Investigating the Mechanisms Underlying the Low Irradiance-Tolerance of the Economically Important Seaweed Species Pyropia haitanensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth Conditions and Treatment
2.2. Measurement of Pigment Contents
2.3. Determination of [Y(II)]
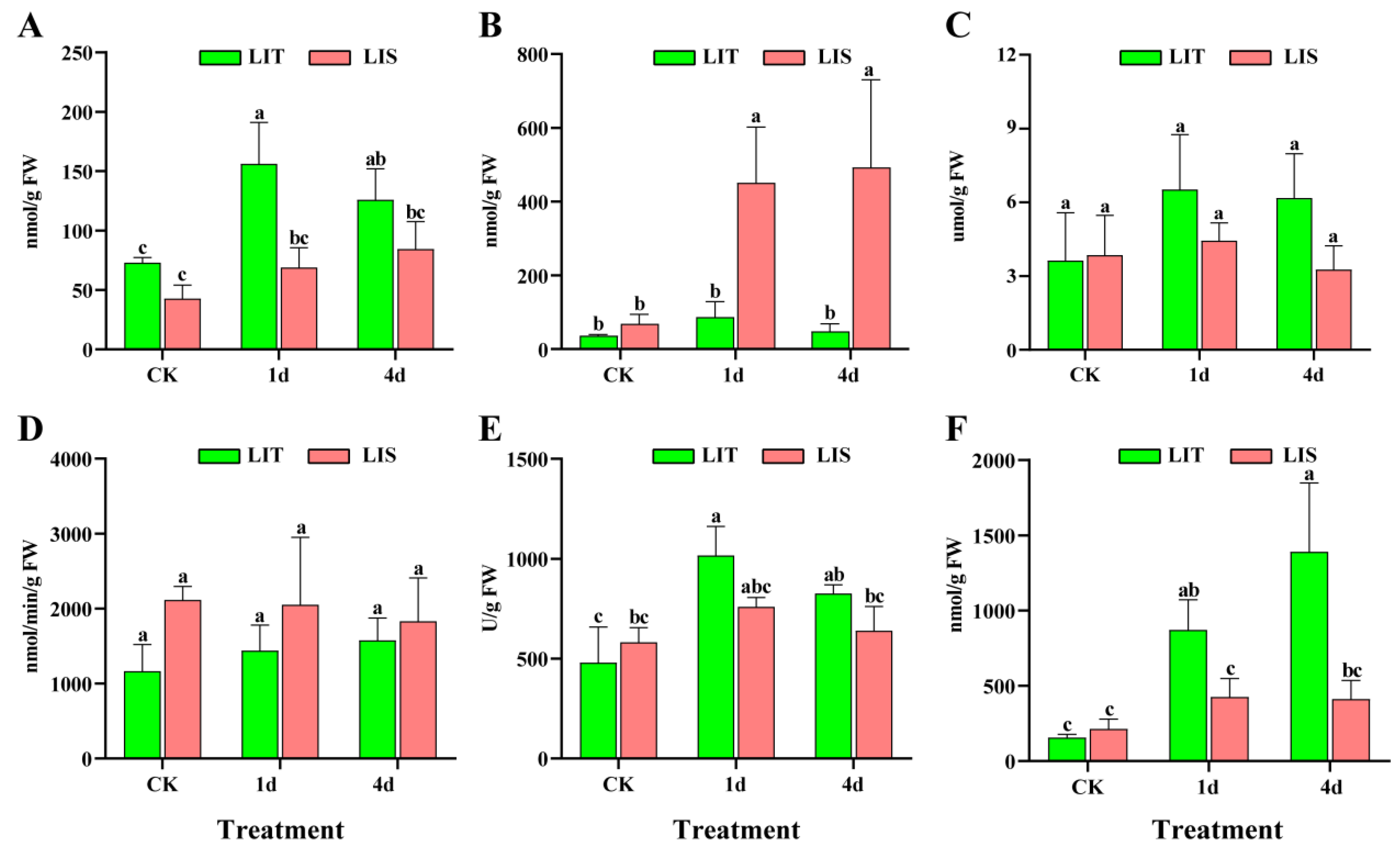
2.4. Measurement ROS, Malondialdehyde (MDA), and Antioxidant Enzyme Activities
2.5. Transcriptome Analysis
2.6. Research Route
