Network Pharmacology Study to Reveal the Potentiality of a Methanol Extract of Caesalpinia sappan L. Wood against Type-2 Diabetes Mellitus
Abstract
:1. Introduction
2. Materials and methods
2.1. Plant Collection and Extraction
2.2. GC-MS Analysis
2.3. Filtration of Bioactive Constituents from MECSW
2.4. Acquisition of Compound and T2DM Associated Targets
2.5. Creating a Network Involving Intersecting Targets
2.6. Network Layout for the Pathway Compound Target (PCT)
2.7. Investigation of the Role of GO and KEGG Pathways in Common Intersected Targets
2.8. Formulation and Purification of the Ligand and Receptor Protein
2.9. Glide Directed Molecular Docking Assay
2.10. Quantum Chemistry of Key Ingredients
3. Results
3.1. Exploration of MECSW Ingredients Using GC-MS
3.2. Drug Candidates Filtering
3.3. Common Intersected Targets of Compounds within SEA and STP Database
3.4. Potential Overlapping Targets between T2DM Targets and Compound Linked 238 Intersecting Targets
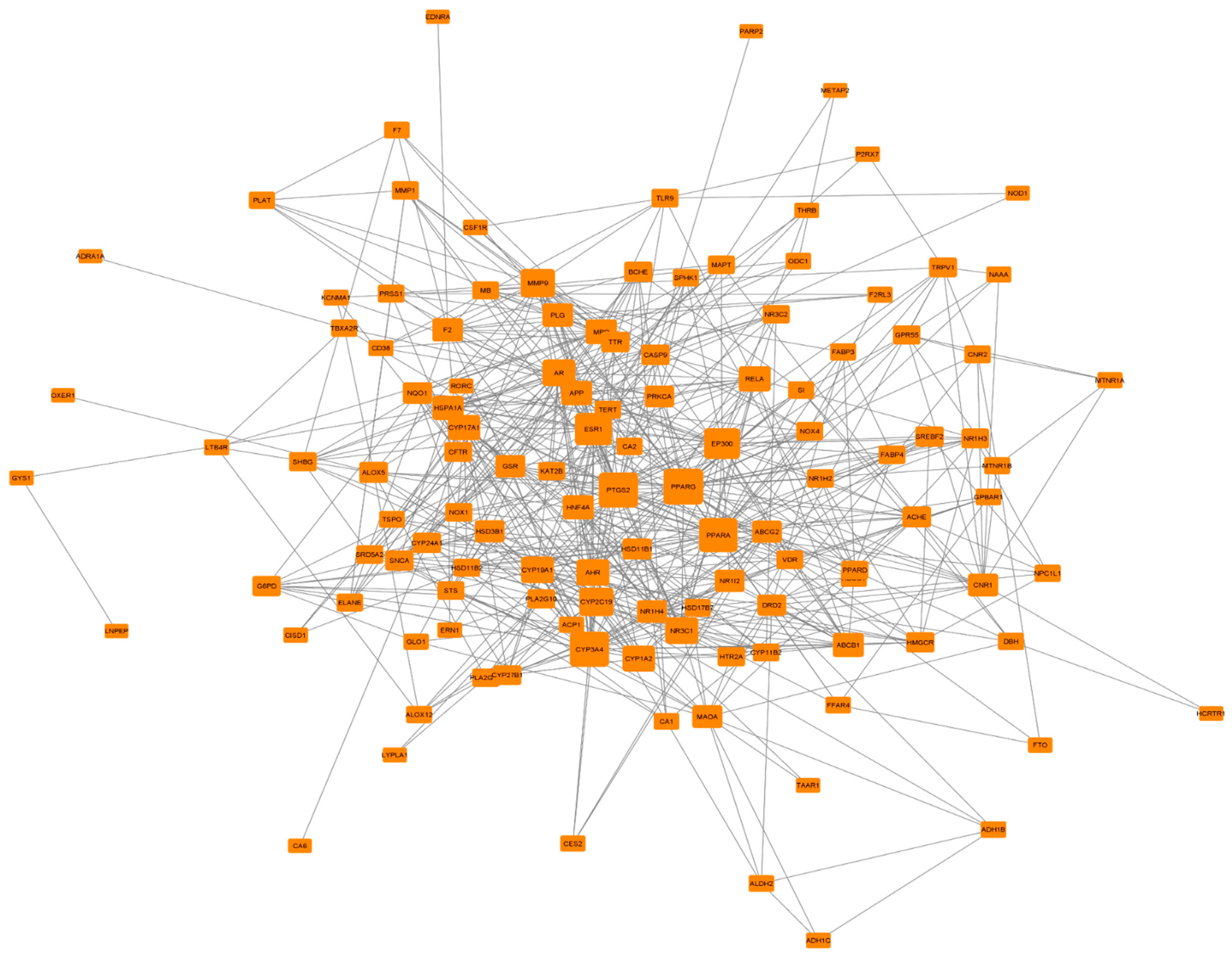
3.5. PPI Network Analysis of 124 Common Targets
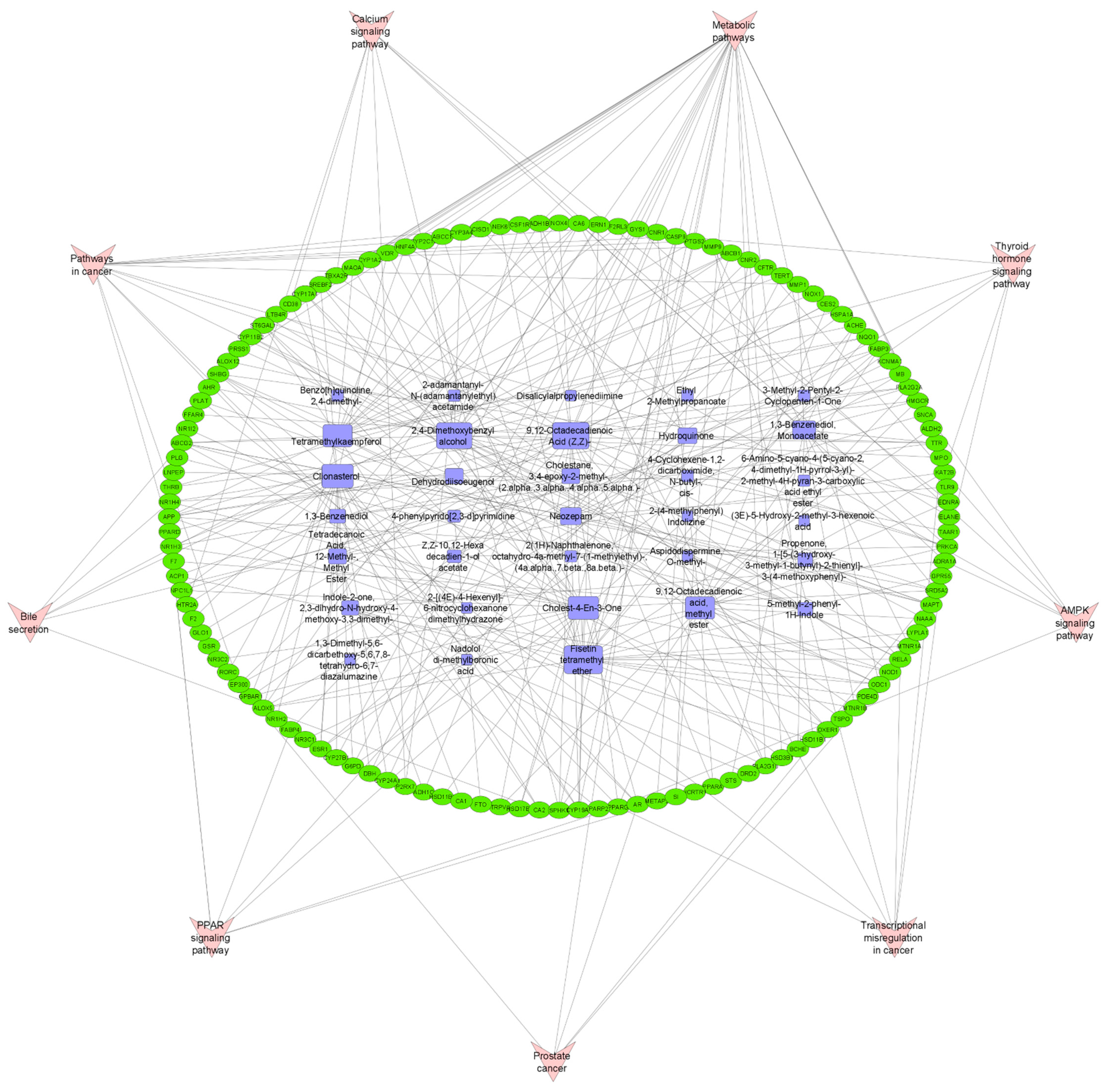
3.6. Pathway Compound Target (PCT) Network Analysis
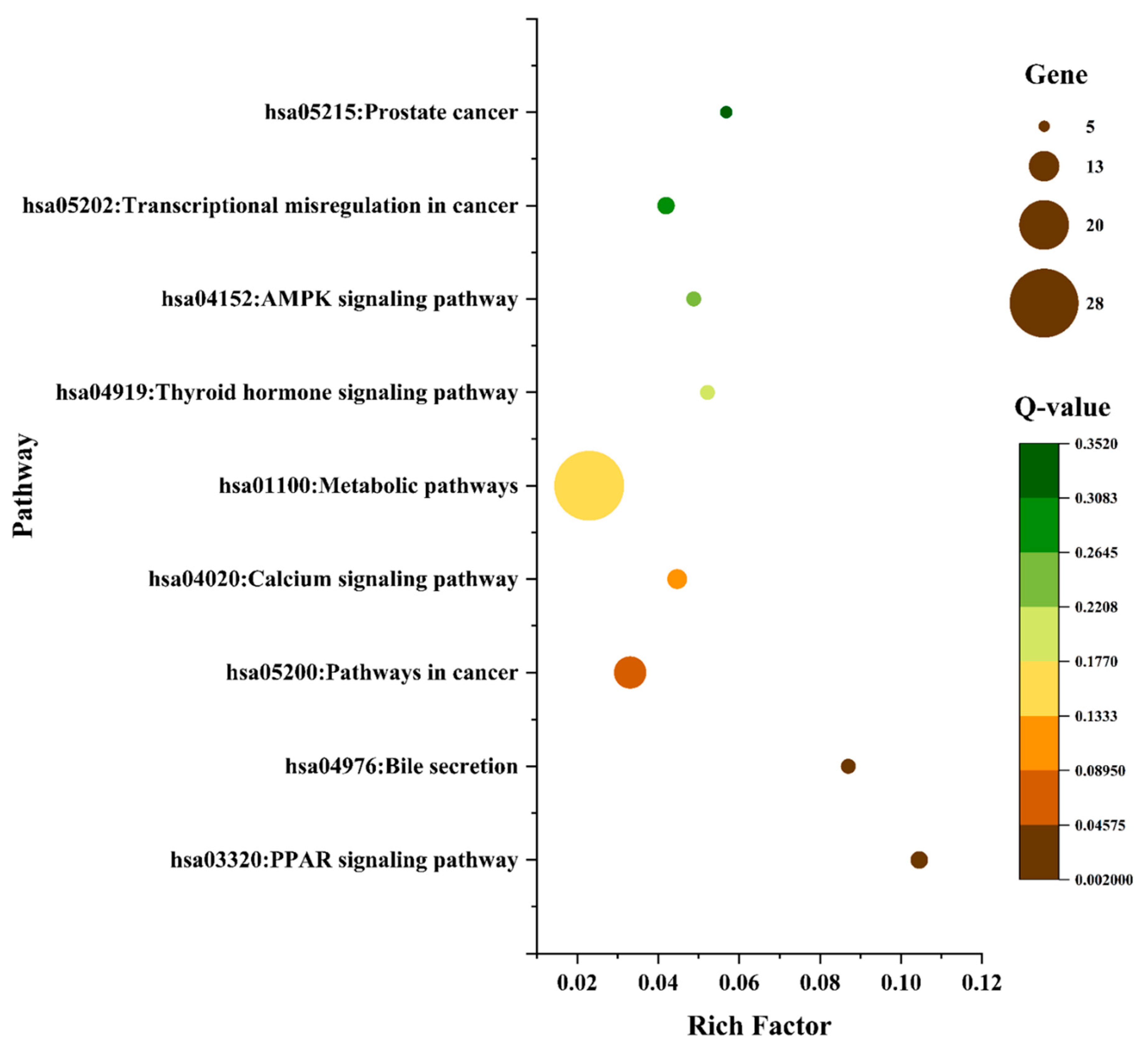
3.7. Gene-Ontology (GO) and KEGG Pathway Enrichment Analysis of 124 Common Targets
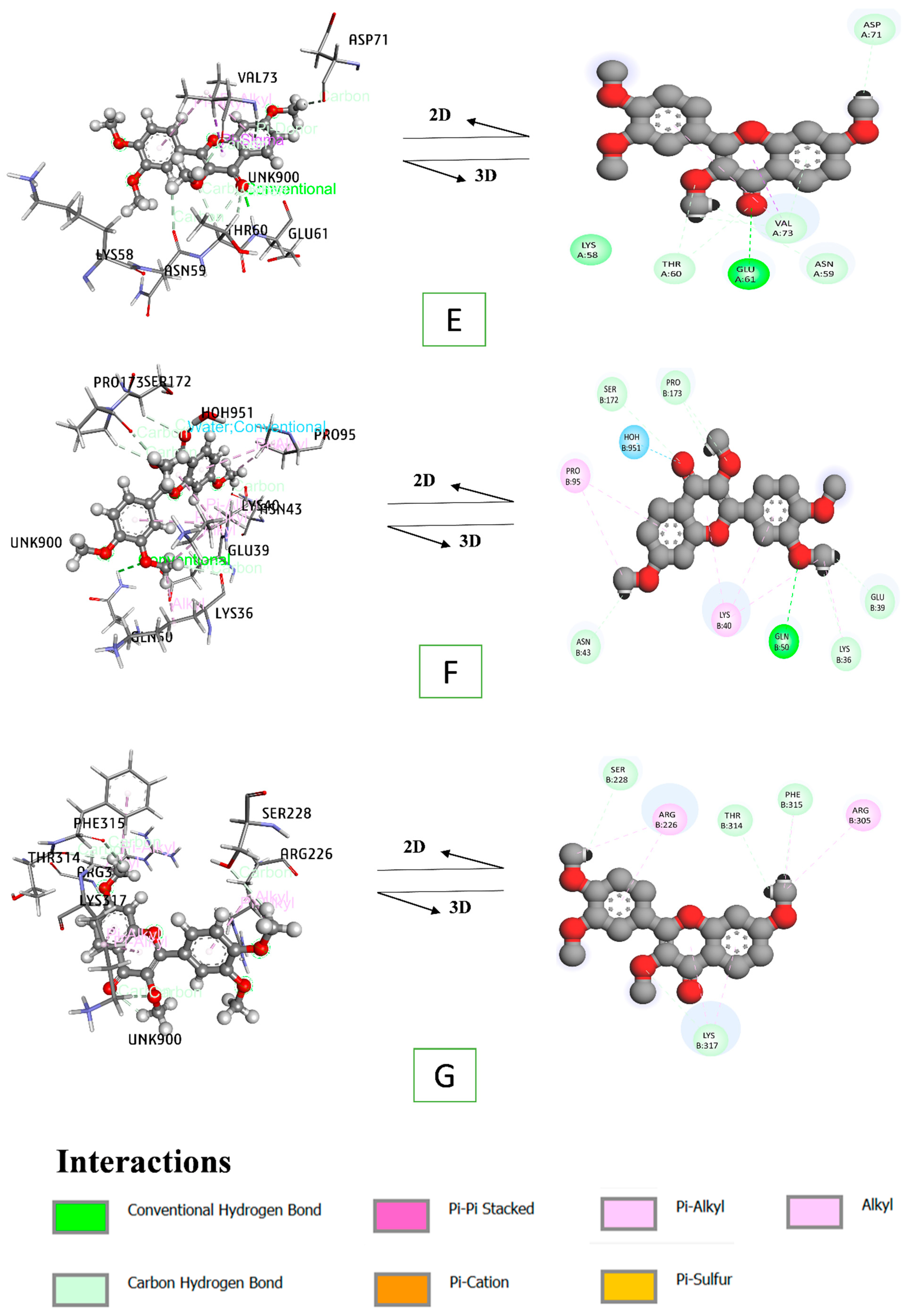
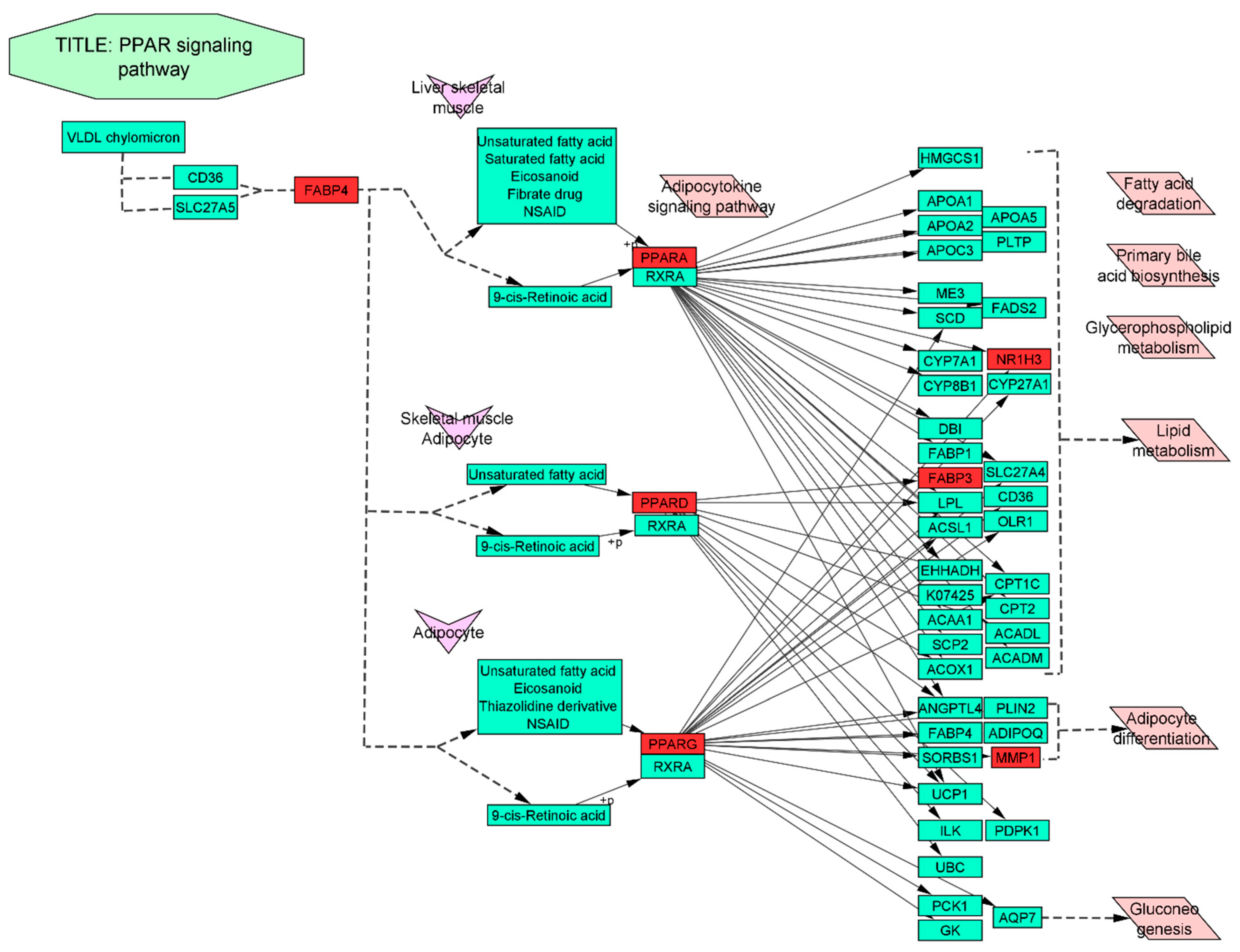
3.8. Docking Interaction of a Key Substance with PPAR Signaling Pathway Enriched Targets
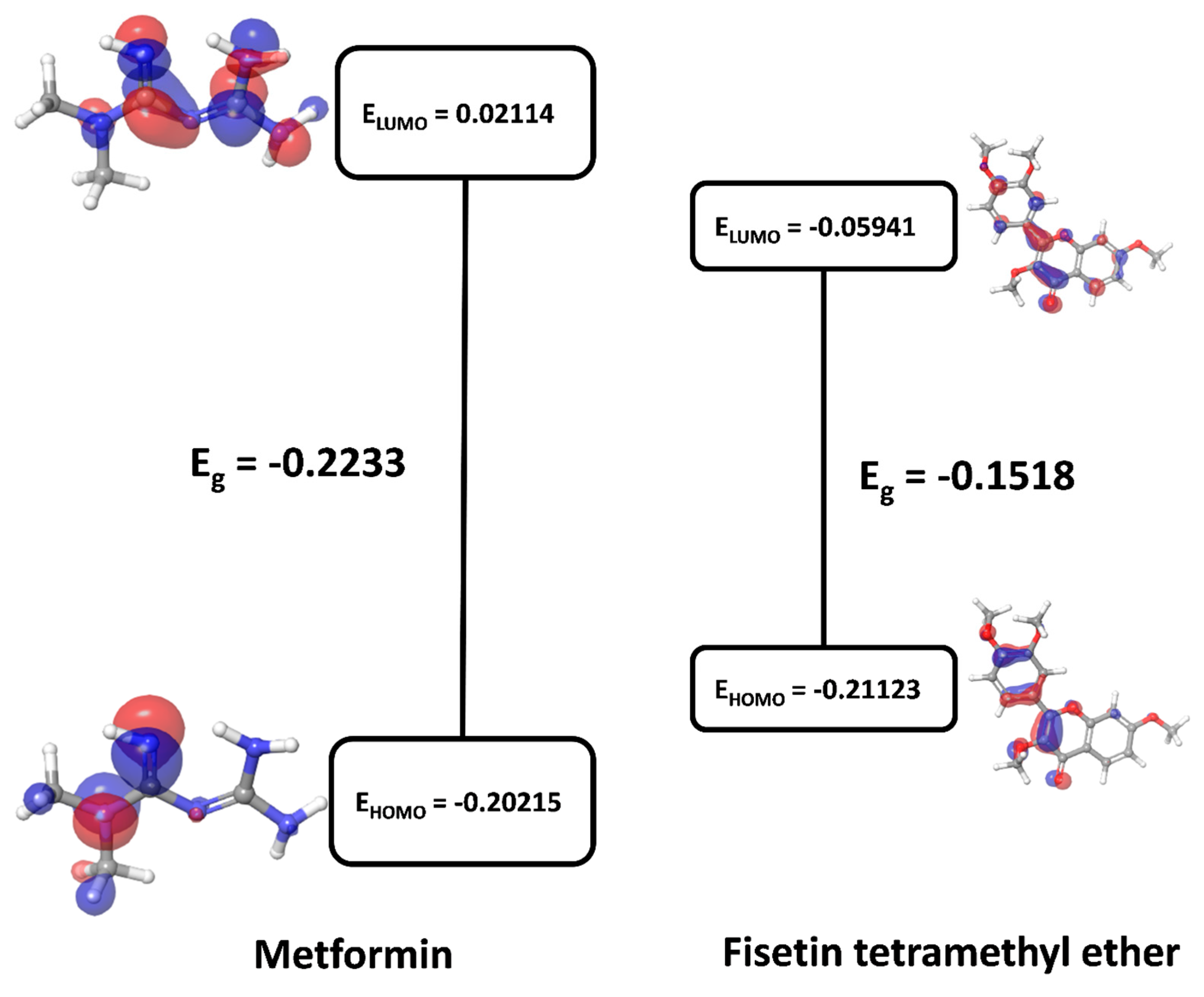
3.9. Quantum Chemistry of Key Ingredients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| T2DM | Type-2 diabetes mellitus |
| PPI | Protein–protein interaction |
| SEA | Similarity ensemble approach |
| SMILES | Simplified molecular input line entry system |
| STP | Swiss target prediction |
| NR1H3 | Nuclear receptor subfamily 1 group H member 3 |
| OMIM | Online mendelian inheritance in man |
| PPARG | Peroxisome proliferator activated receptor gamma |
| MECSW | Methanolic extract of Caesalpinia sappan wood |
| FABP3 | Fatty acid binding protein 3 |
| PPARA | Peroxisome proliferator activated receptor alpha |
| FABP4 | Fatty acid binding protein 4 |
| PPARD | Peroxisome proliferator activated receptor delta |
| MMP1 | Matrix metallopeptidase 1 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| FDR | False discovery rate |
References
- IDF. Atlas IDF Diabetes Atlas, 10th ed.; 2021; Available online: https://diabetesatlas.org/atlas/tenth-edition/ (accessed on 3 August 2020).
