Interaction Analysis of MRP1 with Anticancer Drugs Used in Ovarian Cancer: In Silico Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ligand Preparation
2.2. Protein
2.3. Molecular Docking
3. Results
3.1. Homology Modelling of MRP1
3.2. Analysis of Docking Results
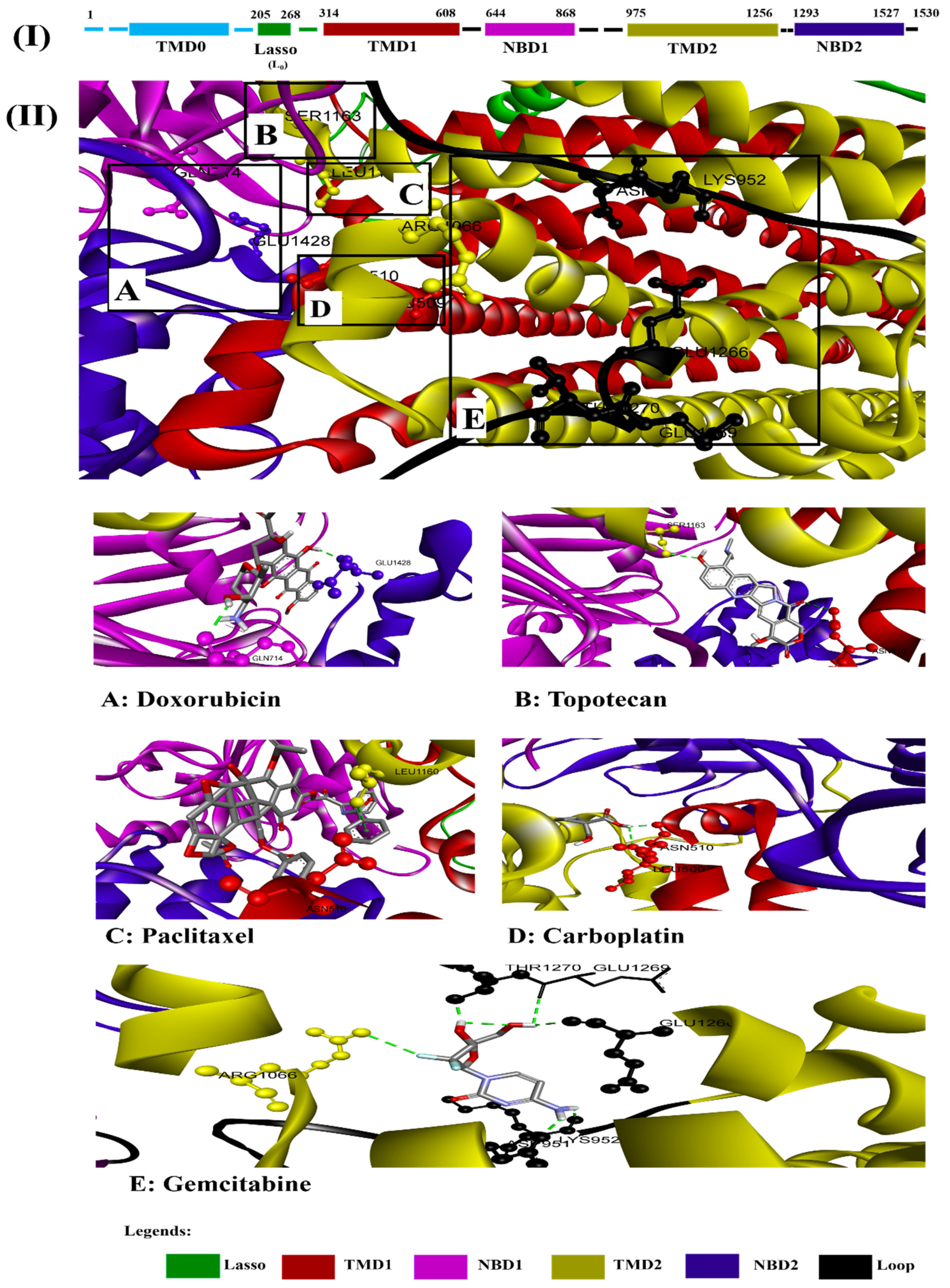
3.2.1. Paclitaxel Demonstrated Strong Interaction with MRP1
3.2.2. Doxorubicin
3.2.3. Topotecan
3.2.4. Gemcitabine
3.2.5. Carboplatin
3.3. Residue Frequencies
3.4. Alignment of Ligands with MRP1 to Predict the Various Binding Sites/Position on Full Protein Structure
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- American Cancer Society. Cancer Facts & Figures; American Cancer Society: Atlanta, GA, USA, 2018; pp. 233–236. [Google Scholar]
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Della Corte, L.; Foreste, V.; Di Filippo, C.; Giampaolino, P.; Bifulco, G. Poly (ADP-ribose) polymerase (PARP) as target for the treatment of epithelial ovarian cancer: What to know. Expert Opin. Investig. Drugs 2021, 30, 543–554. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannistra, S.A. Epithelial Cancer of the Ovary. BMJ 2004, 312, 258. [Google Scholar] [CrossRef]
- Auner, V.; Sehouli, J.; Oskay-Oezcelik, G.; Horvat, R.; Speiser, P.; Zeillinger, R. ABC transporter gene expression in benign and malignant ovarian tissue. Gynecol. Oncol. 2010, 117, 198–201. [Google Scholar] [CrossRef] [PubMed]
- Trimbos, J.B. Surgical treatment of early-stage ovarian cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 41, 60–70. [Google Scholar] [CrossRef]
- Wright, A.A.; Cronin, A.; Milne, D.E.; Bookman, M.A.; Burger, R.A.; Cohn, D.E.; Cristea, M.C.; Griggs, J.J.; Keating, N.L.; Levenback, C.F.; et al. Use and effectiveness of intraperitoneal chemotherapy for treatment of ovarian cancer. J. Clin. Oncol. 2015, 33, 2841–2847. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Antibiotic Adjuvants: Rescuing Antibiotics from Resistance. Trends Microbiol. 2016, 24, 862–871. [Google Scholar] [CrossRef]
- Elias, K.M.; Guo, J.; Bast, R.C.J. Early Detection of Ovarian Cancer. Hematol. Oncol. Clin. N. Am. 2018, 32, 903–914. [Google Scholar] [CrossRef] [Green Version]
- Ren, F.; Shen, J.; Shi, H.; Hornicek, F.J.; Kan, Q.; Duan, Z. Novel mechanisms and approaches to overcome multidrug resistance in the treatment of ovarian cancer. Biochim. Biophys. Acta—Rev. Cancer 2016, 1866, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Tropé, C.; Kaern, J. Primary surgery for ovarian cancer. Eur. J. Surg. Oncol. 2006, 32, 844–852. [Google Scholar] [CrossRef]