2.7. Data Analysis
3. Results
3.1. Changes in the Y(II) of Blades under LI Stress
3.2. Changes in the Pigment Contents of Blades under LI Stress
3.3. Antioxidant Enzyme and ROS Contents in Blades under LI Stress
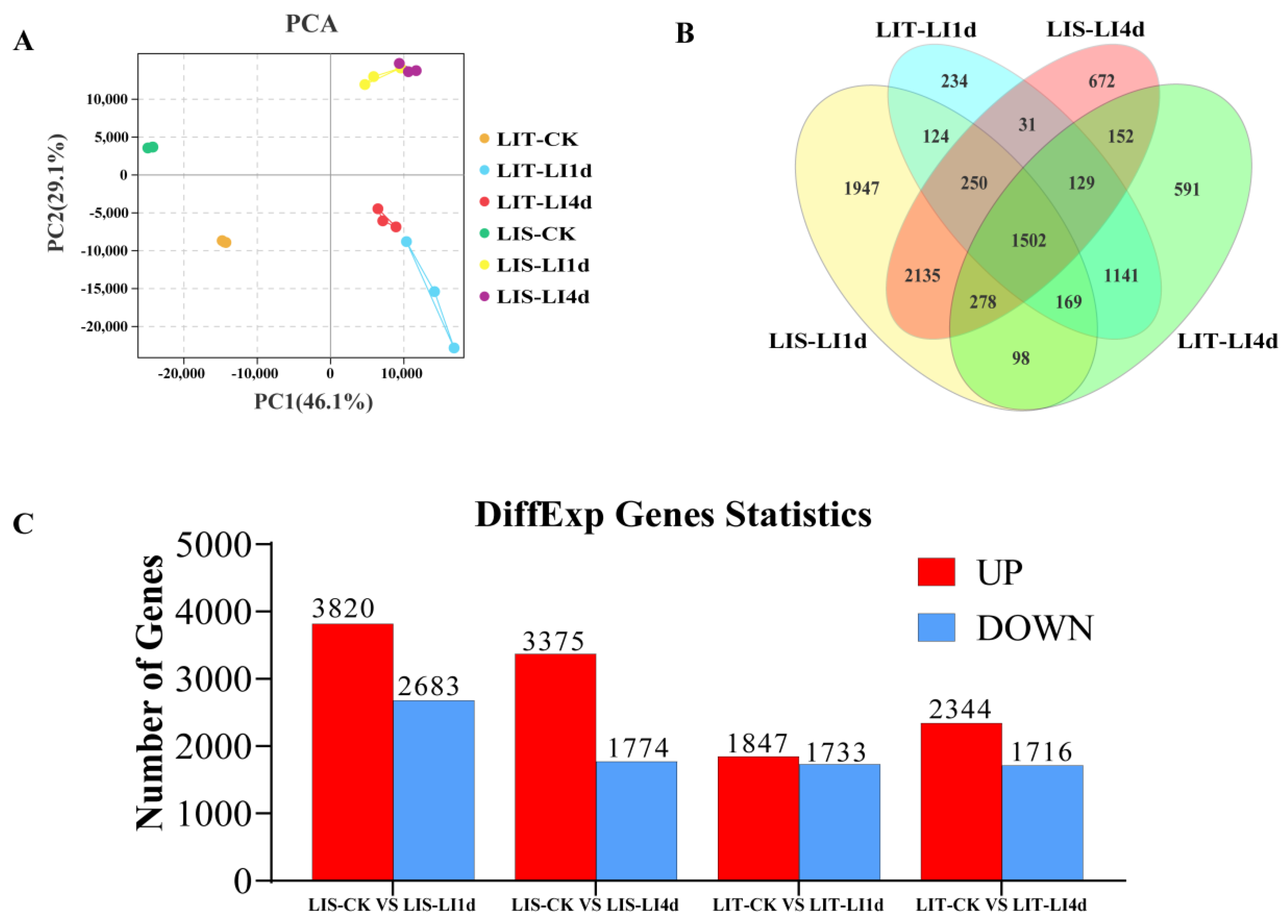
3.4. Principal Component Analysis of Transcriptome Data
3.5. Overall Analysis of Differentially Expressed Genes
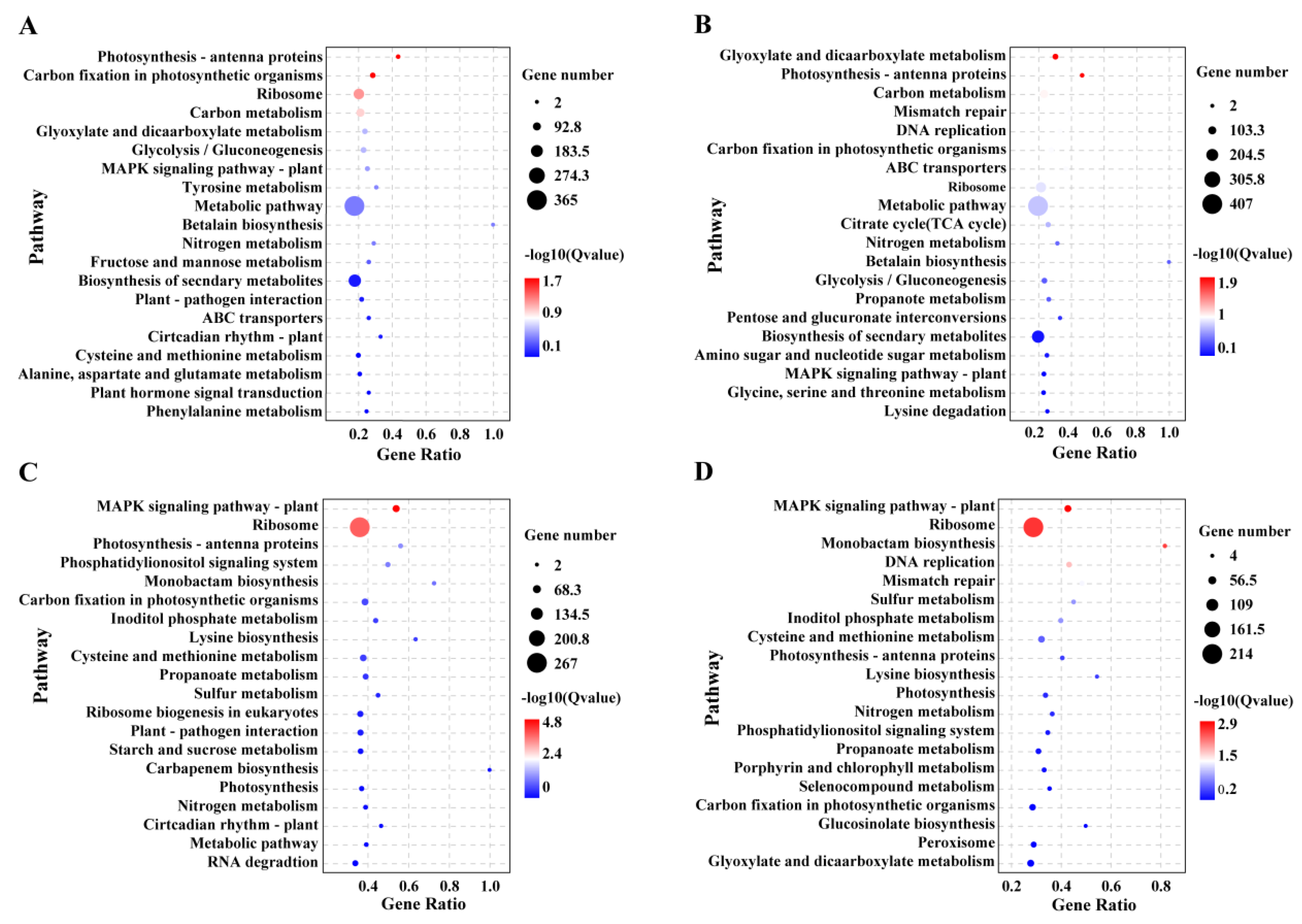
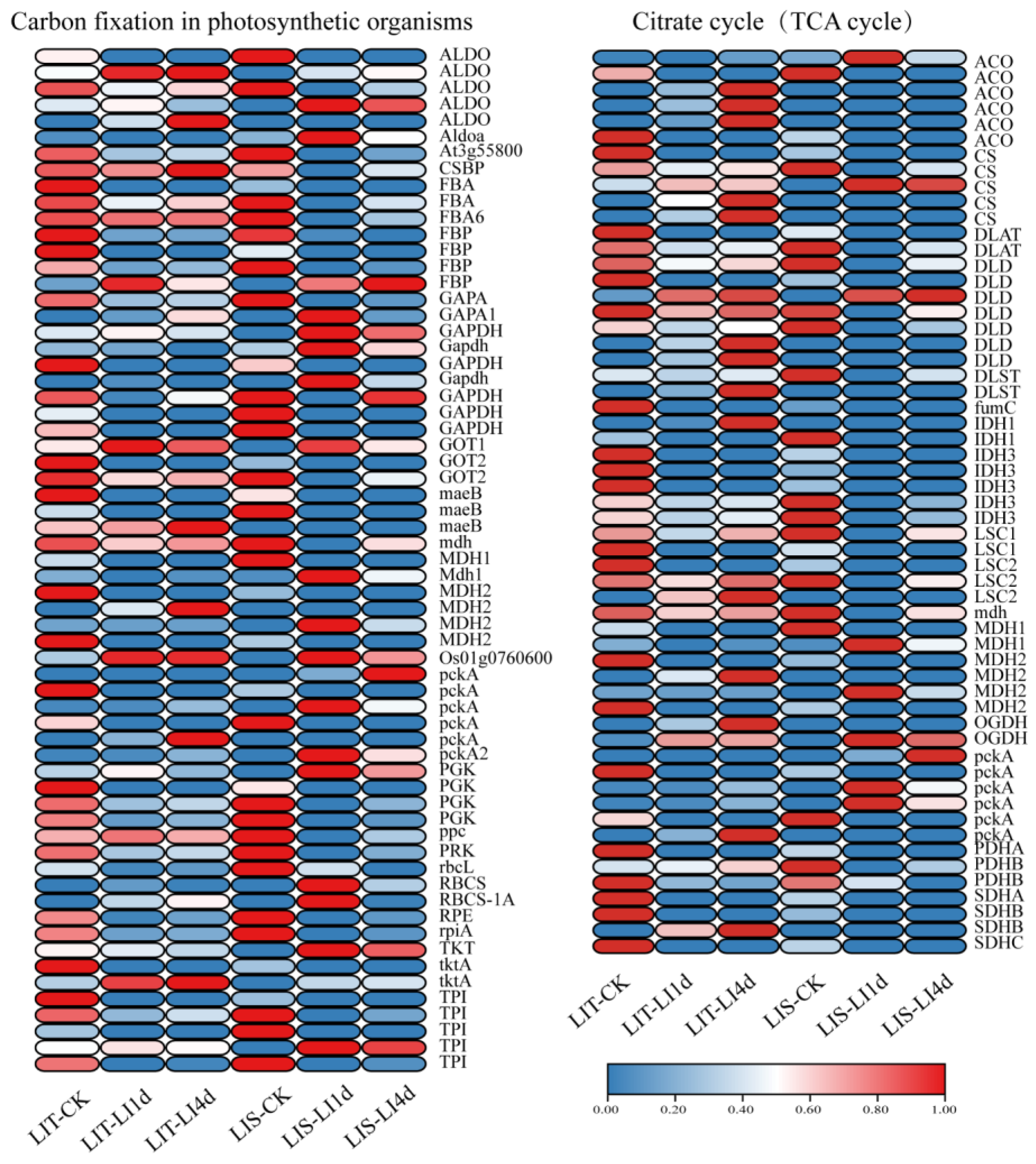