- Ling, C.; Groop, L. Epigenetics: A molecular link between environmental factors and type 2 diabetes. Diabetes 2009, 58, 2718–2725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Annis, A.M.; Caulder, M.S.; Cook, M.L.; Duquette, D. Family history, diabetes, and other demographic and risk factors among participants of the national health and nutrition examination survey 1999–2002. Prev. Chronic Dis. 2005, 2, A19. [Google Scholar] [PubMed]
- Sirdah, M.M.; Reading, N.S. Genetic predisposition in type 2 diabetes: A promising approach toward a personalized management of diabetes. Clin. Genet. 2020, 98, 525–547. [Google Scholar] [CrossRef] [PubMed]
- Stumvoll, M.; Goldstein, B.J.; Van Haeften, T.W. Type 2 diabetes: Principles of pathogenesis and therapy. Lancet 2005, 365, 1333–1346. [Google Scholar] [CrossRef]
- Defronzo, R.A. From the triumvirate to the ominous octet: A new paradigm for the treatment of type 2 diabetes mellitus. Diabetes 2009, 58, 773–795. [Google Scholar] [CrossRef] [Green Version]
- Reaven, G.M. Role of insulin resistance in human disease. Diabetes 1988, 37, 1595–1607. [Google Scholar] [CrossRef]
- Pinti, M.V.; Fink, G.K.; Hathaway, Q.A.; Durr, A.J.; Kunovac, A.; Hollander, J.M. Mitochondrial dysfunction in type 2 diabetes mellitus: An organ-based analysis. Am. J. Physiol. Metab. 2019, 316, E268–E285. [Google Scholar] [CrossRef]
- Wu, P.P.; Kor, C.T.; Hsieh, M.C.; Hsieh, Y.P. Association between End-Stage Renal Disease and Incident Diabetes Mellitus—A Nationwide Population-Based Cohort Study. J. Clin. Med. 2018, 7, 343. [Google Scholar] [CrossRef] [Green Version]
- Hsu, Y.C.; Lin, J.T.; Ho, H.J.; Kao, Y.H.; Huang, Y.T.; Hsiao, N.W.; Wu, M.S.; Liu, Y.Y.; Wu, C.Y. Antiviral treatment for hepatitis C virus infection is associated with improved renal and cardiovascular outcomes in diabetic patients. Hepatology 2014, 59, 1293–1302. [Google Scholar] [CrossRef] [Green Version]
- Gordon-Dseagu, V.L.Z.; Shelton, N.; Mindell, J. Diabetes mellitus and mortality from all-causes, cancer, cardiovascular and respiratory disease: Evidence from the Health Survey for England and Scottish Health Survey cohorts. J. Diabetes Complicat. 2014, 28, 791–797. [Google Scholar] [CrossRef] [Green Version]
- Olokoba, A.B.; Obateru, O.A.; Olokoba, L.B. Type 2 diabetes mellitus: A review of current trends. Oman Med. J. 2012, 27, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Mazzola, N. Review of current and emerging therapies in type 2 diabetes mellitus. Am. J. Manag. Care 2012, 18, S17–S26. [Google Scholar] [PubMed]
- Zintzaras, E.; Miligkos, M.; Ziakas, P.; Balk, E.M.; Mademtzoglou, D.; Doxani, C.; Mprotsis, T.; Gowri, R.; Xanthopoulou, P.; Mpoulimari, I.; et al. Assessment of the relative effectiveness and tolerability of treatments of type 2 diabetes mellitus: A network meta-analysis. Clin. Ther. 2014, 36, 1443–1453.e9. [Google Scholar] [CrossRef] [PubMed]
- El-Kaissi, S.; Sherbeeni, S. Pharmacological Management of Type 2 Diabetes Mellitus: An Update. Curr. Diabetes Rev. 2011, 7, 392–405. [Google Scholar] [CrossRef] [PubMed]
- Tsang, M.-W. The Management of Type 2 Diabetic Patients with Hypoglycaemic Agents. ISRN Endocrinol. 2012, 2012, 478120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, S.C.; Mavridis, D.; Nicolucci, A.; Johnson, D.W.; Tonelli, M.; Craig, J.C.; Maggo, J.; Gray, V.; De Berardis, G.; Ruospo, M.; et al. Comparison of clinical outcomes and adverse events associated with glucose-lowering drugs in patients with type 2 diabetes a meta-analysis. J. Am. Med. Assoc. 2016, 316, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Bolen, S.; Feldman, L.; Vassy, J.; Wilson, L.; Yeh, H.C.; Marinopoulos, S.; Wiley, C.; Selvin, E.; Wilson, R.; Bass, E.B.; et al. Systematic review: Comparative effectiveness and safety of oral medications for type 2 diabetes mellitus. Ann. Intern. Med. 2007, 147, 386–399. [Google Scholar] [CrossRef]
- Hung, H.Y.; Qian, K.; Morris-Natschke, S.L.; Hsu, C.S.; Lee, K.H. Recent discovery of plant-derived anti-diabetic natural products. Nat. Prod. Rep. 2012, 29, 580–606. [Google Scholar] [CrossRef]
- Covington, M.B. Traditional Chinese Medicine in the Treatment of Diabetes. Diabetes Spectr. 2001, 14, 154–159. [Google Scholar] [CrossRef] [Green Version]
- Ye, X.P.; Song, C.Q.; Yuan, P.; Mao, R.G. α-Glucosidase and α-Amylase Inhibitory Activity of Common Constituents from Traditional Chinese Medicine Used for Diabetes Mellitus. Chin. J. Nat. Med. 2010, 8, 349–352. [Google Scholar] [CrossRef]
- Li, J.; Bai, L.; Li, X.; He, L.; Zheng, Y.; Lu, H.; Li, J.; Zhong, L.; Tong, R.; Jiang, Z.; et al. Antidiabetic potential of flavonoids from traditional Chinese medicine: A review. Am. J. Chin. Med. 2019, 47, 933–957. [Google Scholar] [CrossRef]
- Masaenah, E.; Elya, B.; Setiawan, H.; Fadhilah, Z.; Wediasari, F.; Nugroho, G.A.; Elfahmi; Mozef, T. Antidiabetic activity and acute toxicity of combined extract of Andrographis paniculata, Syzygium cumini, and Caesalpinia sappan. Heliyon 2021, 7, e08561. [Google Scholar] [CrossRef] [PubMed]
- Muti, A.F.; Pradana, D.L.C.; Rahmi, E.P. Extract of Caesalpinia sappan L. heartwood as food treatment anti-diabetic: A narrative review. IOP Conf. Ser. Earth Environ. Sci. 2021, 755, 012042. [Google Scholar] [CrossRef]
- Srilakshmi, V.S.; Vijayan, P.; Raj, P.V.; Dhanaraj, S.A.; Chandrashekhar, H.R. Hepatoprotective properties of Caesalpinia sappan Linn. heartwood on carbon tetrachloride induced toxicity. Indian J. Exp. Biol. 2010, 48, 905–910. [Google Scholar]
- Nirmal, N.P.; Rajput, M.S.; Prasad, R.G.S.V.; Ahmad, M. Brazilin from Caesalpinia sappan heartwood and its pharmacological activities: A review. Asian Pac. J. Trop. Med. 2015, 8, 421–430. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.X.; Nguyen, M.T.T.; Nguyen, T.A.; Nguyen, N.Y.T.; Phan, D.A.T.; Thi, P.H.; Nguyen, T.H.P.; Dang, P.H.; Nguyen, N.T.; Ueda, J.Y.; et al. Cleistanthane diterpenes from the seed of Caesalpinia sappan and their antiausterity activity against PANC-1 human pancreatic cancer cell line. Fitoterapia 2013, 91, 148–153. [Google Scholar] [CrossRef]
- Zhu, N.L.; Xu, X.D.; Hu, M.G.; Sun, Z.H.; Wu, T.Y.; Yuan, J.Q.; Wu, H.F.; Tian, Y.; Li, P.F.; Yang, J.S.; et al. Bioactive cassane diterpenoids from the seeds of Caesalpinia sappan. Phytochem. Lett. 2017, 22, 113–116. [Google Scholar] [CrossRef]
- Min, B.S.; Cuong, T.D.; Hung, T.M.; Min, B.K.; Shin, B.S.; Woo, M.H. Compounds from the heartwood of Caesalpinia sappan and their anti-inflammatory activity. Bioorg. Med. Chem. Lett. 2012, 22, 7436–7439. [Google Scholar] [CrossRef]
- Nguyen, H.X.; Nguyen, N.T.; Dang, P.H.; Thi Ho, P.; Nguyen, M.T.T.; Van Can, M.; Dibwe, D.F.; Ueda, J.Y.; Awale, S. Cassane diterpenes from the seed kernels of Caesalpinia sappan. Phytochemistry 2016, 122, 286–293. [Google Scholar] [CrossRef]
- Bae, I.K.; Min, H.Y.; Han, A.R.; Seo, E.K.; Sang, K.L. Suppression of lipopolysaccharide-induced expression of inducible nitric oxide synthase by brazilin in RAW 264. 7 macrophage cells. Eur. J. Pharmacol. 2005, 513, 237–242. [Google Scholar] [CrossRef]