- Kartal-Yandim, M.; Adan-Gokbulut, A.; Baran, Y. Molecular mechanisms of drug resistance and its reversal in cancer. Crit. Rev. Biotechnol. 2016, 36, 716–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jibodh, R.A.; Lagas, J.S.; Nuijen, B.; Beijnen, J.H.; Schellens, J.H.M. Taxanes: Old drugs, new oral formulations. Eur. J. Pharmacol. 2013, 717, 40–46. [Google Scholar] [CrossRef]
- Conseil, G.; Deeley, R.G.; Cole, S.P.C. Role of two adjacent cytoplasmic tyrosine residues in MRP1 (ABCC1) transport activity and sensitivity to sulfonylureas. Biochem. Pharmacol. 2005, 69, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Sirisha, K.; Shekhar, M.C.; Umasankar, K.; Mahendar, P.; Sadanandam, A.; Achaiah, G.; Reddy, V.M. Molecular docking studies and in vitro screening of new dihydropyridine derivatives as human MRP1 inhibitors. Bioorg. Med. Chem. 2011, 19, 3249–3254. [Google Scholar] [CrossRef]
- He, S.-M.; Li, R.; Kanwar, J.R.; Zhou, S.-F. Structural and Functional Properties of Human Multidrug Resistance Protein 1 (MRP1/ABCC1). Curr. Med. Chem. 2012, 18, 439–481. [Google Scholar] [CrossRef]
- Tong, X.; Zhao, J.; Zhang, Y.; Mu, P.; Wang, X. Expression levels of MRP1, GST-π, and GSK3Β in ovarian cancer and the relationship with drug resistance and prognosis of patients. Oncol. Lett. 2019, 18, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Cole, S.P.C. Multidrug resistance protein 1 (mrp1, abcc1), a “multitasking” atp-binding cassette (abc,) transporter. J. Biol. Chem. 2014, 289, 30880–30888. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.X.; Cui, L.; Riordan, J.R.; Chang, X.B. ATP binding to the first nucleotide-binding domain of multidrug resistance protein MRP1 increases binding and hydrolysis of ATP and trapping of ADP at the second domain. J. Biol. Chem. 2002, 277, 5110–5119. [Google Scholar] [CrossRef] [Green Version]
- Obata, H.; Yahata, T.; Quan, J.; Sekine, M.; Tanaka, K. Association between single nucleotide polymorphisms of drug resistance-associated genes and response to chemotherapy in advanced ovarian cancer. Anticancer Res. 2006, 26, 2227–2232. [Google Scholar]
- Wang, Y.; Xiao, J.; Suzek, T.O.; Zhang, J.; Wang, J.; Bryant, S.H. PubChem: A public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009, 37, 623–633. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- BIOVIA; Dassault Systèmes. Discovery Studio Visualizer, v21.1.0.20298; Dassault Systèmes: San Diego, CA, USA, 2021; Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 21 December 2021).