3.6. KEGG Enrichment Analysis
4. Discussion
4.1. Effects of LI Stress on P. haitanensis Blades
4.2. Mechanism Underlying P. haitanensis Resistance to LI Stress
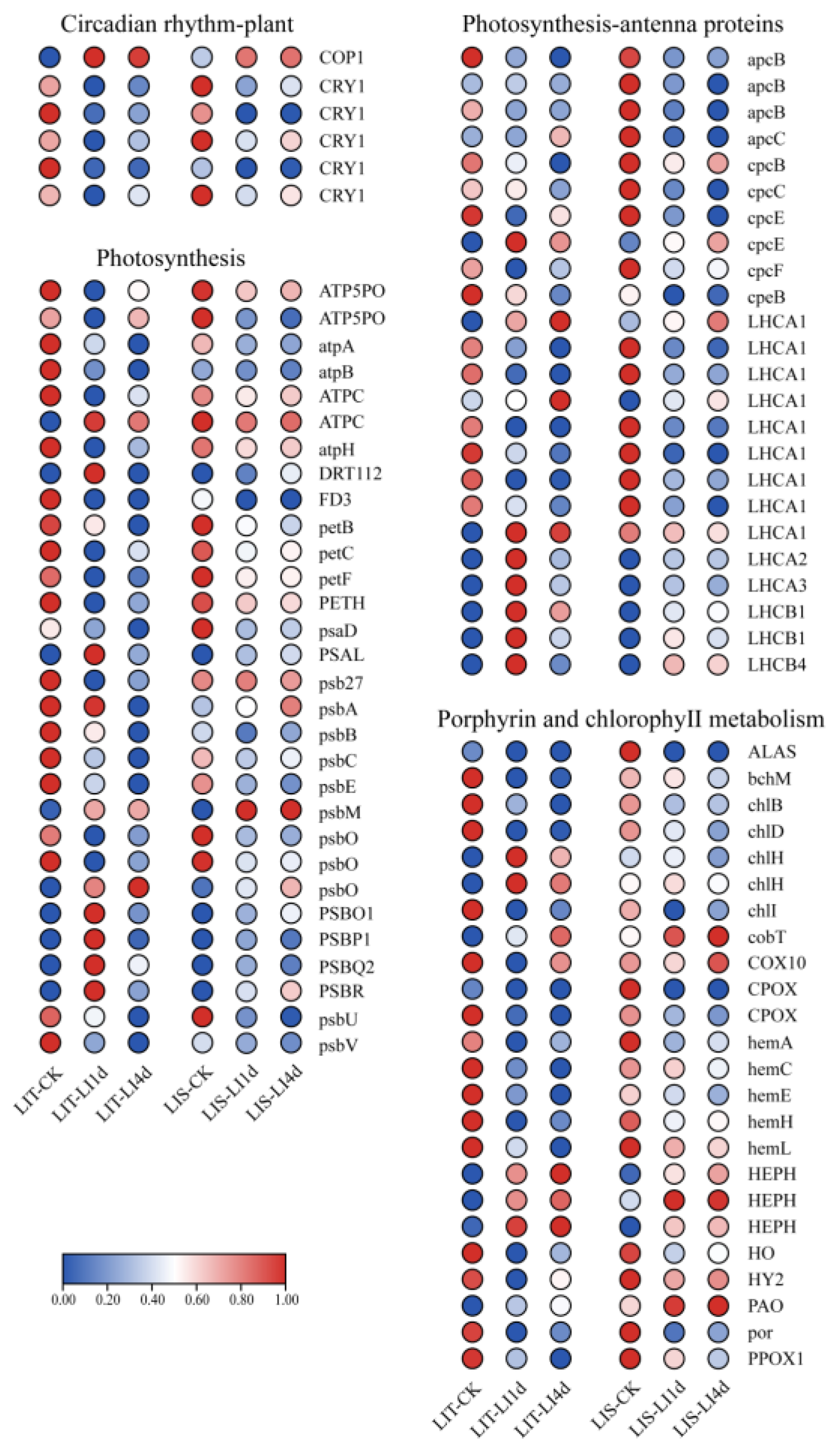
4.2.1. Improved Photosynthetic Efficiency
4.2.2. Enhanced Signal Transduction
4.2.3. Regulation of Redox Homeostasis and Energy Supply
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blouin, N.A.; Brodie, J.A.; Grossman, A.C.; Xu, P.; Brawley, S.H. Porphyra: A marine crop shaped by stress. Trends Plant Sci. 2011, 16, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Stoyneva-Gärtner, M.; Uzunov, B. An ethnobiological glance on globalization impact on the traditional use of algae and fungi as food in Bulgaria. J. Nutr. Food Sci. 2015, 5, 413. [Google Scholar]
- Cho, T.J.; Rhee, M.S. Health functionality and quality control of laver (Porphyra, Pyropia): Current issues and future perspectives as an edible seaweed. Mar. Drugs 2019, 18, 14. [Google Scholar] [CrossRef] [PubMed]
- Duarte, C.M.; Bruhn, A.; Krause-Jensen, D. A seaweed aquaculture imperative to meet global sustainability targets. Nat. Sustain. 2022, 5, 185–193. [Google Scholar] [CrossRef]
- FAO. Fishery and Aquaculture Statistics. Global Aquaculture Production; FAO: Rome, Italy, 2019; Available online: http://www.fao.org/fishery/ (accessed on 18 September 2019).
