Structured, Harmonized, and Interoperable Integration of Clinical Routine Data to Compute Heart Failure Risk Scores
Abstract
:1. Introduction
2. Materials and Methods
2.1. Management
2.2. Data
- (1)
- Age ≥ 18 years;
- (2)
- Established diagnosis of chronic heart failure;
- (3)
- Capacity to consent;
- (4)
- Completed patient information and written informed consent.
- (1)
- Life expectancy less than 6 months due to non-cardiac pre-existing conditions;
- (2)
- Incapacity to consent.
2.3. OpenEHR Archetypes and Templates
2.4. Primary Systems and Data Integration
2.5. Data Retrieval and Preparation
- Personal data: calculate age from birth date.
- Medical history: calculate BMI from weight and height, calculate HF duration and HF duration ≥ 18 months, assign levels to categorical variables.
- Medication: (1) we use information on medication that is closest to the date of patient recruitment. (2) If applicable, we map medication groups from the openEHR template to synonymous groups required for score calculation.
- Echocardiography: again, we use the information closest to the date on which the patient was recruited.
- Laboratory Data: again, we use the information closest to the date on which the patient was recruited.
2.6. Plausibility Checks
2.7. Score Calculation
2.8. Further Assumptions
- Personal data—birth date: Both scores require the age of the patient. Due to privacy restrictions, the year of birth, but not the exact birth date, was oftentimes only available for many patients. In the absence of an exact birth date, we set the birth date to the 1st July of the respective year.
- Medical history—COPD: The MAGGIC score requires the diagnosis of COPD, which is by definition based on a spirometry measurement [12]. This was replaced with information extracted from anamnesis.
- Patient history—first diagnosis date of HF: The Barcelona BioHF V1 and the MAGGIC score require the HF duration and the HF duration ≥ 18 months on monthly precision, respectively. For patients who had only the year of the first diagnosis of HF documented, we set the date of HF onset to the 1st July of the respective year.
- Medication: Information that a drug was not prescribed is not documented in routine clinical practice. If the drug was not recorded in the medication list closest to the medical history date, it was assumed that it was not taken.