- Zeng, K.W.; Yu, Q.; Song, F.J.; Liao, L.X.; Zhao, M.B.; Dong, X.; Jiang, Y.; Tu, P.F. Deoxysappanone B, a homoisoflavone from the Chinese medicinal plant Caesalpinia sappan L., protects neurons from microglia-mediated inflammatory injuries via inhibition of IκB kinase (IKK)-NF-κB and p38/ERK MAPK pathways. Eur. J. Pharmacol. 2015, 748, 18–29. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, H.; Lin, H.; Su, H.; Xing, D.; Du, L. Brazilein protects the brain against focal cerebral ischemia reperfusion injury correlating to inflammatory response suppression. Eur. J. Pharmacol. 2007, 558, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.J.; Jeong, G.H.; Jeong, Y.H.; Kim, T.H. Evaluation of Antioxidant and Anti-diabetic Effects of Sappan Lignum by Extraction Method. Korea J. Herbol. 2017, 32, 1–7. [Google Scholar]
- Chinnala, K.M.; Elsani, M.M.; Nalla, M.K.; Chinnala, K.M. Anti diabetic activity of methanolic extract of Caesalpinia sappan Linn. on alloxan induced diabetes mellitus in rats. Int. J. Exp. Pharmacol. 2015, 5, 65–69. [Google Scholar]
- You, E.J.; Khil, L.Y.; Kwak, W.J.; Won, H.S.; Chae, S.H.; Lee, B.H.; Moon, C.K. Effects of brazilin on the production of fructose-2,6-bisphosphate in rat hepatocytes. J. Ethnopharmacol. 2005, 102, 53–57. [Google Scholar] [CrossRef]
- Holidah, D.; Dewi, I.P.; Christianty, F.M.; Muhammadiy, N.S.; Huda, N. Antidiabetic and antidyslipidemic activity of secang (Caesalpinia sappan l.) wood extract on diabetic rat. Res. J. Pharm. Technol. 2021, 14, 2801–2806. [Google Scholar] [CrossRef]
- Jia, Y.; Wang, H.; Song, Y.; Liu, K.; Dou, F.; Lu, C.; Ge, J.; Chi, N.; Ding, Y.; Hai, W.; et al. Application of a liquid chromatography-tandem mass spectrometry method to the pharmacokinetics, tissue distribution and excretion studies of brazilin in rats. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 931, 61–67. [Google Scholar] [CrossRef]
- Mueller, M.; Weinmann, D.; Toegel, S.; Holzer, W.; Unger, F.M.; Viernstein, H. Compounds from Caesalpinia sappan with anti-inflammatory properties in macrophages and chondrocytes. Food Funct. 2016, 7, 1671–1679. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Guan, S.; Feng, R.; Tang, R.; Guo, D. Separation of four homoisoflavonoids from Caesalpinia sappan by high-speed counter-current chromatography. Phytochem. Anal. 2012, 23, 228–231. [Google Scholar] [CrossRef]
- Atangwho, I.J.; Ebong, P.E.; Egbung, G.E.; Obi, A.U. Extract of Vernonia amygdalina Del. (African bitter leaf) can reverse pancreatic cellular lesion after alloxan damage in the rat. Aust. J. Basic Appl. Sci. 2010, 4, 711–716. [Google Scholar]
- Meliani, N.; Dib, M.E.A.; Allali, H.; Tabti, B. Hypoglycaemic effect of Berberis vulgaris L. in normal and streptozotocin-induced diabetic rats. Asian Pac. J. Trop. Biomed. 2011, 1, 468–471. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hu, B.; Feng, S.; Wang, J.; Zhang, F. Target recognition and network pharmacology for revealing anti-diabetes mechanisms of natural product. J. Comput. Sci. 2020, 45, 101186. [Google Scholar] [CrossRef]
- Guo, W.; Huang, J.; Wang, N.; Tan, H.Y.; Cheung, F.; Chen, F.; Feng, Y. Integrating network pharmacology and pharmacological evaluation for deciphering the action mechanism of herbal formula Zuojin Pill in suppressing hepatocellular carcinoma. Front. Pharmacol. 2019, 10, 1185. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology. Nat. Biotechnol. 2007, 25, 1110–1111. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Li, S.; Zhang, B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin. J. Nat. Med. 2013, 11, 110–120. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, X.; Li, S. An integrative platform of TCM network pharmacology and its application on a herbal formula, Qing-Luo-Yin. Evid.-Based Complement. Altern. Med. 2013, 2013, 456747. [Google Scholar] [CrossRef] [Green Version]
- Berger, S.I.; Iyengar, R. Network analyses in systems pharmacology. Bioinformatics 2009, 25, 2466–2472. [Google Scholar] [CrossRef]
- Oh, K.; Adnan, M.; Cho, D. Uncovering Mechanisms of Zanthoxylum piperitum Fruits for the Alleviation of Rheumatoid Arthritis Based on Network Pharmacology. Biology 2021, 10, 703. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rappaport, N.; Twik, M.; Plaschkes, I.; Nudel, R.; Iny Stein, T.; Levitt, J.; Gershoni, M.; Morrey, C.P.; Safran, M.; Lancet, D. MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017, 45, D877–D887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamosh, A.; Scott, A.F.; Amberger, J.S.; Bocchini, C.A.; McKusick, V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005, 33, D514. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Shi, X.; Li, X.; Zhang, L.; Wu, P.; Mao, J.; Xing, R.; Zhang, N.; Wang, P. Network Pharmacology Approach to Uncover the Mechanism Governing the Effect of Simiao Powder on Knee Osteoarthritis. BioMed Res. Int. 2020, 2020, 6971503. [Google Scholar] [CrossRef]
- Xiong, Y.; Yang, Y.; Xiong, W.; Yao, Y.; Wu, H.; Zhang, M. Network pharmacology-based research on the active component and mechanism of the antihepatoma effect of Rubia cordifolia L. J. Cell. Biochem. 2019, 120, 12461–12472. [Google Scholar] [CrossRef]
- Auniq, R.; Chy, M.; Adnan, M.; Roy, A.; Islam, M.; Khan, T.; Hasan, M.; Ahmed, S.; Khan, M.; Islam, N.; et al. Assessment of anti-nociceptive and anthelmintic activities of Vitex Peduncularis Wall. leaves and in silico molecular docking, ADME/T, and PASS prediction studies of its isolated compounds. J. Complement. Med. Res. 2019, 10, 170. [Google Scholar] [CrossRef]
- Adnan, M.; Nazim Uddin Chy, M.; Mostafa Kamal, A.T.M.; Azad, M.O.K.; Paul, A.; Uddin, S.B.; Barlow, J.W.; Faruque, M.O.; Park, C.H.; Cho, D.H. Investigation of the biological activities and characterization of bioactive constituents of ophiorrhiza rugosa var. prostrata (D.Don) & Mondal leaves through in vivo, in vitro, and in silico approaches. Molecules 2019, 24, 1367. [Google Scholar] [CrossRef] [Green Version]
- Shovo, M.A.R.B.; Tona, M.R.; Mouah, J.; Islam, F.; Chowdhury, M.H.U.; Das, T.; Paul, A.; Ağagündüz, D.; Rahman, M.M.; Emran, T.B.; et al. Computational and Pharmacological Studies on the Antioxidant, Thrombolytic, Anti-Inflammatory, and Analgesic Activity of Molineria capitulata. Curr. Issues Mol. Biol. 2021, 43, 35. [Google Scholar] [CrossRef]
- Adnan, M.; Chy, M.; Uddin, N.; Kama, A.T.M.; Azad, M.; Kalam, O.; Chowdhury, K.A.A.; Kabir, M.S.H.; Gupta, S.D.; Chowdhury, M. Comparative Study of Piper sylvaticum Roxb. Leaves and Stems for Anxiolytic and Antioxidant Properties Through in vivo, in vitro, and in silico Approaches. Biomedicines 2020, 8, 68. [Google Scholar] [CrossRef] [Green Version]
- Bochevarov, A.D.; Harder, E.; Hughes, T.F.; Greenwood, J.R.; Braden, D.A.; Philipp, D.M.; Rinaldo, D.; Halls, M.D.; Zhang, J.; Friesner, R.A. Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013, 113, 2110–2142. [Google Scholar] [CrossRef]