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Bagnoli, M.; Beretta, G.L.; Gatti, L.; Pilotti, S.; Alberti, P.; Tarantino, E.; Barbareschi, M.; Canevari, S.; Mezzanzanica, D.; Perego, P. Clinicopathological impact of ABCC1/MRP1 and ABCC4/MRP4 in epithelial ovarian carcinoma. BioMed Res. Int. 2013, 2013, 143202. [Google Scholar] [CrossRef]
- Gillet, J.P.; Efferth, T.; Remacle, J. Chemotherapy-induced resistance by ATP-binding cassette transporter genes. Biochim. Biophys. Acta—Rev. Cancer 2007, 1775, 237–262. [Google Scholar] [CrossRef]
- Shuang, T.; Wang, M.; Zhou, Y.; Shi, C. Over-expression of nuclear NF-κB1 and c-Rel correlates with chemoresistance and prognosis of serous epithelial ovarian cancer. Exp. Mol. Pathol. 2016, 100, 139–144. [Google Scholar] [CrossRef]
- Ye, Q.; Liu, K.; Shen, Q.; Li, Q.; Hao, J.; Han, F.; Jiang, R.W. Reversal of multidrug resistance in cancer by multi-functional flavonoids. Front. Oncol. 2019, 9, 487. [Google Scholar] [CrossRef] [Green Version]
- Kathawala, R.J.; Gupta, P.; Ashby, C.R.; Chen, Z.S. The modulation of ABC transporter-mediated multidrug resistance in cancer: A review of the past decade. Drug Resist. Updates 2015, 18, 1–17. [Google Scholar] [CrossRef]
- Dhasmana, D.; Singh, A.; Shukla, R.; Tripathi, T.; Garg, N. Targeting Nucleotide Binding Domain of Multidrug Resistance-associated Protein-1 (MRP1) for the Reversal of Multi Drug Resistance in Cancer. Sci. Rep. 2018, 8, 11973. [Google Scholar] [CrossRef] [PubMed]
- Podolski-Renić, A.; Anpelković, T.; Banković, J.; Tanić, N.; Ruždijić, S.; Pešić, M. The role of paclitaxel in the development and treatment of multidrug resistant cancer cell lines. Biomed. Pharmacother. 