- Medication—loop diuretic: The Barcelona BioHF Score requires dosing information in terms of furosemide and torasemide equivalents. In case of absence of this information, we equated documented loop diuretic use as the lower dosage of furosemide, because this was the usual dosage at all participating hospitals.
3. Results
3.1. The OpenEHR Approach, Data Queries, Processing, and Score Calculation Steps
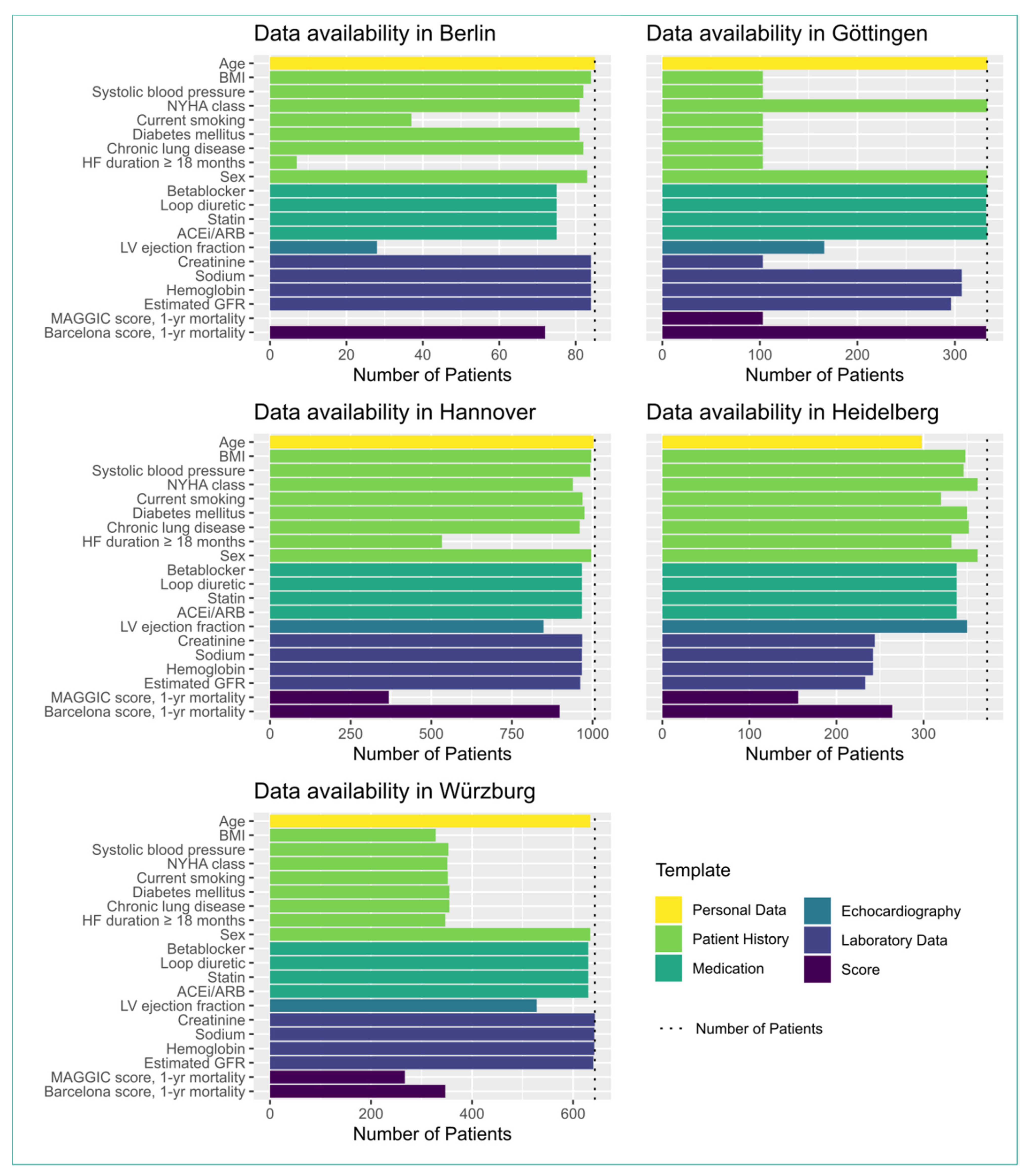
3.2. Data Integration
- If a necessary data type was rarely collected during routine visits or was not collected at all, these data were lacking for the study. In Berlin, the HF duration ≥ 18 months and the smoking status were taken from discharge letters, which seldom contained this information or the information was asked from the patients themselves, who often could not properly remember the exact point in time of their first heart failure event. Therefore, those data types had a low availability in Berlin.
- At some sites, the collection of data within some source systems began after the start of the study. This is the case for Würzburg, where the patient history was collected via a dedicated patient history form. This form was implemented after the start of the study. Therefore, the patient history data were only available for patients for which the anamnesis data were collected using this form, or for returning patients who visited the clinic another time.
- We tried to automate the transfer to the target openEHR systems as much as possible. However, due to technical or organizational reasons, there were parts of the ETL processes where manual work was needed to integrate the data into the target systems. In Berlin, the echocardiography report was transcribed manually from the source system to the openEHR template. This led to a reduced availability in the LVEF values in Berlin.
- In an ideal scenario, ETL processes directly retrieve data from streams originating from the source systems. This ensures that the target system (i.e., MeDIC) is in sync with the source system (e.g., by listening to an HL7-communication server). Unfortunately, some partner sites use data snapshots rather than data streams to fill their openEHR platform. This may lead to very different proportions of recruited vs. documented patients at the respective partner sites.