- Azam, F.; Alabdullah, N.H.; Ehmedat, H.M.; Abulifa, A.R.; Taban, I.; Upadhyayula, S. NSAIDs as potential treatment option for preventing amyloid β toxicity in Alzheimer’s disease: An investigation by docking, molecular dynamics, and DFT studies. J. Biomol. Struct. Dyn. 2017, 36, 2099–2117. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dai, W.; Li, W.; Zhang, Y.; Wu, Y.; Guan, C.; Zhang, A.; Huang, H.; Li, Y. Integrated Network Pharmacology and Metabonomics to Reveal the Myocardial Protection Effect of Huang-Lian-Jie-Du-Tang on Myocardial Ischemia. Front. Pharmacol. 2020, 11, 2246. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Qiu, P.; Zhu, R.; Zhao, L.; Zhang, P.; Wang, Y.; Li, C.; Chai, K.; Shou, D.; Zhao, H. A Combined Phytochemistry and Network Pharmacology Approach to Reveal the Potential Antitumor Effective Substances and Mechanism of Phellinus igniarius. Front. Pharmacol. 2019, 10, 266. [Google Scholar] [CrossRef] [PubMed]
- Kasper, J.S.; Liu, Y.; Pollak, M.N.; Rifai, N.; Giovannucci, E. Hormonal profile of diabetic men and the potential link to prostate cancer. Cancer Causes Control 2008, 19, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Travis, R.C.; Appleby, P.N.; Martin, R.M.; Holly, J.M.P.; Albanes, D.; Black, A.; Bueno-de-Mesquita, H.B.A.; Chan, J.M.; Chen, C.; Chirlaque, M.-D.; et al. A Meta-analysis of Individual Participant Data Reveals an Association between Circulating Levels of IGF-I and Prostate Cancer Risk. Cancer Res. 2016, 76, 2288–2300. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.; Song, M.; Preston, M.A.; Ma, W.; Hu, Y.; Pernar, C.H.; Stopsack, K.H.; Ebot, E.M.; Fu, B.C.; Zhang, Y.; et al. The association of diabetes with risk of prostate cancer defined by clinical and molecular features. Br. J. Cancer 2020, 123, 657–665. [Google Scholar] [CrossRef]
- Frasca, F.; Pandini, G.; Scalia, P.; Sciacca, L.; Mineo, R.; Costantino, A.; Goldfine, I.D.; Belfiore, A.; Vigneri, R. Insulin Receptor Isoform A, a Newly Recognized, High-Affinity Insulin-Like Growth Factor II Receptor in Fetal and Cancer Cells. Mol. Cell. Biol. 1999, 19, 3278–3288. [Google Scholar] [CrossRef] [Green Version]
- Sciacca, L.; Mineo, R.; Pandini, G.; Murabito, A.; Vigneri, R.; Belfiore, A. In IGF-I receptor-deficient leiomyosarcoma cells autocrine IGF-II induces cell invasion and protection from apoptosis via the insulin receptor isoform A. Oncogene 2002, 21, 8240–8250. [Google Scholar] [CrossRef] [Green Version]
- Vella, V.; Pandini, G.; Sciacca, L.; Mineo, R.; Vigneri, R.; Pezzino, V.; Belfiore, A. A novel autocrine loop involving IGF-II and the insulin receptor isoform-A stimulates growth of thyroid cancer. J. Clin. Endocrinol. Metab. 2002, 87, 245–254. [Google Scholar] [CrossRef]
- Kalli, K.R.; Falowo, O.I.; Bale, L.K.; Zschunke, M.A.; Roche, P.C.; Conover, C.A. Functional insulin receptors on human epithelial ovarian carcinoma cells: Implications for IGF-II mitogenic signaling. Endocrinology 2002, 143, 3259–3267. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.E.; Gleave, M.E.; Zakikhani, M.; Bell, R.H.; Piura, E.; Vickers, E.; Cunningham, M.; Larsson, O.; Fazli, L.; Pollak, M. Insulin receptor expression by human prostate cancers. Prostate 2009, 69, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Zelenko, Z.; Gallagher, E.J.; Antoniou, I.M.; Sachdev, D.; Nayak, A.; Yee, D.; LeRoith, D. EMT reversal in human cancer cells after IR knockdown in hyperinsulinemic mice. Endocr. Relat. Cancer 2016, 23, 747–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Detimary, P.; Gilon, P.; Henquin, J.C. Interplay between cytoplasmic Ca2+ and the ATP/ADP ratio: A feedback control mechanism in mouse pancreatic islets. Biochem. J. 1998, 333, 269–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salt, I.P.; Johnson, G.; Ashcroft, S.J.H.; Hardie, D.G. AMP-activated protein kinase is activated by low glucose in cell lines derived from pancreatic β cells, and may regulate insulin release. Biochem. J. 1998, 335, 533–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misra, P.; Chakrabarti, R. The role of AMP kinase in diabetes. Indian J. Med. Res. 2007, 125, 389–398. [Google Scholar] [PubMed]
- Jeon, S.M. Regulation and function of AMPK in physiology and diseases. Exp. Mol. Med. 2016, 48, e245. [Google Scholar] [CrossRef]
- Potenza, M.; Via, M.A.; Yanagisawa, R.T. Excess thyroid hormone and carbohydrate meta bolism. Endocr. Pract. 2009, 15, 254–262. [Google Scholar] [CrossRef]
- Ray, S.; Ghosh, S. Thyroid Disorders and Diabetes Mellitus: Double Trouble. J. Diabetes Res. Ther. 2016, 2, 1–7. [Google Scholar] [CrossRef]
- Hameed, I.; Masoodi, S.R.; Mir, S.A.; Nabi, M.; Ghazanfar, K.; Ganai, B.A. Type 2 diabetes mellitus: From a metabolic disorder to an inflammatory condition. World J. Diabetes 2015, 6, 598. [Google Scholar] [CrossRef]
- Lanner, J.T.; Katz, A.; Tavi, P.; Sandström, M.E.; Zhang, S.J.; Wretman, C.; James, S.; Fauconnier, J.; Lännergren, J.; Bruton, J.D.; et al. The role of Ca2+ influx for insulin-mediated glucose uptake in skeletal muscle. Diabetes 2006, 55, 2077–2083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.H.; Wei, Y.H. Role of mitochondrial dysfunction and dysregulation of Ca2+ homeostasis in the pathophysiology of insulin resistance and type 2 diabetes. J. Biomed. Sci. 2017, 24, 70. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Kanazawa, I.; Takaoka, S.; Sugimoto, T. Serum calcium is positively correlated with fasting plasma glucose and insulin resistance, independent of parathyroid hormone, in male patients with type 2 diabetes mellitus. Metabolism 2011, 60, 1334–1339. [Google Scholar] [CrossRef] [PubMed]
- Calle, E.E.; Kaaks, R. Overweight, obesity and cancer: Epidemiological evidence and proposed mechanisms. Nat. Rev. Cancer 2004, 4, 579–591. [Google Scholar] [CrossRef]
- Ricart, W.; Fernández-Real, J.M. No decrease in free IGF-I with increasing insulin in obesity-related insulin resistance. Obes. Res. 2001, 9, 631–636. [Google Scholar] [CrossRef]
- Masur, K.; Vetter, C.; Hinz, A.; Tomas, N.; Henrich, H.; Niggemann, B.; Zänker, K.S. Diabetogenic glucose and insulin concentrations modulate transcriptom and protein levels involved in tumour cell migration, adhesion and proliferation. Br. J. Cancer 2011, 104, 345–352. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, P.; Cariou, B.; Lien, F.; Kuipers, F.; Staels, B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 2009, 89, 147–191. [Google Scholar] [CrossRef] [Green Version]
- Prawitt, J.; Caron, S.; Staels, B. Bile acid metabolism and the pathogenesis of type 2 diabetes. Curr. Diabetes Rep. 2011, 11, 160–166. [Google Scholar] [CrossRef] [Green Version]
- Hauner, H. The mode of action of thiazolidinediones. Diabetes Metab. Res. Rev. 2002, 18, S10–S15. [Google Scholar] [CrossRef]
- Kim, H.I.; Ahn, Y.H. Role of Peroxisome Proliferator-Activated Receptor-γ in the Glucose-Sensing Apparatus of Liver and β-Cells. Diabetes 2004, 53, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, H.U.; Adnan, M.; Oh, K.K.; Cho, D.H. Decrypting molecular mechanism insight of Phyllanthus emblica L. fruit in the treatment of type 2 diabetes mellitus by network pharmacology. Phytomedicine Plus 2021, 1, 100144. [Google Scholar] [CrossRef]