- Xu, K.; Li, M.; Wang, W.; Xu, Y.; Ji, D.; Chen, C.; Xie, C. Differences in organic carbon release between conchocelis and thalli of Pyropia haitanensis and responses to changes in light intensity and pH. Algal Res. 2022, 61, 102574. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, W.; Yu, H. Effects of exogenous epibrassinolide on photosynthetic characteristics in tomato (Lycopersicon esculentum Mill) seedlings under weak light stress. J. Agric. Food Chem. 2010, 58, 3642–3645. [Google Scholar] [CrossRef]
- Zivcak, M.; Brestic, M.; Kalaji, H.M. Photosynthetic responses of sun-and shade-grown barley leaves to high light: Is the lower PSII connectivity in shade leaves associated with protection against excess of light? Photosynth. Res. 2014, 119, 339–354. [Google Scholar] [CrossRef]
- Lu, T.; Yu, H.; Li, Q.; Chai, L.; Jiang, W. Improving plant growth and alleviating photosynthetic inhibition and oxidative stress from low-light stress with exogenous GR24 in tomato (Solanum lycopersicum L.) seedlings. Front. Plant Sci. 2019, 10, 490. [Google Scholar] [CrossRef]
- Li, H.; Jiang, D.; Wollenweber, B.; Dai, T.; Cao, W. Effects of shading on morphology, physiology and grain yield of winter wheat. Eur. J. Agron. 2010, 33, 267–275. [Google Scholar] [CrossRef]
- Wang, Q.; Hou, F.; Dong, S.; Xie, B.; Li, A.; Zhang, H.; Zhang, L. Effects of shading on the photosynthetic capacity, endogenous hormones and root yield in purple-fleshed sweetpotato (Ipomoea batatas (L.) Lam). Plant Growth Regul. 2014, 72, 113–122. [Google Scholar] [CrossRef]
- Murchie, E.; Horton, P. Acclimation of photosynthesis to irradiance and spectral quality in British plant species: Chlorophyll content, photosynthetic capacity and habitat preference. Plant Cell Environ. 1997, 20, 438–448. [Google Scholar] [CrossRef]
- Fan, Y.; Chen, J.; Wang, Z.; Tan, T.; Li, S.; Li, J.; Wang, B.; Zhang, J.; Cheng, Y.; Wu, X. Soybean (Glycine max L. Merr.) seedlings response to shading: Leaf structure, photosynthesis and proteomic analysis. BMC Plant Biol. 2019, 19, 34. [Google Scholar] [CrossRef]
- Favaretto, V.F.; Martinez, C.A.; Soriani, H.H.; Furriel, R.P. Differential responses of antioxidant enzymes in pioneer and late-successional tropical tree species grown under sun and shade conditions. Environ. Exp. Bot. 2011, 70, 20–28. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, K.; Wang, W.; Xu, Y.; Ji, D.; Chen, C.; Xie, C. Physiological differences in photosynthetic inorganic carbon utilization between gametophytes and sporophytes of the economically important red algae Pyropia haitanensis. Algal Res. 2019, 39, 101436. [Google Scholar] [CrossRef]
- Jensen, A. Chlorophylls and Carotenoids; Cambridge University Press: London, UK, 1978; pp. 59–70. [Google Scholar]
- Fang-Yuan, Y.; Guy, R.D. Variable chlorophyll fluorescence in response to water plus heat stress treatments in three coniferous tree seedlings. J. For. Res. 2004, 15, 24–28. [Google Scholar] [CrossRef]
- Zhang, Y.; Xie, C.; Chen, C.; Ji, D.; Zhou, W. Physiological responses of gametophytic blades of Porphyra haitanensis to rising temperature stresses. J. Fish. China 2011, 35, 379–386. [Google Scholar]
- Li, B.; Chen, C.; Xu, Y.; Ji, D.; Xie, C. Validation of housekeeping genes as internal controls for studying the gene expression in Pyropia haitanensis (Bangiales, Rhodophyta) by quantitative real-time PCR. Acta Oceanol. Sin. 2014, 33, 152–159. [Google Scholar] [CrossRef]
- Bell, G.; Danneberger, T.; McMahon, M. Spectral irradiance available for turfgrass growth in sun and shade. Crop Sci. 2000, 40, 189–195. [Google Scholar] [CrossRef]