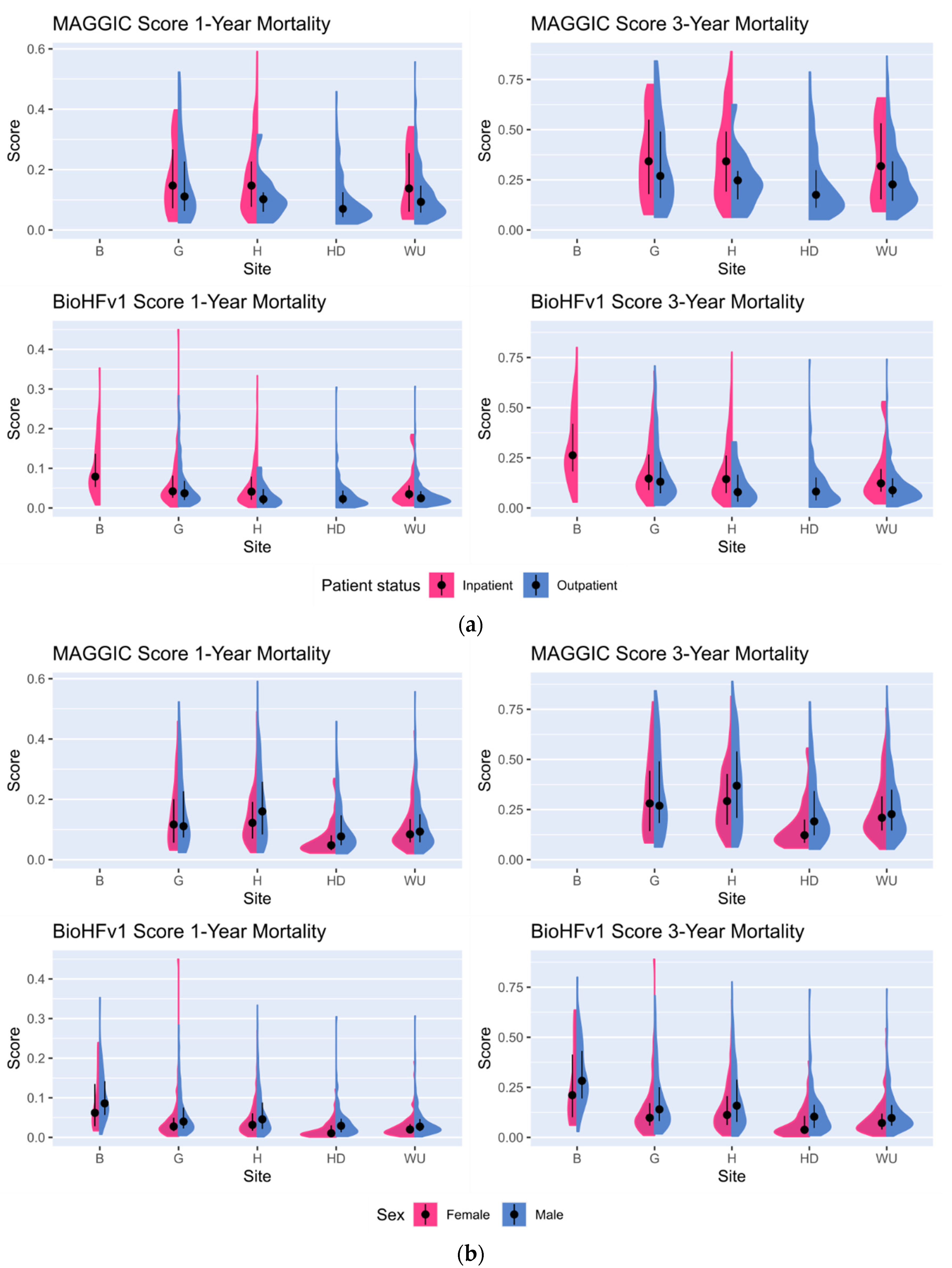
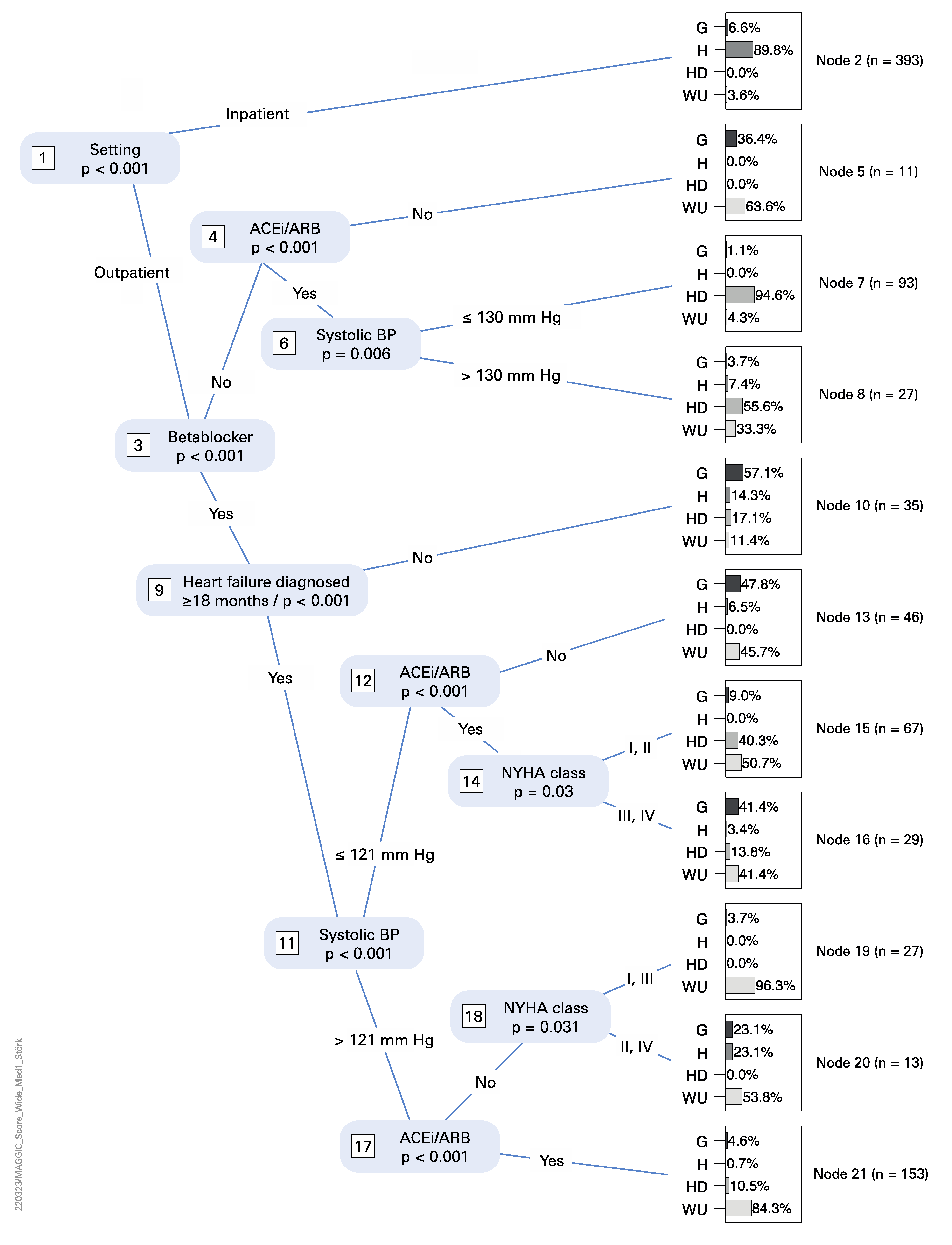
3.3. Statistical Analysis
4. Discussion
4.1. The OpenEHR Approach, Data Queries, Processing, and Score Calculation Steps
4.2. Data Integration
- 1.
- In the case of existing data, availability issues need to be solved, such as:
- (a)
- More ETL routes have to be established from the primary systems to the data integration centers.
- (b)
- Vendors of medical devices need to be encouraged to enable reusability of recorded data by integrating export APIs into their products.
- (c)
- Source systems, which currently provide data in unstructured free text form, have to be transformed into structured data, providing systems, so that no error-prone manual or automatic information extraction is needed.
- 2.
- In the case of missing data (e.g., the “HF duration ≥ 18 month” in the present study), reasons for its absence need to be identified. Potential reasons may include:
- (a)
- Time constraints on detailed documentation in routine care, e.g., it is too time consuming for the documenting physician to capture the data item.
- (b)
- Alternatively, the physician considers data as not necessary for a particular patient, despite its value in secondary use (e.g., research).
4.3. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. AQL Queries
References
- Seferović, P.M.; Vardas, P.; Jankowska, E.A.; Maggioni, A.P.; Timmis, A.; Milinković, I.; Polovina, M.; Gale, C.P.; Lund, L.H.; Lopatin, Y.; et al. The Heart Failure Association Atlas: Heart Failure Epidemiology and Management Statistics 2019. Eur. J. Heart Fail. 2021, 23, 906–914. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Böhm, M.; Burri, H.; Butler, J.; Čelutkienė, J.; Chioncel, O.; et al. 2021 ESC Guidelines for the Diagnosis and Treatment of Acute and Chronic Heart Failure: Developed by the Task Force for the Diagnosis and Treatment of Acute and Chronic Heart Failure of the European Society of Cardiology (ESC). With the Special Contribution of the Heart Failure Association (HFA) of the ESC. Eur. J. Heart Fail. 2022, 24, 4–131. [Google Scholar] [CrossRef]
- Christ, M.; Störk, S.; Dörr, M.; Heppner, H.J.; Müller, C.; Wachter, R.; Riemer, U. Trend HF Germany Project Heart Failure Epidemiology 2000–2013: Insights from the German Federal Health Monitoring System. Eur. J. Heart Fail. 2016, 18, 1009–1018. [Google Scholar] [CrossRef] [Green Version]