- Ahmadian, M.; Suh, J.M.; Hah, N.; Liddle, C.; Atkins, A.R.; Downes, M.; Evans, R.M. PPARγ signaling and metabolism: The good, the bad and the future. Nat. Med. 2013, 19, 557–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wargent, E.; Sennitt, M.V.; Stocker, C.; Mayes, A.E.; Brown, L.; O’Dowd, J.; Wang, S.; Einerhand, A.W.C.; Mohede, I.; Arch, J.R.S.; et al. Prolonged treatment of genetically obese mice with conjugated linoleic acid improves glucose tolerance and lowers plasma insulin concentration: Possible involvement of PPAR activation. Lipids Health Dis. 2005, 4, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barroso, I.; Gurnell, M.; Crowley, V.E.F.; Agostini, M.; Schwabe, J.W.; Soos, M.A.; Maslen, G.L.; Williams, T.D.M.; Lewis, H.; Schafer, A.J.; et al. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature 1999, 402, 880–883. [Google Scholar] [CrossRef]
- Pappa, K.I.; Gazouli, M.; Anastasiou, E.; Iliodromiti, Z.; Antsaklis, A.; Anagnou, N.P. Circadian clock gene expression is impaired in gestational diabetes mellitus. Gynecol. Endocrinol. 2013, 29, 331–335. [Google Scholar] [CrossRef]
- Prasath, G.S.; Pillai, S.I.; Subramanian, S.P. Fisetin improves glucose homeostasis through the inhibition of gluconeogenic enzymes in hepatic tissues of streptozotocin induced diabetic rats. Eur. J. Pharmacol. 2014, 740, 248–254. [Google Scholar] [CrossRef]
- Prasath, G.S.; Subramanian, S.P. Antihyperlipidemic Effect of Fisetin, a Bioflavonoid of Strawberries, Studied in Streptozotocin-Induced Diabetic Rats. J. Biochem. Mol. Toxicol. 2014, 28, 442–449. [Google Scholar] [CrossRef]
- Prasath, G.S.; Subramanian, S.P. Fisetin, a tetra hydroxy flavone recuperates antioxidant status and protects hepatocellular ultrastructure from hyperglycemia mediated oxidative stress in streptozotocin induced experimental diabetes in rats. Food Chem. Toxicol. 2013, 59, 249–255. [Google Scholar] [CrossRef]
- Chiang, C.-K.; Ho, T.-I.; Peng, Y.-S.; Hsu, S.-P.; Pai, M.-F.; Yang, S.-Y.; Hung, K.-Y.; Wu, K.-D. Rosiglitazone in Diabetes Control in Hemodialysis Patients With and Without Viral Hepatitis Infection. Diabetes Care 2007, 30, 3–7. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.M.; Shawon, J.; Halim, M.A. Multiple receptor conformers based molecular docking study of fluorine enhanced ethionamide with mycobacterium enoyl ACP reductase (InhA). J. Mol. Graph. Model. 2017, 77, 386–398. [Google Scholar] [CrossRef]
- Shahinozzaman, M.; Taira, N.; Ishii, T.; Halim, M.; Hossain, M.; Tawata, S. Anti-Inflammatory, Anti-Diabetic, and Anti-Alzheimer’s Effects of Prenylated Flavonoids from Okinawa Propolis: An Investigation by Experimental and Computational Studies. Molecules 2018, 23, 2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearson, R.G. Absolute electronegativity and hardness correlated with molecular orbital theory. Proc. Natl. Acad. Sci. USA 1986, 83, 8440–8441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucido, M.J.; Orlando, B.J.; Vecchio, A.J.; Malkowski, M.G. Crystal Structure of Aspirin-Acetylated Human Cyclooxygenase-2: Insight into the Formation of Products with Reversed Stereochemistry. Biochemistry 2016, 55, 1226–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranga Rao, R.; Tiwari, A.K.; Prabhakar Reddy, P.; Suresh Babu, K.; Ali, A.Z.; Madhusudana, K.; Madhusudana Rao, J. New furanoflavanoids, intestinal α-glucosidase inhibitory and free-radical (DPPH) scavenging, activity from antihyperglycemic root extract of Derris indica (Lam.). Bioorg. Med. Chem. 2009, 17, 5170–5175. [Google Scholar] [CrossRef]
- Maher, P.; Dargusch, R.; Ehren, J.L.; Okada, S.; Sharma, K.; Schubert, D. Fisetin Lowers Methylglyoxal Dependent Protein Glycation and Limits the Complications of Diabetes. PLoS ONE 2011, 6, e21226. [Google Scholar] [CrossRef]
- Miyata, T.; Van Ypersele de Strihou, C.; Imasawa, T.; Yoshino, A.; Ueda, Y.; Ogura, H.; Kominami, K.; Onogi, H.; Inagi, R.; Nangaku, M.; et al. Glyoxalase I deficiency is associated with an unusual level of advanced glycation end products in a hemodialysis patient. Kidney Int. 2001, 60, 2351–2359. [Google Scholar] [CrossRef] [Green Version]





| SL. No. | RT Time (min) | Area (%) | PubChem CID | Chemical Formula | Compound Class | Compounds Name |
|---|---|---|---|---|---|---|
| 1 | 5.366 | 0.22 | 5363448 | C7H12O3 | Fatty acyls | (3E)-5-Hydroxy-2-methyl-3-hexenoic acid |
| 2 | 5.683 | 1.3 | 5055 | C8H8O3 | Phenol esters | 1,3-Benzenediol, Monoacetate |
| 3 | 6.606 | 0.53 | 7342 | C6H12O2 | Carboxylic acids and derivatives | Ethyl 2-Methylpropanoate |
| 4 | 6.856 | 0.7 | 5054 | C6H6O2 | Phenols | 1,3-Benzenediol |
| 5 | 7.529 | 2.14 | 62378 | C11H18O | Organooxygen compounds | 3-Methyl-2-Pentyl-2-Cyclopenten-1-One |
| 6 | 8.116 | 0.52 | 81750 | C9H12O3 | Benzene and substituted derivatives | 2,4-Dimethoxybenzyl alcohol |
| 7 | 8.231 | 0.92 | 91477 | C27H44O | Steroids and steroid derivatives | Cholest-4-En-3-One |
| 8 | 8.51 | 1.26 | 785 | C6H6O2 | Phenols | Hydroquinone |
| 9 | 8.731 | 0.96 | 5284421 | C19H34O2 | Fatty acyls | 9,12-Octadecadienoic acid, methyl ester |
| 10 | 8.818 | 0.15 | 21206 | C16H32O2 | Fatty acyls | Tetradecanoic acid, 12-Methyl-, Methyl Ester |
| 11 | 8.914 | 3.95 | 5280450 | C18H32O2 | Fatty acyls | 9,12-Octadecanoic Acid(Z,Z)- |
| 12 | 8.981 | 3.34 | 5363372 | C18H32O2 | Fatty acyls | Z,Z-10,12-Hexadecadien-1-ol acetate |
| 13 | 9.375 | 0.18 | 41133 | C14H24O | Organooxygen compounds | 2(1H)-Naphthalenone, octahydro-4a-methyl-7-(1-methylethyl)-, (4a.alpha.,7.beta.,8a.beta.)- |
| 14 | 10.01 | 0.22 | 7210 | C17H18N2O2 | Phenols | 2-((E)-[((E)-2-([(E)-(2-Hydroxyphenyl)Methylidene]Amino)Propyl)Imino]Phenol or, Disalicylalpropylenediimine |
| 15 | 10.19 | 0.47 | 5364132 | C14H25N3O2 | Allyl-type 1,3-dipolar organic compounds | 2-[(4E)-4-Hexenyl]-6-nitrocyclohexanone dimethylhydrazone |
| 16 | 10.65 | 0.66 | 4506 | C15H11N3O3 | Benzodiazepines | 7-Nitro-1,3-dihydro-5-phenyl-2H-1,4-benzodiazepin-2-one or, Neozepam |
| 17 | 10.73 | 0.52 | 610113 | C11H13NO3 | Indoles and derivatives | Indole-2-one, 2,3-dihydro-N-hydroxy-4-methoxy-3,3-dimethyl- |
| 18 | 10.88 | 3.94 | 610177 | C13H9N3 | Diazines | Pyrido[2,3-d]pyrimidine, 4-phenyl- |
| 19 | 11.19 | 2.93 | 5116643 | C24H37NO | Carboxylic acids and derivatives | Acetamide, 2-(adamantan-1-yl)-N-(1-adamantan-1-ylethyl)- or, 2-adamantanyl-N-(adamantanylethyl)acetamide |
| 20 | 11.28 | 1.22 | 5379033 | C20H22O4 | 2-arylbenzofuran flavonoids | Phenol, 4-[2,3-dihydro-7-methoxy-3-methyl-5-(1-propenyl)-2-benzofuranyl]-2-methoxy- or, Dehydrodiisoeugenol |
| 21 | 11.65 | 13.93 | 5379034 | C19H18O3S | Phenol ethers | Propenone, 1-[5-(3-hydroxy-3-methyl-1-butynyl)-2-thienyl]-3-(4-methoxyphenyl)- |
| 22 | 11.9 | 1.78 | 346948 | C15H13N | Pyrroles | Indolizine, 2-(4-methylphenyl)- |
| 23 | 11.99 | 6.15 | 610018 | C17H18N4O3 | Pyrans | 6-Amino-5-cyano-4-(5-cyano-2,4-dimethyl-1H-pyrrol-3-yl)-2-methyl-4H-pyran-3-carboxylic acid ethyl ester |
