learning Approach to Coronary Computed Tomographic Angiography-Based Fractional Flow Reserve: Result From the MACHINE Consortium. Circ. Cardiovasc. Imaging 2018, 11, e007217. [Google Scholar] [CrossRef]
- Tesche, C.; De Cecco, C.N.; Baumann, S.; Renker, M.; McLaurin, T.W.; Duguay, T.M.; Bayer, R.R., 2nd; Steinberg, D.H.; Grant, K.L.; Canstein, C.; et al. Coronary CT Angiography-derived Fractional Flow Reserve: Machine Learning Algorithm versus Computational Fluid Dynamics Modeling. Radiology 2018, 288, 64–72. [Google Scholar] [CrossRef]
- Morais, T.C.; Assuncao-Jr, A.N.; Dantas Junior, R.N.; Silva, C.; Paula, C.B.; Torres, R.A.; Magalhaes, T.A.; Nomura, C.H.; Avila, L.F.R.; Parga Filho, J.R. Diagnostic Performance of a Machine Learning-Based CT-Derived FFR in Detecting Flow-Limiting Stenosis. Arq. Bras. Cardiol. 2021, 116, 1091–1098. [Google Scholar] [CrossRef]
- Tang, C.X.; Guo, B.J.; Schoepf, J.U.; Bayer, R.R., 2nd; Liu, C.Y.; Qiao, H.Y.; Zhou, F.; Lu, G.M.; Zhou, C.S.; Zhang, L.J. Feasibility and prognostic role of machine learning-based FFR(CT) in patients with stent implantation. Eur. Radiol. 2021, 31, 6592–6604. [Google Scholar] [CrossRef]
- Qiao, H.Y.; Tang, C.X.; Schoepf, U.J.; Bayer, R.R., 2nd; Tesche, C.; Di Jiang, M.; Yin, C.Q.; Zhou, C.S.; Zhou, F.; Lu, M.J.; et al. One-year outcomes of CCTA alone versus machine learning-based FFR(CT) for coronary artery disease: A single-center, prospective study. Eur. Radiol. 2022, 32, 5179–5188. [Google Scholar] [CrossRef]
- Branch, K.R.; Haley, R.D.; Bittencourt, M.S.; Patel, A.R.; Hulten, E.; Blankstein, R. Myocardial computed tomography perfusion. Cardiovasc. Diagn. Ther. 2017, 7, 452–462. [Google Scholar] [CrossRef]
- D’Angelo, T.; Martin, S.; Micari, A.; Booz, C.; Steyer, A.; Blandino, A.; Lanzafame, L.R.; Koch, V.; Ascenti, G.; Mazziotti, S. Coronary angiography using spectral detector dual-energy CT: Is it the time to assess myocardial first-pass perfusion? Eur. Radiol. Exp. 2022, 6, 60. [Google Scholar] [CrossRef]
- Xiong, T.Y.; Redwood, S.; Prendergast, B.; Chen, M. Coronaviruses and the cardiovascular system: Acute and long-term implications. Eur. Heart J. 2020, 41, 1798–1800. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Liu, J.; Sun, Z.; Cui, Y.; He, Y.; Yang, Z. Deep learning analysis in coronary computed tomographic angiography imaging for the assessment of patients with coronary artery stenosis. Comput. Methods Programs Biomed. 2020, 196, 105651. [Google Scholar] [CrossRef]
- Muscogiuri, G.; Chiesa, M.; Baggiano, A.; Spadafora, P.; De Santis, R.; Guglielmo, M.; Scafuri, S.; Fusini, L.; Mushtaq, S.; Conte, E.; et al. Diagnostic performance of deep learning algorithm for analysis of computed tomography myocardial perfusion. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 3119–3128. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Emanuel, E.J. Predicting the Future—Big Data, Machine Learning, and Clinical Medicine. N. Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef]