- Störk, S.; Handrock, R.; Jacob, J.; Walker, J.; Calado, F.; Lahoz, R.; Hupfer, S.; Klebs, S. Epidemiology of Heart Failure in Germany: A Retrospective Database Study. Clin. Res. Cardiol. 2017, 106, 913–922. [Google Scholar] [CrossRef] [Green Version]
- Lloyd-Jones, D.M.; Braun, L.T.; Ndumele, C.E.; Smith, S.C.; Sperling, L.S.; Virani, S.S.; Blumenthal, R.S. Use of Risk Assessment Tools to Guide Decision-Making in the Primary Prevention of Atherosclerotic Cardiovascular Disease: A Special Report From the American Heart Association and American College of Cardiology. Circulation 2019, 139, e1162–e1177. [Google Scholar] [CrossRef]
- Parcha, V.; Malla, G.; Kalra, R.; Patel, N.; Sanders-van Wijk, S.; Pandey, A.; Shah, S.J.; Arora, G.; Arora, P. Diagnostic and Prognostic Implications of Heart Failure with Preserved Ejection Fraction Scoring Systems. ESC Heart Fail. 2021, 8, 2089–2102. [Google Scholar] [CrossRef] [PubMed]
- Toumpourleka, M.; Patoulias, D.; Katsimardou, A.; Doumas, M.; Papadopoulos, C. Risk Scores and Prediction Models in Chronic Heart Failure: A Comprehensive Review. Curr. Pharm. Des. 2021, 27, 1289–1297. [Google Scholar] [CrossRef] [PubMed]
- Haarbrandt, B.; Schreiweis, B.; Rey, S.; Sax, U.; Scheithauer, S.; Rienhoff, O.; Knaup-Gregori, P.; Bavendiek, U.; Dieterich, C.; Brors, B.; et al. HiGHmed—An Open Platform Approach to Enhance Care and Research across Institutional Boundaries. Methods Inf. Med. 2018, 57, e66–e81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wulff, A.; Sommer, K.K.; Ballout, S.; Haarbrandt, B.; Gietzelt, M.; HiGHmed Consortium; Haarbrandt, B.; Gietzelt, M. A Report on Archetype Modelling in a Nationwide Data Infrastructure Project. Stud. Health Technol. Inform. 2019, 258, 146–150. [Google Scholar] [CrossRef]
- Pocock, S.J.; Ariti, C.A.; McMurray, J.J.V.; Maggioni, A.; Køber, L.; Squire, I.B.; Swedberg, K.; Dobson, J.; Poppe, K.K.; Whalley, G.A.; et al. Predicting Survival in Heart Failure: A Risk Score Based on 39 372 Patients from 30 Studies. Eur. Heart J. 2013, 34, 1404–1413. [Google Scholar] [CrossRef]