2011, 65, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, K.; Igishi, T.; Kawasaki, Y.; Kato, K.; Matsumoto, S.; Katayama, S.; Sako, T.; Shigeoka, Y.; Suyama, H.; Sugitani, A.; et al. Phase II study of weekly paclitaxel in patients with non-small cell lung cancer who have failed previous treatments. Oncology 2004, 66, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.F.; Wang, X.Y.; Fu, Z.Q.; Peng, Q.H.; Zhang, J.Y.; Ye, F.; Fu, Y.F.; Zhou, C.Y.; Lu, W.G.; Cheng, X.D.; et al. TXNDC17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy 2015, 11, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Norouzi, M.; Yathindranath, V.; Thliveris, J.A.; Kopec, B.M.; Siahaan, T.J.; Miller, D.W. Doxorubicin-loaded iron oxide nanoparticles for glioblastoma therapy: A combinational approach for enhanced delivery of nanoparticles. Sci. Rep. 2020, 10, 11292. [Google Scholar] [CrossRef]
- Cagel, M.; Grotz, E.; Bernabeu, E.; Moretton, M.A.; Chiappetta, D.A. Doxorubicin: Nanotechnological overviews from bench to bedside. Drug Discov. Today 2017, 22, 270–281. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Yu, L.; Liu, B.L.; He, X.J.; Zhang, B.Y. Downregulation of P-gp, Ras and p-ERK1/2 contributes to the arsenic trioxide-induced reduction in drug resistance towards doxorubicin in gastric cancer cell lines. Mol. Med. Rep. 2015, 12, 7335–7343. [Google Scholar] [CrossRef] [Green Version]
- Tian, Q.; Zhang, J.; Chan, S.; Chin Tan, T.; Duan, W.; Huang, M.; Zhu, Y.; Chan, E.; Yu, Q.; Nie, Y.; et al. Topotecan Is a Substrate for Multidrug Resistance Associated Protein 4. Curr. Drug Metab. 2005, 7, 105–118. [Google Scholar] [CrossRef]
- Lee, M.W.; Ryu, H.; Song, I.C.; Yun, H.J.; Jo, D.Y.; Ko, Y.B.; Lee, H.J. Efficacy of cisplatin combined with topotecan in patients with advanced or recurrent ovarian cancer as second- or higher-line palliative chemotherapy. Medicine 2020, 99, e19931. [Google Scholar] [CrossRef]