- Kramer, C.M.; Barkhausen, J.; Bucciarelli-Ducci, C.; Flamm, S.D.; Kim, R.J.; Nagel, E. Standardized cardiovascular magnetic resonance imaging (CMR) protocols: 2020 update. J. Cardiovasc. Magn. Reason. 2020, 22, 17. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Jolly, M.P.; Georgescu, B.; Haye, C.; Speier, P.; Schmidt, M.; Bi, X.; Kroeker, R.; Comaniciu, D.; Kellman, P.; et al. Automatic view planning for cardiac MRI acquisition. Med. Image Comput. Comput. Assist. Interv. 2011, 6893, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Nitta, S.; Takeguchi, T.; Matsumoto, N.; Kuhara, S.; Yokoyama, K.; Ishimura, R.; Nitatori, T. Automatic slice alignment method for cardiac magnetic resonance imaging. MAGMA 2013, 26, 451–461. [Google Scholar] [CrossRef]
- Yokoyama, K.; Nitta, S.; Kuhara, S.; Ishimura, R.; Kariyasu, T.; Imai, M.; Nitatori, T.; Takeguchi, T.; Shiodera, T. Automatic slice-alignment method in cardiac magnetic resonance imaging for evaluation of the right ventricle in patients with pulmonary hypertension. AIP Adv. 2015, 5, 097182. [Google Scholar] [CrossRef]
- Oktay, O.; Bai, W.; Guerrero, R.; Rajchl, M.; de Marvao, A.; O’Regan, D.P.; Cook, S.A.; Heinrich, M.P.; Glocker, B.; Rueckert, D. Stratified Decision Forests for Accurate Anatomical Landmark Localization in Cardiac Images. IEEE Trans. Med. Imaging 2017, 36, 332–342. [Google Scholar] [CrossRef]
- Blansit, K.; Retson, T.; Masutani, E.; Bahrami, N.; Hsiao, A. Deep Learning-based Prescription of Cardiac MRI Planes. Radiol. Artif. Intell. 2019, 1, e180069. [Google Scholar] [CrossRef]
- Fotaki, A.; Fuin, N.; Nordio, G.; Velasco Jimeno, C.; Qi, H.; Emmanuel, Y.; Pushparajah, K.; Botnar, R.M.; Prieto, C. Accelerating 3D MTC-BOOST in patients with congenital heart disease using a joint multi-scale variational neural network reconstruction. Magn. Reason. Imaging 2022, 92, 120–132. [Google Scholar] [CrossRef]
- Weine, J.; van Gorkum, R.J.H.; Stoeck, C.T.; Vishnevskiy, V.; Kozerke, S. Synthetically trained convolutional neural networks for improved tensor estimation from free-breathing cardiac DTI. Comput. Med. Imaging Graph. 2022, 99, 102075. [Google Scholar] [CrossRef]
- Fratz, S.; Chung, T.; Greil, G.F.; Samyn, M.M.; Taylor, A.M.; Valsangiacomo Buechel, E.R.; Yoo, S.J.; Powell, A.J. Guidelines and protocols for cardiovascular magnetic resonance in children and adults with congenital heart disease: SCMR expert consensus group on congenital heart disease. J. Cardiovasc. Magn. Reason. 2013, 15, 51. [Google Scholar] [CrossRef] [PubMed]
- Muscogiuri, G.; Suranyi, P.; Eid, M.; Varga-Szemes, A.; Griffith, L.; Pontone, G.; Schoepf, U.J.; De Cecco, C.N. Pediatric Cardiac MR Imaging: Practical Preoperative Assessment. Magn. Reason. Imaging Clin. N. Am. 2019, 27, 243–262. [Google Scholar] [CrossRef]
- Bluemke, D.A.; Kronmal, R.A.; Lima, J.A.; Liu, K.; Olson, J.; Burke, G.L.; Folsom, A.R. The relationship of left ventricular mass and geometry to incident cardiovascular events: The MESA (Multi-Ethnic Study of Atherosclerosis) study. J. Am. Coll. Cardiol. 2008, 52, 2148–2155. [Google Scholar] [CrossRef] [PubMed]