- Lupón, J.; de Antonio, M.; Vila, J.; Peñafiel, J.; Galán, A.; Zamora, E.; Urrutia, A.; Bayes-Genis, A. Development of a Novel Heart Failure Risk Tool: The Barcelona Bio-Heart Failure Risk Calculator (BCN Bio-HF Calculator). PLoS ONE 2014, 9, e85466. [Google Scholar] [CrossRef] [PubMed]
- Rabe, K.F.; Hurd, S.; Anzueto, A.; Barnes, P.J.; Buist, S.A.; Calverley, P.; Fukuchi, Y.; Jenkins, C.; Rodriguez-Roisin, R.; van Weel, C.; et al. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Pulmonary Disease: GOLD Executive Summary. Am. J. Respir. Crit. Care Med. 2007, 176, 532–555. [Google Scholar] [CrossRef] [PubMed]
- Hothorn, T.; Zeileis, A. Partykit: A Modular Toolkit for Recursive Partytioning in R. J. Mach. Learn. Res. 2015, 16, 3905–3909. [Google Scholar]
- Dietrich, G.; Krebs, J.; Liman, L.; Fette, G.; Ertl, M.; Kaspar, M.; Störk, S.; Puppe, F. Replicating Medication Trend Studies Using Ad Hoc Information Extraction in a Clinical Data Warehouse. BMC Med. Inform. Decis. Mak. 2019, 19, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| OpenEHR Template | Data Items * |
|---|---|
| Personal Data | Year of birth (B, M) |
| Patient history | Sex (B, M) Weight (M) Height (M) Systolic blood pressure (M) NYHA dysfunctional class (B, M) Current smoking (M) Time of first diagnosis of HF (M) Diabetes mellitus (M) Chronic lung disease (M) |
| Medication | Betablocker (B, M) ACEi/ARB (B, M) Statin (B) Loop diuretic (B) |
| Echocardiography | LV ejection fraction (B, M) |
| Laboratory Data | Creatinine (M) [LOINC 14682-9, 2160-0] Sodium (B) [LOINC 2951-2] Hemoglobin (B) [LOINC 718-7, 30350-3] Estimated GFR (B) [LOINC 62238-1] |
| - | Inpatient/Outpatient |
| Template | Hannover | Heidelberg | Göttingen | Würzburg | Berlin |
|---|---|---|---|---|---|
| Personal Data | SAP i.s.h.med | SAP i.s.h.med | SAP IS-H | SAP i.s.h.med | SAP IS-H, Custom 1 |
| Patient History | Custom 1 | Custom 1 | Custom 1 | SAP i.s.h.med, ignimed 1 | Custom 1 |
| Medication | Custom 1 | Custom 1 | Custom 1 | SAP i.s.h.med, Meona | Custom 1 |
| Echocardiography | Philips IntelliSpace | MySQL DB solution | GE Healthcare Carddas | KardioText EchoText | Custom 1 |
| Laboratory Data | OSM Opus::L | Nexus Swisslab | OSM Opus::L | Nexus Swisslab | Medat; GLIMS |
| Variable | Lower Limit | Upper Limit | Unit |
|---|---|---|---|
| Age | 18 | 110 | years |
| BMI | 14 | 60 | kg/m2 |