| 24 | 12.51 | 11.82 | 631121 | C20H26N2O3 | Plumeran-type alkaloids | Aspidodispermine, O-methyl- |
| 25 | 12.63 | 3.08 | 631095 | C19H18O6 | Flavonoids | 4H-1-Benzopyran-4-one, 3,5,7-trimethoxy-2-(4-methoxyphenyl)- or, Tetramethylkaempferol |
| 26 | 12.73 | 3.38 | 631112 | C19H29B2NO4 | Tetralins | Nadolol di-methylboronic acid |
| 27 | 12.85 | 5.34 | 631171 | C19H18O6 | Flavonoids | 4H-1-Benzopyran-4-one, 2-(3,4-dimethoxyphenyl)-3,7-dimethoxy- or, Fisetin tetramethyl ether |
| 28 | 12.95 | 12.81 | 284060 | C12H18N6O6 | Diazines | 1,3-Dimethyl-5,6-dicarbethoxy-5,6,7,8-tetrahydro-6,7-diazalumazine |
| 29 | 13.89 | 1.75 | 541560 | C28H48O | Steroids and steroid derivatives | Cholestane, 3,4-epoxy-2-methyl-, (2.alpha.,3.alpha.,4.alpha.,5.alpha.)- |
| 30 | 14.09 | 1.65 | 91733922 | C12H17NO2 | Isoindoles and derivatives | 4-Cyclohexene-1,2-dicarboximide, N-butyl-, cis- |
| 31 | 14.65 | 5.69 | 457801 | C29H50O | Steroids and steroid derivatives | Stigmast-5-en-3-ol, (3beta,24S)- or, Clionasterol |
| 32 | 15.27 | 0.4 | 83247 | C15H13N | Indoles and derivatives | 1H-Indole, 5-methyl-2-phenyl- |
| 33 | 15.66 | 0.68 | 610182 | C15H13N | Quinolines and derivatives | Benzo[h]quinoline, 2,4-dimethyl- |
| Compounds Name | MW | HBA | HBD | MLogP | Number of Violations | Bioavailability |
|---|---|---|---|---|---|---|
| <500 | <10 | ≤5 | ≤4.15 | ≤1 | >0.1 | |
| (3E)-5-Hydroxy-2-methyl-3-hexenoic acid | 144.17 | 3 | 2 | 0.64 | 0 | 0.85 |
| 1,3-Benzenediol, Monoacetate | 152.15 | 3 | 1 | 1.32 | 0 | 0.55 |
| Ethyl 2-Methylpropanoate | 116.16 | 2 | 0 | 1.27 | 0 | 0.55 |
| 1,3-Benzenediol | 110.11 | 2 | 2 | 0.79 | 0 | 0.55 |
| 3-Methyl-2-Pentyl-2-Cyclopenten-1-One | 166.26 | 1 | 0 | 2.49 | 0 | 0.55 |
| 2,4-Dimethoxybenzyl alcohol | 168.19 | 3 | 1 | 0.92 | 0 | 0.55 |
| Cholest-4-En-3-One | 384.64 | 1 | 0 | 6.23 | 1 | 0.55 |
| Hydroquinone | 110.11 | 2 | 2 | 0.79 | 0 | 0.55 |
| 9,12-Octadecadienoic acid, methyl ester | 294.47 | 2 | 0 | 4.7 | 1 | 0.55 |
| Tetradecanoic acid, 12-Methyl-, Methyl Ester | 256.42 | 2 | 0 | 4.19 | 1 | 0.55 |
| 9,12-Octadecanoic Acid (Z,Z)- | 280.45 | 2 | 1 | 4.47 | 1 | 0.85 |
| Z,Z-10,12-Hexadecadien-1-ol acetate | 280.45 | 2 | 0 | 4.47 | 1 | 0.55 |
| 2(1H)-Naphthalenone, octahydro-4a-methyl-7-(1-methylethyl)-, (4a.alpha.,7.beta.,8a.beta.)- | 208.34 | 1 | 0 | 3.41 | 0 | 0.55 |
| Disalicylalpropylenediimine | 282.34 | 4 | 2 | 1.81 | 0 | 0.55 |
| 2-[(4E)-4-Hexenyl]-6-nitrocyclohexanone dimethylhydrazone | 267.37 | 3 | 0 | 1.82 | 0 | 0.55 |
| Neozepam | 281.27 | 4 | 1 | 0.9 | 0 | 0.55 |
| Indole-2-one, 2,3-dihydro-N-hydroxy-4-methoxy-3,3-dimethyl- | 207.23 | 3 | 1 | 1.4 | 0 | 0.55 |
| 4-phenylpyrido[2,3-d]pyrimidine | 207.23 | 3 | 0 | 2.03 | 0 | 0.55 |
| 2-adamantanyl-N-(adamantanylethyl)acetamide | 355.56 | 1 | 1 | 5.2 | 1 | 0.55 |
| Dehydrodiisoeugenol | 326.39 | 4 | 1 | 2.73 | 0 | 0.55 |
| Propenone, 1-[5-(3-hydroxy-3-methyl-1-butynyl)-2-thienyl]-3-(4-methoxyphenyl)- | 326.41 | 3 | 1 | 2.43 | 0 | 0.55 |
| 2-(4-methylphenyl) Indolizine | 207.27 | 0 | 0 | 3.32 | 0 | 0.55 |
| 6-Amino-5-cyano-4-(5-cyano-2,4-dimethyl-1H-pyrrol-3-yl)-2-methyl-4H-pyran-3-carboxylic acid ethyl ester | 326.35 | 5 | 2 | -0.3 | 0 | 0.56 |
| Aspidodispermine, O-methyl- | 342.43 | 4 | 1 | 2.02 | 0 | 0.55 |
| Tetramethylkaempferol | 342.34 | 6 | 0 | 0.94 | 0 | 0.55 |
| Nadolol di-methylboronic acid | 357.06 | 5 | 0 | 1.21 | 0 | 0.55 |
| Fisetin tetramethyl ether | 342.34 | 6 | 0 | 0.94 | 0 | 0.55 |
| 1,3-Dimethyl-5,6-dicarbethoxy-5,6,7,8-tetrahydro-6,7-diazalumazine | 342.31 | 7 | 2 | 1.22 | 1 | 0.55 |
| Cholestane, 3,4-epoxy-2-methyl-, (2.alpha.,3.alpha.,4.alpha.,5.alpha.)- | 400.68 | 1 | 0 | 6.68 | 1 | 0.55 |
| 4-Cyclohexene-1,2-dicarboximide, N-butyl-, cis- | 207.27 | 2 | 0 | 1.68 | 0 | 0.55 |
| Clionasterol | 414.71 | 1 | 1 | 6.73 | 1 | 0.55 |
| 5-methyl-2-phenyl-1H-Indole | 207.27 | 0 | 1 | 3.32 | 0 | 0.55 |
| Benzo[h]quinoline, 2,4-dimethyl- | 207.27 | 1 | 0 | 3.32 | 0 | 0.55 |
| Gene | PDB ID | Compound | Docking Score |
|---|---|---|---|
| PPARG | 3E00 | Fisetin Tetramethyl Ether | −6.092 |
| Rosiglitazone | −4.73 | ||
| Metformin * | −3.761 | ||
| 2-Chloro-5-Nitro-N-Phenylbenzamide | −4.381 | ||
| PPARA | 1K7L | Fisetin Tetramethyl Ether | −5.563 |
| Bezfibrate | −5.204 | ||
| Rosiglitazone | −4.156 | ||
| Metformin * | −3.145 | ||
| 2-(1-Methyl-3-Oxo-3-Phenyl-Propylamino)-3-{4-[2-(5-Methyl-2-Phenyl-Oxazol-4-Yl)-Ethoxy]-Phenyl}-Propionic acid | −5.279 | ||
| PPARD | 5U3Q | Fisetin Tetramethyl Ether | −5.58 |
| Bezfibrate | −4.06 | ||
| Rosiglitazone | −3.372 | ||
| Metformin * | −3.319 | ||
| 6-(2-{[([1,1′-Biphenyl]-4-Carbonyl)(propan-2-Yl) amino]methyl}phenoxy)hexanoic acid | −2.845 | ||
| FABP3 | 5HZ9 | Fisetin Tetramethyl Ether | −3.924 |
| Rosiglitazone | −3.339 | ||
| Metformin * | −2.908 | ||
| 6-Chloranyl-2-Methyl-4-Phenyl-Quinoline-3-Carboxylic acid | −3.329 | ||
| FABP4 | 3P6D | Fisetin Tetramethyl Ether | −3.816 |
| Rosiglitazone | −3.196 | ||
| Metformin * | −2.81 | ||
| 3-(4-Methoxy-3-Methylphenyl)propanoic acid | −3.75 | ||
| MMP1 | 1SU3 | Fisetin Tetramethyl Ether | −4.043 |
| Rosiglitazone | −3.79 | ||
| Metformin * | −2.74 | ||
| 4-(2-Hydroxyethyl)-1-Piperazine ethanesulfonic acid | −4.365 | ||
| NR1H3 | 1UHL | Fisetin Tetramethyl Ether | −2.967 |
| Rosiglitazone | −4.983 | ||
| Metformin * | −4.431 | ||
| N-(2,2,2-Trifluoroethyl)-N-{4-[2,2,2-Trifluoro-1-Hydroxy-1-(Trifluoromethyl) ethyl]phenyl}benzenesulfonamide | −2.22 |
| Compound | Enthalpy | Gibbs Free Energy | HOMO | LUMO | Eg | η | S |
|---|---|---|---|---|---|---|---|
| Fisetin Tetramethyl Ether | −1186.191 | −1186.19096 | −0.21123 | −0.05941 | −0.1518 | −0.0759 | −13.173 |
| Metformin | −432.845 | −432.844572 | −0.20215 | 0.02114 | −0.2233 | −0.1116 | −8.957 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adnan, M.; Jeon, B.-B.; Chowdhury, M.H.U.; Oh, K.-K.; Das, T.; Chy, M.N.U.; Cho, D.-H. Network Pharmacology Study to Reveal the Potentiality of a Methanol Extract of Caesalpinia sappan L. Wood against Type-2 Diabetes Mellitus. Life 2022, 12, 277. https://doi.org/10.3390/life12020277
Adnan M, Jeon B-B, Chowdhury MHU, Oh K-K, Das T, Chy MNU, Cho D-H. Network Pharmacology Study to Reveal the Potentiality of a Methanol Extract of Caesalpinia sappan L. Wood against Type-2 Diabetes Mellitus. Life. 2022; 12(2):277. https://doi.org/10.3390/life12020277
Chicago/Turabian StyleAdnan, Md., Byeong-Bae Jeon, Md. Helal Uddin Chowdhury, Ki-Kwang Oh, Tuhin Das, Md. Nazim Uddin Chy, and Dong-Ha Cho. 2022. "Network Pharmacology Study to Reveal the Potentiality of a Methanol Extract of Caesalpinia sappan L. Wood against Type-2 Diabetes Mellitus" Life 12, no. 2: 277. https://doi.org/10.3390/life12020277
APA StyleAdnan, M., Jeon, B.-B., Chowdhury, M. H. U., Oh, K.-K., Das, T., Chy, M. N. U., & Cho, D.-H. (2022). Network Pharmacology Study to Reveal the Potentiality of a Methanol Extract of Caesalpinia sappan L. Wood against Type-2 Diabetes Mellitus. Life, 12(2), 277. https://doi.org/10.3390/life12020277