- Varga-Szemes, A.; Muscogiuri, G.; Schoepf, U.J.; Wichmann, J.L.; Suranyi, P.; De Cecco, C.N.; Cannao, P.M.; Renker, M.; Mangold, S.; Fox, M.A.; et al. Clinical feasibility of a myocardial signal intensity threshold-based semi-automated cardiac magnetic resonance segmentation method. Eur. Radiol. 2016, 26, 1503–1511. [Google Scholar] [CrossRef]
- Petitjean, C.; Dacher, J.N. A review of segmentation methods in short axis cardiac MR images. Med. Image Anal. 2011, 15, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Suinesiaputra, A.; Bluemke, D.A.; Cowan, B.R.; Friedrich, M.G.; Kramer, C.M.; Kwong, R.; Plein, S.; Schulz-Menger, J.; Westenberg, J.J.; Young, A.A.; et al. Quantification of LV function and mass by cardiovascular magnetic resonance: Multi-center variability and consensus contours. J. Cardiovasc. Magn. Reason. 2015, 17, 63. [Google Scholar] [CrossRef]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.A.; Cetin, I.; Lekadir, K.; Camara, O.; Gonzalez Ballester, M.A.; et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef]
- Bai, W.; Sinclair, M.; Tarroni, G.; Oktay, O.; Rajchl, M.; Vaillant, G.; Lee, A.M.; Aung, N.; Lukaschuk, E.; Sanghvi, M.M.; et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J. Cardiovasc. Magn. Reason. 2018, 20, 65. [Google Scholar] [CrossRef]
- Tao, Q.; Yan, W.; Wang, Y.; Paiman, E.H.M.; Shamonin, D.P.; Garg, P.; Plein, S.; Huang, L.; Xia, L.; Sramko, M.; et al. Deep Learning-based Method for Fully Automatic Quantification of Left Ventricle Function from Cine MR Images: A Multivendor, Multicenter Study. Radiology 2019, 290, 81–88. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Z.; Lai, Q.; Li, S.; Guo, Y.; Wang, Y.; Dai, Z.; Huang, J. ESA-UNet for assisted diagnosis of cardiac magnetic resonance image based on the semantic segmentation of the heart. Front. Cardiovasc. Med. 2022, 9, 1012450. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.K.L.; Zhang, A.; Yang, K.; Wu, S.; Ghista, D.N. GCW-UNet segmentation of cardiac magnetic resonance images for evaluation of left atrial enlargement. Comput. Methods Programs Biomed. 2022, 221, 106915. [Google Scholar] [CrossRef] [PubMed]
- Edwards, N.C.; Routledge, H.; Steeds, R.P. T2-weighted magnetic resonance imaging to assess myocardial oedema in ischaemic heart disease. Heart 2009, 95, 1357–1361. [Google Scholar] [CrossRef]
- Kellman, P.; Hansen, M.S. T1-mapping in the heart: Accuracy and precision. J. Cardiovasc. Magn. Reason. 2014, 16, 2. [Google Scholar] [CrossRef]
- Satoh, H.; Sano, M.; Suwa, K.; Saitoh, T.; Nobuhara, M.; Saotome, M.; Urushida, T.; Katoh, H.; Hayashi, H. Distribution of late gadolinium enhancement in various types of cardiomyopathies: Significance in differential diagnosis, clinical features and prognosis. World J. Cardiol. 2014, 6, 585–601. [Google Scholar] [CrossRef] [PubMed]
- Kammerlander, A.A.; Marzluf, B.A.; Zotter-Tufaro, C.; Aschauer, S.; Duca, F.; Bachmann, A.; Knechtelsdorfer, K.; Wiesinger, M.; Pfaffenberger, S.; Greiser, A.; et al. T1 Mapping by CMR Imaging: From Histological Validation to Clinical Implication. JACC Cardiovasc. Imaging 2016, 9, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Muscogiuri, G.; Suranyi, P.; Schoepf, U.J.; De Cecco, C.N.; Secinaro, A.; Wichmann, J.L.; Fuller, S.R.; Lesslie, V.W.; Varga-Szemes, A. Cardiac Magnetic Resonance T1-Mapping of the Myocardium: Technical Background and Clinical Relevance. J. Thorac. Imaging 2018, 33, 71–80. [Google Scholar] [CrossRef]