- Podolec, R.; Ulm, R. Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 2018, 45, 18–25. [Google Scholar] [CrossRef]
- Lau, O.S.; Deng, X.W. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012, 17, 584–593. [Google Scholar] [CrossRef]
- Osterlund, M.T.; Hardtke, C.S.; Wei, N.; Deng, X.W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 2000, 405, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zuo, Z.; Liu, H.; Liu, X.; Lin, C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011, 25, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Gommers, C.M.; Visser, E.J.; St Onge, K.R.; Voesenek, L.A.; Pierik, R. Shade tolerance: When growing tall is not an option. Trends Plant Sci. 2013, 18, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Valladares, F.; Niinemets, Ü. Shade tolerance, a key plant feature of complex nature and consequences. Annu. Rev.Ecol.Evol. Syst. 2008, 39, 237–257. [Google Scholar] [CrossRef]
- Lin, Y.; Xu, K.; Xu, Y.; Ji, D.; Chen, C.; Wang, W.; Xie, C. Transcriptome Co-expression Network Analysis Identifies Key Genes Regulating Conchosporangia Maturation of Pyropia haitanensis. Front. Genet. 2021, 12, 680120. [Google Scholar] [CrossRef]
- Wang, W.; Teng, F.; Lin, Y.; Ji, D.; Xu, Y.; Chen, C.; Xie, C. Transcriptomic study to understand thermal adaptation in a high temperature-tolerant strain of Pyropia haitanensis. PLoS ONE 2018, 13, e0195842. [Google Scholar]
- Zheng, H.; Xu, Y.; Ji, D.; Xu, K.; Chen, C.; Wang, W.; Xie, C. Calcium-Calmodulin-Involved Heat Shock Response of Neoporphyra haitanensis. Front. Mar. Sci. 2022, 9, 875308. [Google Scholar] [CrossRef]
- Xu, P.; Lloyd, C.W.; Staiger, C.J.; Drobak, B. Association of phosphatidylinositol 4-kinase with the plant cytoskeleton. Plant Cell 1992, 4, 941–951. [Google Scholar] [CrossRef]
- Arisz, S.A.; Testerink, C.; Munnik, T. Plant PA signaling via diacylglycerol kinase. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2009, 1791, 869–875. [Google Scholar] [CrossRef]
- Kue Foka, I.C.; Ketehouli, T.; Zhou, Y.; Li, X.-W.; Wang, F.-W.; Li, H. The emerging roles of diacylglycerol kinase (DGK) in plant stress tolerance, growth, and development. Agronomy 2020, 10, 1375. [Google Scholar] [CrossRef]
- Ohlrogge, J.; Browse, J. Lipid biosynthesis. Plant Cell 1995, 7, 957. [Google Scholar]
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2009, 2, ra45. [Google Scholar] [CrossRef]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef]
- Chen, J.; Chen, Y.; Yang, R.; Luo, Q.; Chen, H.; Xu, J.; Yan, X.; Shen, B. Response to wound-activated stress through a lipid oxidative metabolic pathway in Pyropia haitanensis. Algal Res. 2018, 35, 206–214. [Google Scholar] [CrossRef]
- Wang, W.; Xing, L.; Xu, K.; Ji, D.; Xu, Y.; Chen, C.; Xie, C. Salt stress-induced H2O2 and Ca2+ mediate K+/Na+ homeostasis in Pyropia haitanensis. J. Appl. Phycol. 2020, 32, 4199–4210. [Google Scholar] [CrossRef]
- Feng, Z.; Wu, L.; Sun, Z.; Yang, J.; Liu, G.; Niu, J.; Wang, G. Control of Reactive Oxygen Species through Antioxidant Enzymes Plays a Pivotal Role during the Cultivation of Neopyropia yezoensis. J. Mar. Sci. Eng. 2022, 10, 109. [Google Scholar] [CrossRef]
- Li, C.; Kong, F.; Sun, P.; Bi, G.; Li, N.; Mao, Y.; Sun, M. Genome-wide identification and expression pattern analysis under abiotic stress of mitogen-activated protein kinase genes in Pyropia yezoensis. J. Appl. Phycol. 2018, 30, 2561–2572. [Google Scholar] [CrossRef]
- Kong, F.; Cao, M.; Li, N.; Sun, B.; Sun, M.; Mao, Y. Genome-wide identification, phylogeny, and expressional profiles of the mitogen-activated protein kinase kinase kinase (MAPKKK) gene family in Pyropia yezoensis. Front. Mar. Sci. 2020, 7, 193. [Google Scholar] [CrossRef]