- Berg, T.; Nøttrup, T.J.; Roed, H. Gemcitabine for recurrent ovarian cancer—A systematic review and meta-analysis. Gynecol. Oncol. 2019, 155, 530–537. [Google Scholar] [CrossRef]
- Pfisterer, J.; Plante, M.; Vergote, I.; Du Bois, A.; Wagner, U.; Hirte, H.; Lacave, A.J.; Stähle, A.; Kimmig, R.; Eisenhauer, E. Gemcitabine/carboplatin (GC) vs. carboplatin (C) in platinum sensitive recurrent ovarian cancer (OVCA). Results of a Gynecologic Cancer Intergroup randomized phase III trial of the AGO OVAR, the NCIC CTG and the EORTC GCG. J. Clin. Oncol. 2004, 22, 5005. [Google Scholar] [CrossRef]
- Lorusso, D.; Di Stefano, A.; Fanfani, F.; Scambia, G. Role of gemcitabine in ovarian cancer treatment. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2006, 17 (Suppl. 5), v188–v194. [Google Scholar] [CrossRef] [PubMed]
- Pfisterer, J.; Vergote, I.; Du Bois, A.; Eisenhauer, E. Combination therapy with gemcitabine and carboplatin in recurrent ovarian cancer. Int. J. Gynecol. Cancer Off. J. Int. Gynecol. Cancer Soc. 2005, 15 (Suppl. 1), 36–41. [Google Scholar] [CrossRef]
- Villella, J.; Marchetti, D.; Odunsi, K.; Rodabaugh, K.; Driscoll, D.L.; Lele, S. Response of combination platinum and gemcitabine chemotherapy for recurrent epithelial ovarian carcinoma. Gynecol. Oncol. 2004, 95, 539–545. [Google Scholar] [CrossRef]
- Mutch, D.G. Gemcitabine combination chemotherapy of ovarian cancer. Gynecol. Oncol. 2003, 90, S16–S20. [Google Scholar] [CrossRef]
- Nasr, F.L.; Chahine, G.Y.; Kattan, J.G.; Farhat, F.S.; Mokaddem, W.T.; Tueni, E.A.; Dagher, J.E.; Ghosn, M.G. Gemcitabine plus carboplatin combination therapy as second-line treatment in patients with relapsed breast cancer. Clin. Breast Cancer 2004, 5, 117–122. [Google Scholar] [CrossRef] [PubMed]