- Haslbauer, J.D.; Lindner, S.; Valbuena-Lopez, S.; Zainal, H.; Zhou, H.; D’Angelo, T.; Pathan, F.; Arendt, C.A.; Bug, G.; Serve, H.; et al. CMR imaging biosignature of cardiac involvement due to cancer-related treatment by T1 and T2 mapping. Int. J. Cardiol 2019, 275, 179–186. [Google Scholar] [CrossRef]
- Gatti, M.; D’Angelo, T.; Muscogiuri, G.; Dell’aversana, S.; Andreis, A.; Carisio, A.; Darvizeh, F.; Tore, D.; Pontone, G.; Faletti, R. Cardiovascular magnetic resonance of cardiac tumors and masses. World J. Cardiol. 2021, 13, 628–649. [Google Scholar] [CrossRef]
- Siebermair, J.; Kholmovski, E.G.; Marrouche, N. Assessment of Left Atrial Fibrosis by Late Gadolinium Enhancement Magnetic Resonance Imaging: Methodology and Clinical Implications. JACC Clin. Electrophysiol. 2017, 3, 791–802. [Google Scholar] [CrossRef]
- Muscogiuri, G.; Guaricci, A.I.; Cau, R.; Saba, L.; Senatieri, A.; Chierchia, G.; Pontone, G.; Volpato, V.; Palmisano, A.; Esposito, A.; et al. Multimodality imaging in acute myocarditis. J. Clin. Ultrasound. 2022, 50, 1097–1109. [Google Scholar] [CrossRef]
- Moon, J.C.; Messroghli, D.R.; Kellman, P.; Piechnik, S.K.; Robson, M.D.; Ugander, M.; Gatehouse, P.D.; Arai, A.E.; Friedrich, M.G.; Neubauer, S.; et al. Myocardial T1 mapping and extracellular volume quantification: A Society for Cardiovascular Magnetic Resonance (SCMR) and CMR Working Group of the European Society of Cardiology consensus statement. J. Cardiovasc. Magn. Reason. 2013, 15, 92. [Google Scholar] [CrossRef]
- h-Ici, D.O.; Jeuthe, S.; Al-Wakeel, N.; Berger, F.; Kuehne, T.; Kozerke, S.; Messroghli, D.R. T1 mapping in ischaemic heart disease. Eur. Heart J. Cardiovasc. Imaging 2014, 15, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, V.M.; Piechnik, S.K.; Robson, M.D.; Neubauer, S.; Karamitsos, T.D. Myocardial tissue characterization by magnetic resonance imaging: Novel applications of T1 and T2 mapping. J. Thorac. Imaging 2014, 29, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Messroghli, D.R.; Moon, J.C.; Ferreira, V.M.; Grosse-Wortmann, L.; He, T.; Kellman, P.; Mascherbauer, J.; Nezafat, R.; Salerno, M.; Schelbert, E.B.; et al. Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2* and extracellular volume: A consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imaging (EACVI). J. Cardiovasc. Magn. Reason. 2017, 19, 75. [Google Scholar] [CrossRef]
- D’Angelo, T.; Grigoratos, C.; Mazziotti, S.; Bratis, K.; Pathan, F.; Blandino, A.; Elen, E.; Puntmann, V.O.; Nagel, E. High-throughput gadobutrol-enhanced CMR: A time and dose optimization study. J. Cardiovasc. Magn. Reason. 2017, 19, 83. [Google Scholar] [CrossRef] [PubMed]
- Puntmann, V.O.; Carr-White, G.; Jabbour, A.; Yu, C.Y.; Gebker, R.; Kelle, S.; Rolf, A.; Zitzmann, S.; Peker, E.; D’Angelo, T.; et al. Native T1 and ECV of Noninfarcted Myocardium and Outcome in Patients with Coronary Artery Disease. J. Am. Coll. Cardiol. 2018, 71, 766–778. [Google Scholar] [CrossRef]