- Zhang, S.; Klessig, D.F. MAPK cascades in plant defense signaling. Trends Plant Sci. 2001, 6, 520–527. [Google Scholar] [CrossRef]
- Zhang, M.; Su, J.; Zhang, Y.; Xu, J.; Zhang, S. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 2018, 45, 1–10. [Google Scholar] [CrossRef]
- Kong, F.; Dong, D.; Li, N.; Sun, B.; Sun, M. Characterization of PyMAPK2, a D group mitogen-activated protein kinase gene from Pyropia yezoensis responding to various abiotic stress. Algal Res. 2021, 59, 102445. [Google Scholar] [CrossRef]
- Wang, W.; Zheng, H.; Wen, J.; Xu, K.; Xu, Y.; Ji, D.; Chen, C.; Xie, C. Early signaling events in the heat stress response of Pyropia haitanensis revealed by phosphoproteomic and lipidomic analyses. Algal Res. 2022, 67, 102837. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Xu, Y.; Xiao, H.; Chen, C.; Xu, K.; Xie, C. Superoxide dismutase genes in Pyropia haitanensis: Molecular cloning, characterization and mRNA expression. Acta Oceanol. Sin. 2016, 35, 101–111. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Chen, T.; Xing, L.; Xu, K.; Xu, Y.; Ji, D.; Chen, C.; Xie, C. Regulatory mechanisms underlying the maintenance of homeostasis in Pyropia haitanensis under hypersaline stress conditions. Sci. Total Environ. 2019, 662, 168–179. [Google Scholar] [CrossRef]
- Wang, W.; Chen, T.; Xu, Y.; Xu, K.; Ji, D.; Chen, C.; Xie, C. Investigating the mechanisms underlying the hyposaline tolerance of intertidal seaweed, Pyropia haitanensis. Algal Res. 2020, 47, 101886. [Google Scholar] [CrossRef]
- Chen, H.; Chu, J.S.-C.; Chen, J.; Luo, Q.; Wang, H.; Lu, R.; Zhu, Z.; Yuan, G.; Yi, X.; Mao, Y. Insights into the ancient adaptation to intertidal environments by red algae based on a genomic and multiomics investigation of Neoporphyra haitanensis. Mol. Biol. Evol. 2022, 39, msab315. [Google Scholar] [CrossRef]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef]
- Li, X.C.; Xing, Y.Z.; Jiang, X.; Qiao, J.; Tan, H.L.; Tian, Y.; Zhou, B. Identification and characterization of the catalase gene pycat from the red alga Pyropia yezoensis (Bangiales, Rhodophyta) 1. J. Phycol. 2012, 48, 664–669. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, C.; Ji, D.; Hang, N.; Xie, C. Proteomic profile analysis of Pyropia haitanensis in response to high-temperature stress. J. Appl. Phycol. 2014, 26, 607–618. [Google Scholar] [CrossRef]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, D.; Zhang, Y.; Zhang, B.; Xu, Y.; Xu, K.; Chen, C.; Xie, C. Investigating the Mechanisms Underlying the Low Irradiance-Tolerance of the Economically Important Seaweed Species Pyropia haitanensis. Life 2023, 13, 481. https://doi.org/10.3390/life13020481
Ji D, Zhang Y, Zhang B, Xu Y, Xu K, Chen C, Xie C. Investigating the Mechanisms Underlying the Low Irradiance-Tolerance of the Economically Important Seaweed Species Pyropia haitanensis. Life. 2023; 13(2):481. https://doi.org/10.3390/life13020481
Chicago/Turabian StyleJi, Dehua, Yichi Zhang, Bao Zhang, Yan Xu, Kai Xu, Changsheng Chen, and Chaotian Xie. 2023. "Investigating the Mechanisms Underlying the Low Irradiance-Tolerance of the Economically Important Seaweed Species Pyropia haitanensis" Life 13, no. 2: 481. https://doi.org/10.3390/life13020481
APA StyleJi, D., Zhang, Y., Zhang, B., Xu, Y., Xu, K., Chen, C., & Xie, C. (2023). Investigating the Mechanisms Underlying the Low Irradiance-Tolerance of the Economically Important Seaweed Species Pyropia haitanensis. Life, 13(2), 481. https://doi.org/10.3390/life13020481