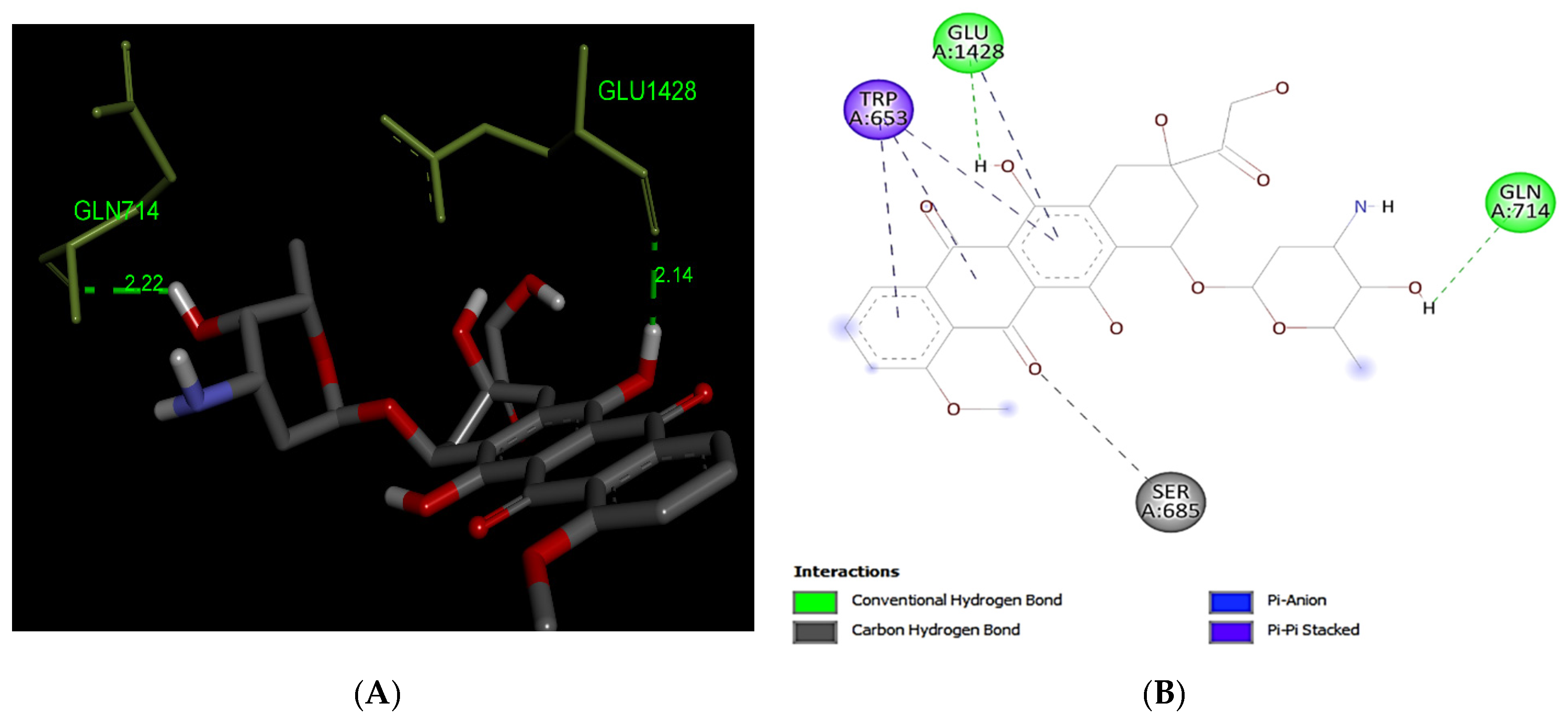


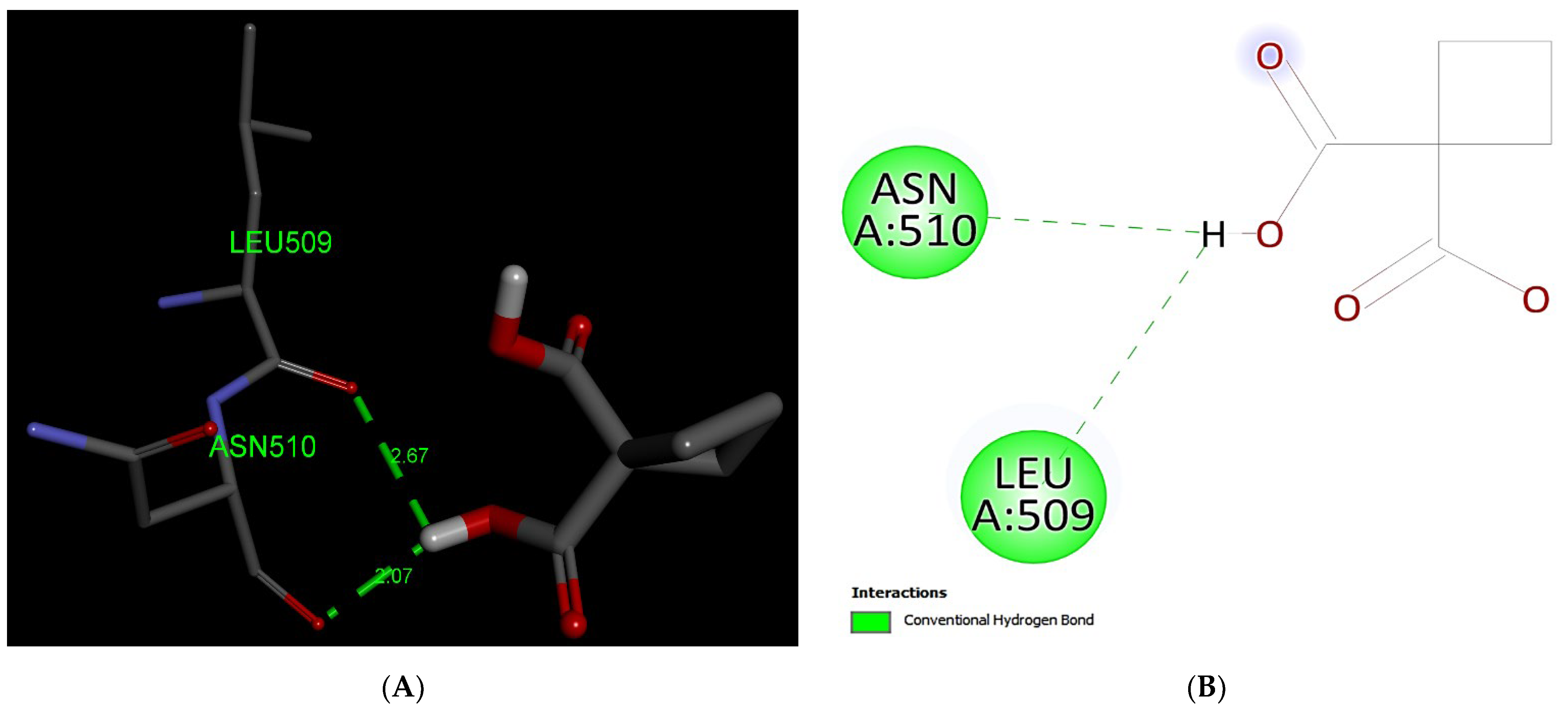
| Interaction of MRP1 | Binding Affinity (kcal/mol) | *A.a Residues Forming H-Bond (s) |
|---|---|---|
| Paclitaxel | −11.3 | 2 (Asn510, Leu1160) |
| Doxorubicin | −10.0 | 2 (Gln714, Glu1428) |
| Topotecan | −9.1 | 2 (Asn510, Ser1163) |
| Gemcitabine | −6.6 | 6 (Asp951, Lys952, Arg1066, Glu1266, Glu1269, Thr1270) |
| Carboplatin | −4.8 | 2 (Leu509, Asn510) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haque, A.; Baig, G.A.; Alshawli, A.S.; Sait, K.H.W.; Hafeez, B.B.; Tripathi, M.K.; Alghamdi, B.S.; Mohammed Ali, H.S.H.; Rasool, M. Interaction Analysis of MRP1 with Anticancer Drugs Used in Ovarian Cancer: In Silico Approach. Life 2022, 12, 383. https://doi.org/10.3390/life12030383
Haque A, Baig GA, Alshawli AS, Sait KHW, Hafeez BB, Tripathi MK, Alghamdi BS, Mohammed Ali HSH, Rasool M. Interaction Analysis of MRP1 with Anticancer Drugs Used in Ovarian Cancer: In Silico Approach. Life. 2022; 12(3):383. https://doi.org/10.3390/life12030383
Chicago/Turabian StyleHaque, Absarul, Ghazanfar Ali Baig, Abdulelah Saleh Alshawli, Khalid Hussain Wali Sait, Bilal Bin Hafeez, Manish Kumar Tripathi, Badrah Saeed Alghamdi, Hani S. H. Mohammed Ali, and Mahmood Rasool. 2022. "Interaction Analysis of MRP1 with Anticancer Drugs Used in Ovarian Cancer: In Silico Approach" Life 12, no. 3: 383. https://doi.org/10.3390/life12030383
APA StyleHaque, A., Baig, G. A., Alshawli, A. S., Sait, K. H. W., Hafeez, B. B., Tripathi, M. K., Alghamdi, B. S., Mohammed Ali, H. S. H., & Rasool, M. (2022). Interaction Analysis of MRP1 with Anticancer Drugs Used in Ovarian Cancer: In Silico Approach. Life, 12(3), 383. https://doi.org/10.3390/life12030383