- Schelbert, E.B.; Messroghli, D.R. State of the Art: Clinical Applications of Cardiac T1 Mapping. Radiology 2016, 278, 658–676. [Google Scholar] [CrossRef]
- Radenkovic, D.; Weingartner, S.; Ricketts, L.; Moon, J.C.; Captur, G. T1 mapping in cardiac MRI. Heart Fail Rev. 2017, 22, 415–430. [Google Scholar] [CrossRef]
- Taylor, A.J.; Salerno, M.; Dharmakumar, R.; Jerosch-Herold, M. T1 Mapping: Basic Techniques and Clinical Applications. JACC Cardiovasc. Imaging 2016, 9, 67–81. [Google Scholar] [CrossRef]
- Captur, G.; Manisty, C.; Moon, J.C. Cardiac MRI evaluation of myocardial disease. Heart 2016, 102, 1429–1435. [Google Scholar] [CrossRef] [PubMed]
- Verhaert, D.; Thavendiranathan, P.; Giri, S.; Mihai, G.; Rajagopalan, S.; Simonetti, O.P.; Raman, S.V. Direct T2 quantification of myocardial edema in acute ischemic injury. JACC Cardiovasc. Imaging 2011, 4, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Chan, R.H.; Maron, B.J.; Olivotto, I.; Pencina, M.J.; Assenza, G.E.; Haas, T.; Lesser, J.R.; Gruner, C.; Crean, A.M.; Rakowski, H.; et al. Prognostic value of quantitative contrast-enhanced cardiovascular magnetic resonance for the evaluation of sudden death risk in patients with hypertrophic cardiomyopathy. Circulation 2014, 130, 484–495. [Google Scholar] [CrossRef]
- Chaikriangkrai, K.; Abbasi, M.A.; Sarnari, R.; Dolan, R.; Lee, D.; Anderson, A.S.; Ghafourian, K.; Khan, S.S.; Vorovich, E.E.; Rich, J.D.; et al. Prognostic Value of Myocardial Extracellular Volume Fraction and T2-mapping in Heart Transplant Patients. JACC Cardiovasc. Imaging 2020, 13, 1521–1530. [Google Scholar] [CrossRef]
- Argentiero, A.; Muscogiuri, G.; Rabbat, M.G.; Martini, C.; Soldato, N.; Basile, P.; Baggiano, A.; Mushtaq, S.; Fusini, L.; Mancini, M.E.; et al. The Applications of Artificial Intelligence in Cardiovascular Magnetic Resonance-A Comprehensive Review. J. Clin. Med. 2022, 11, 2866. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Han, K.; Lee, S.; Yang, Y.J.; Kim, P.K.; Choi, B.W.; Suh, Y.J. Automated Measurement of Native T1 and Extracellular Volume Fraction in Cardiac Magnetic Resonance Imaging Using a Commercially Available Deep Learning Algorithm. Korean J. Radiol. 2022, 23, 1251–1259. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, C.; Qi, H.; Si, D.; Ding, H.; Chen, H. Motion correction for native myocardial T1 mapping using self-supervised deep learning registration with contrast separation. NMR Biomed. 2022, 35, e4775. [Google Scholar] [CrossRef]
- Moccia, S.; Banali, R.; Martini, C.; Muscogiuri, G.; Pontone, G.; Pepi, M.; Caiani, E.G. Development and testing of a deep learning-based strategy for scar segmentation on CMR-LGE images. MAGMA 2019, 32, 187–195. [Google Scholar] [CrossRef]
- Zabihollahy, F.; Rajchl, M.; White, J.A.; Ukwatta, E. Fully automated segmentation of left ventricular scar from 3D late gadolinium enhancement magnetic resonance imaging using a cascaded multi-planar U-Net (CMPU-Net). Med. Phys. 2020, 47, 1645–1655. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, G.; Gao, Z.; Xu, C.; Zhang, Y.; Shi, R.; Keegan, J.; Xu, L.; Zhang, H.; Fan, Z.; et al. Deep Learning for Diagnosis of Chronic Myocardial Infarction on Nonenhanced Cardiac Cine MRI. Radiology 2019, 291, 606–617. [Google Scholar] [CrossRef]