| Blood pressure systolic | 70 | 250 | mm [Hg] |
| HF duration * | 0 | Age * 12 | months |
| LV-EF | 4 | 85 | % |
| Creatinine | 26.526 | 1326.3 | µmol/L |
| Sodium | 120 | 150 | mmol/L |
| Hemoglobin | 5 | 20 | g/dL |
| Estimated GFR | 5 | 120 | mL/min/1.73 |
| Partner Sites | |||||
|---|---|---|---|---|---|
| Patient Type | Berlin * | Göttingen | Hannover | Heidelberg | Würzburg |
| Inpatient | 0 | 26 | 353 | 0 | 14 |
| Outpatient | 0 | 77 | 15 | 156 | 253 |
| Total sum | 0 | 103 | 368 | 156 | 267 |
| Partner Sites (Complete/Imputed) | |||||
|---|---|---|---|---|---|
| Patient Type | Berlin * | Göttingen | Hannover | Heidelberg | Würzburg |
| Inpatient | 20/38 | 35/45 | 701/172 | 0 | 14/4 |
| Outpatient | (1) | 105/148 | 22/3 | 179/85 | 277/52 |
| Total sum | 20/38 | 139/193 | 723/175 | 179/85 | 291/56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sommer, K.K.; Amr, A.; Bavendiek, U.; Beierle, F.; Brunecker, P.; Dathe, H.; Eils, J.; Ertl, M.; Fette, G.; Gietzelt, M.; et al. Structured, Harmonized, and Interoperable Integration of Clinical Routine Data to Compute Heart Failure Risk Scores. Life 2022, 12, 749. https://doi.org/10.3390/life12050749
Sommer KK, Amr A, Bavendiek U, Beierle F, Brunecker P, Dathe H, Eils J, Ertl M, Fette G, Gietzelt M, et al. Structured, Harmonized, and Interoperable Integration of Clinical Routine Data to Compute Heart Failure Risk Scores. Life. 2022; 12(5):749. https://doi.org/10.3390/life12050749
Chicago/Turabian StyleSommer, Kim K., Ali Amr, Udo Bavendiek, Felix Beierle, Peter Brunecker, Henning Dathe, Jürgen Eils, Maximilian Ertl, Georg Fette, Matthias Gietzelt, and et al. 2022. "Structured, Harmonized, and Interoperable Integration of Clinical Routine Data to Compute Heart Failure Risk Scores" Life 12, no. 5: 749. https://doi.org/10.3390/life12050749
APA StyleSommer, K. K., Amr, A., Bavendiek, U., Beierle, F., Brunecker, P., Dathe, H., Eils, J., Ertl, M., Fette, G., Gietzelt, M., Heidecker, B., Hellenkamp, K., Heuschmann, P., Hoos, J. D. E., Kesztyüs, T., Kerwagen, F., Kindermann, A., Krefting, D., Landmesser, U., ... Dieterich, C. (2022). Structured, Harmonized, and Interoperable Integration of Clinical Routine Data to Compute Heart Failure Risk Scores. Life, 12(5), 749. https://doi.org/10.3390/life12050749