- Zhang, Q.; Burrage, M.K.; Shanmuganathan, M.; Gonzales, R.A.; Lukaschuk, E.; Thomas, K.E.; Mills, R.; Leal Pelado, J.; Nikolaidou, C.; Popescu, I.A.; et al. Artificial Intelligence for Contrast-Free MRI: Scar Assessment in Myocardial Infarction Using Deep Learning-Based Virtual Native Enhancement. Circulation 2022, 146, 1492–1503. [Google Scholar] [CrossRef]
- Leiner, T. Deep Learning for Detection of Myocardial Scar Tissue: Goodbye to Gadolinium? Radiology 2019, 291, 618–619. [Google Scholar] [CrossRef]
- Sendra-Balcells, C.; Campello, V.M.; Martin-Isla, C.; Vilades, D.; Descalzo, M.L.; Guala, A.; Rodriguez-Palomares, J.F.; Lekadir, K. Domain generalization in deep learning for contrast-enhanced imaging. Comput. Biol. Med. 2022, 149, 106052. [Google Scholar] [CrossRef]
- Vergara, G.R.; Marrouche, N.F. Tailored management of atrial fibrillation using a LGE-MRI based model: From the clinic to the electrophysiology laboratory. J. Cardiovasc. Electrophysiol. 2011, 22, 481–487. [Google Scholar] [CrossRef]
- Fochler, F.; Yamaguchi, T.; Kheirkahan, M.; Kholmovski, E.G.; Morris, A.K.; Marrouche, N.F. Late Gadolinium Enhancement Magnetic Resonance Imaging Guided Treatment of Post-Atrial Fibrillation Ablation Recurrent Arrhythmia. Circ. Arrhythm. Electrophysiol. 2019, 12, e007174. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wu, F.; Yang, G.; Xu, L.; Wong, T.; Mohiaddin, R.; Firmin, D.; Keegan, J.; Zhuang, X. Atrial scar quantification via multi-scale CNN in the graph-cuts framework. Med. Image Anal. 2020, 60, 101595. [Google Scholar] [CrossRef]
- Cho, Y.; Cho, H.; Shim, J.; Choi, J.I.; Kim, Y.H.; Kim, N.; Oh, Y.W.; Hwang, S.H. Efficient Segmentation for Left Atrium With Convolution Neural Network Based on Active Learning in Late Gadolinium Enhancement Magnetic Resonance Imaging. J. Korean Med. Sci. 2022, 37, e271. [Google Scholar] [CrossRef]
- Halliday, B.P.; Baksi, A.J.; Gulati, A.; Ali, A.; Newsome, S.; Izgi, C.; Arzanauskaite, M.; Lota, A.; Tayal, U.; Vassiliou, V.S.; et al. Outcome in Dilated Cardiomyopathy Related to the Extent, Location, and Pattern of Late Gadolinium Enhancement. JACC Cardiovasc. Imaging 2019, 12 Pt 2, 1645–1655. [Google Scholar] [CrossRef] [PubMed]
- Halliday, B.P.; Gulati, A.; Ali, A.; Guha, K.; Newsome, S.; Arzanauskaite, M.; Vassiliou, V.S.; Lota, A.; Izgi, C.; Tayal, U.; et al. Association Between Midwall Late Gadolinium Enhancement and Sudden Cardiac Death in Patients with Dilated Cardiomyopathy and Mild and Moderate Left Ventricular Systolic Dysfunction. Circulation 2017, 135, 2106–2115. [Google Scholar] [CrossRef] [PubMed]
- Arian, F.; Amini, M.; Mostafaei, S.; Rezaei Kalantari, K.; Haddadi Avval, A.; Shahbazi, Z.; Kasani, K.; Bitarafan Rajabi, A.; Chatterjee, S.; Oveisi, M.; et al. Myocardial Function Prediction After Coronary Artery Bypass Grafting Using MRI Radiomic Features and Machine Learning Algorithms. J. Digit Imaging 2022, 35, 1708–1718. [Google Scholar] [CrossRef] [PubMed]
- Dawes, T.J.W.; de Marvao, A.; Shi, W.; Fletcher, T.; Watson, G.M.J.; Wharton, J.; Rhodes, C.J.; Howard, L.; Gibbs, J.S.R.; Rueckert, D.; et al. Machine Learning of Three-dimensional Right Ventricular Motion Enables Outcome Prediction in Pulmonary Hypertension: A Cardiac MR Imaging Study. Radiology 2017, 283, 381–390. [Google Scholar] [CrossRef]
- Chen, R.; Lu, A.; Wang, J.; Ma, X.; Zhao, L.; Wu, W.; Du, Z.; Fei, H.; Lin, Q.; Yu, Z.; et al. Using machine learning to predict one-year cardiovascular events in patients with severe dilated cardiomyopathy. Eur. J. Radiol. 2019, 117, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Pocock, S.J.; Ariti, C.A.; McMurray, J.J.; Maggioni, A.; Kober, L.; Squire, I.B.; Swedberg, K.; Dobson, J.; Poppe, K.K.; Whalley, G.A.; et al. Predicting survival in heart failure: A risk score based on 39 372 patients from 30 studies. Eur. Heart J. 2013, 34, 1404–1413. [Google Scholar] [CrossRef]
- Diller, G.P.; Orwat, S.; Vahle, J.; Bauer, U.M.M.; Urban, A.; Sarikouch, S.; Berger, F.; Beerbaum, P.; Baumgartner, H.; German Competence Network for Congenital Heart Defects Investigators. Prediction of prognosis in patients with tetralogy of Fallot based on deep learning imaging analysis. Heart 2020, 106, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.J. Henderson, R.D.E.; Yi, X.; Babyn, P. Artificial Intelligence Solutions for Analysis of X-ray Images. Can. Assoc. Radiol. J. 2021, 72, 60–72. [Google Scholar] [CrossRef]
- Tsetsos, N.; Poutoglidis, A.; Arsos, G.; Tsentemeidou, A.; Kilmpasanis, A.; Katsampoukas, D.; Fyrmpas, G. 18F-FDG-PET/CT interpretation pitfalls in patients with head and neck cancer. Am. J. Otolaryngol. Head Neck Med. Surg. 2022, 43, 103209. [Google Scholar] [CrossRef] [PubMed]
- Sadaghiani, M.S.; Rowe, S.P.; Sheikhbahaei, S. Applications of artificial intelligence in oncologic 18F-FDG PET/CT imaging: A systematic review. Ann. Transl. Med. 2021, 9, 823. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lanzafame, L.R.M.; Bucolo, G.M.; Muscogiuri, G.; Sironi, S.; Gaeta, M.; Ascenti, G.; Booz, C.; Vogl, T.J.; Blandino, A.; Mazziotti, S.; et al. Artificial Intelligence in Cardiovascular CT and MR Imaging. Life 2023, 13, 507. https://doi.org/10.3390/life13020507
Lanzafame LRM, Bucolo GM, Muscogiuri G, Sironi S, Gaeta M, Ascenti G, Booz C, Vogl TJ, Blandino A, Mazziotti S, et al. Artificial Intelligence in Cardiovascular CT and MR Imaging. Life. 2023; 13(2):507. https://doi.org/10.3390/life13020507
Chicago/Turabian StyleLanzafame, Ludovica R. M., Giuseppe M. Bucolo, Giuseppe Muscogiuri, Sandro Sironi, Michele Gaeta, Giorgio Ascenti, Christian Booz, Thomas J. Vogl, Alfredo Blandino, Silvio Mazziotti, and et al. 2023. "Artificial Intelligence in Cardiovascular CT and MR Imaging" Life 13, no. 2: 507. https://doi.org/10.3390/life13020507
APA StyleLanzafame, L. R. M., Bucolo, G. M., Muscogiuri, G., Sironi, S., Gaeta, M., Ascenti, G., Booz, C., Vogl, T. J., Blandino, A., Mazziotti, S., & D’Angelo, T. (2023). Artificial Intelligence in Cardiovascular CT and MR Imaging. Life, 13(2), 507. https://doi.org/10.3390/life13020507






